Effects of Genetic Polymorphisms of Cathepsin A on Metabolism of Tenofovir Alafenamide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Quantitative Reverse Transcription-PCR Analysis
2.2. PCR-SSCP Analysis
2.3. Luciferase Reporter Assay
2.4. CatA-Stable Expressing Cells
2.5. Western Blotting
2.6. LC/MS/MS Condition on the Detection of TFV-Ala
2.7. CatA Metabolic Activity
3. Results
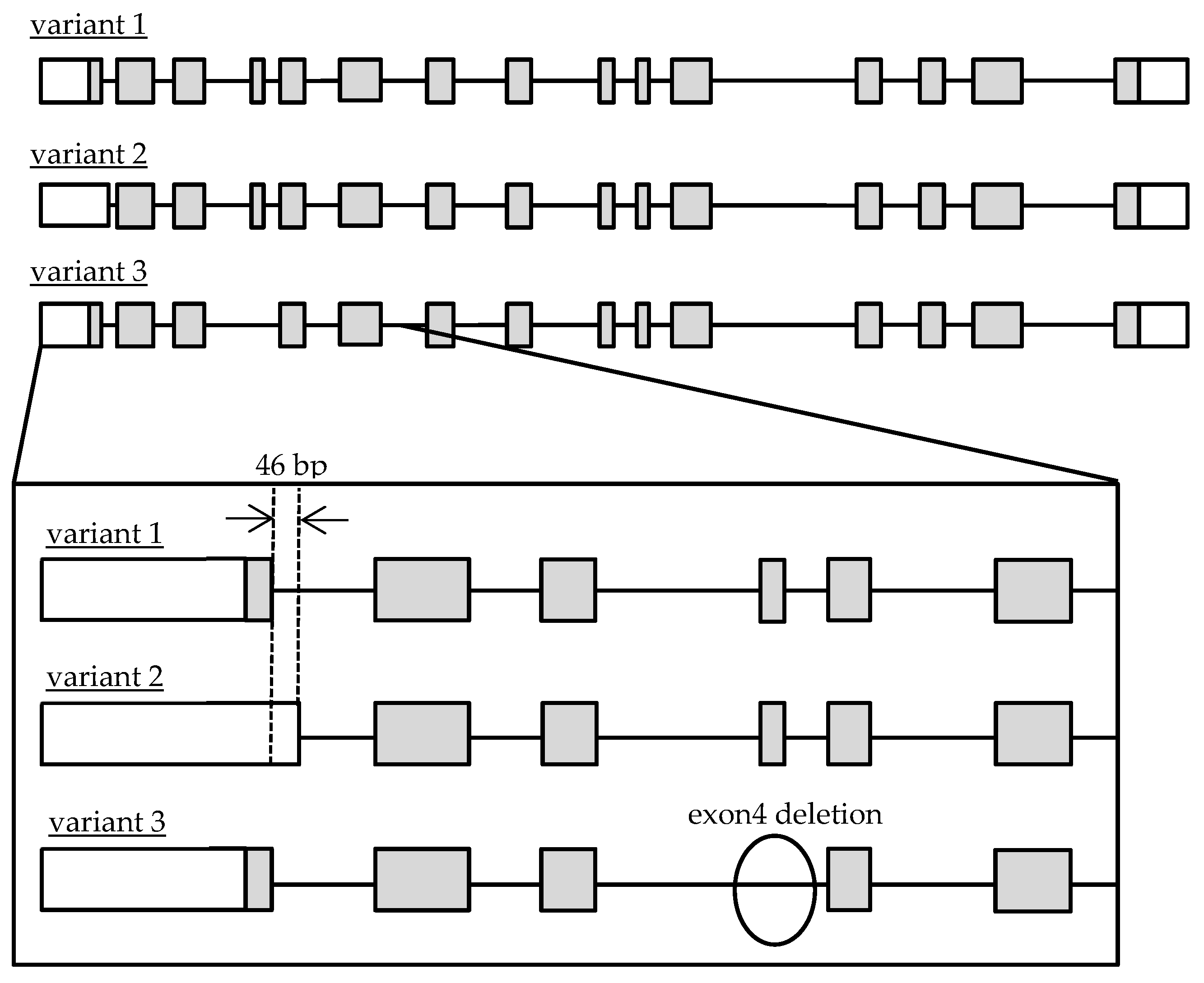
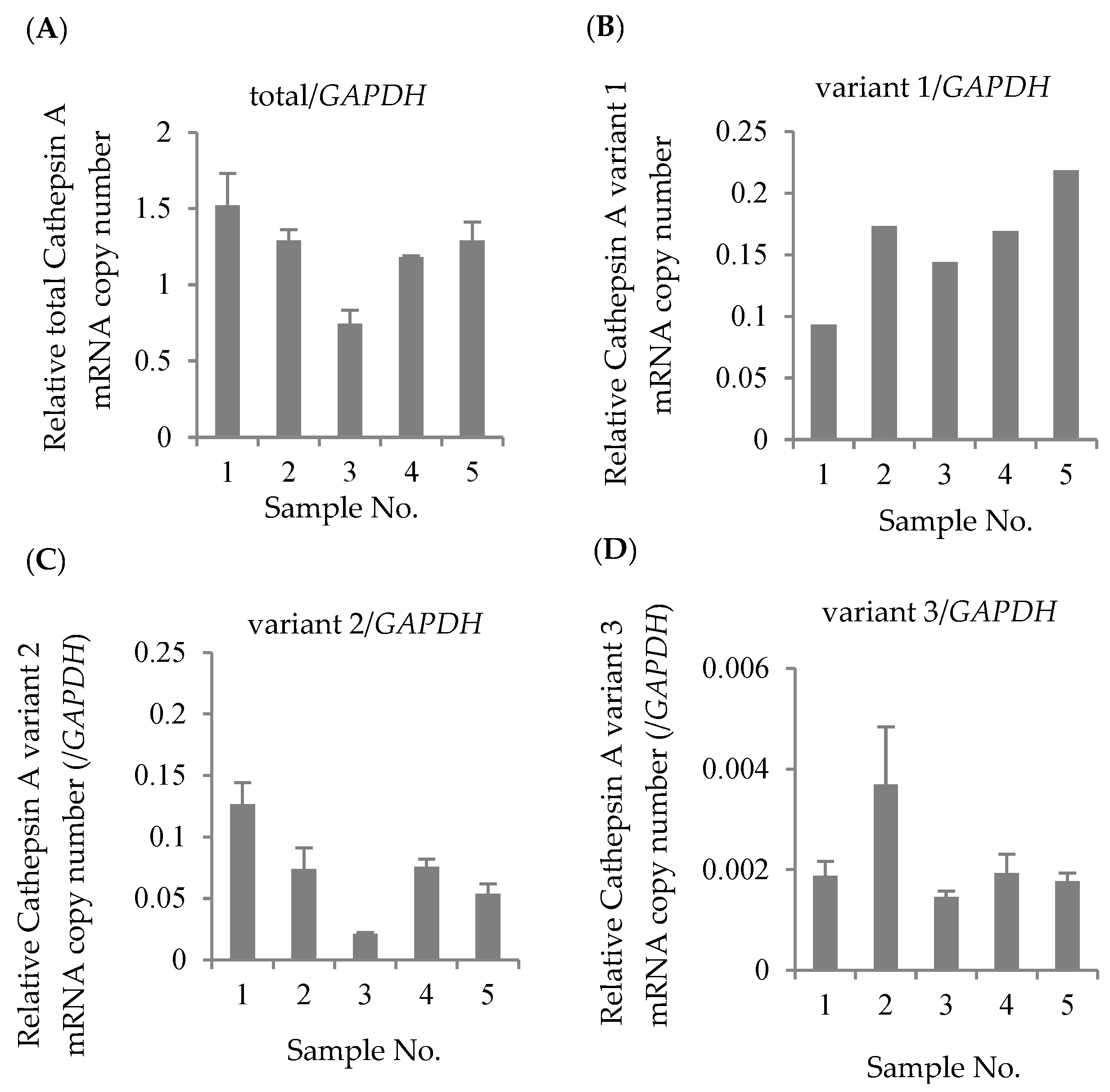
3.1. Quantification of Transcriptional Variants of CatA in Lymphocytes
3.2. Genetic Polymorphism of CatA Gene
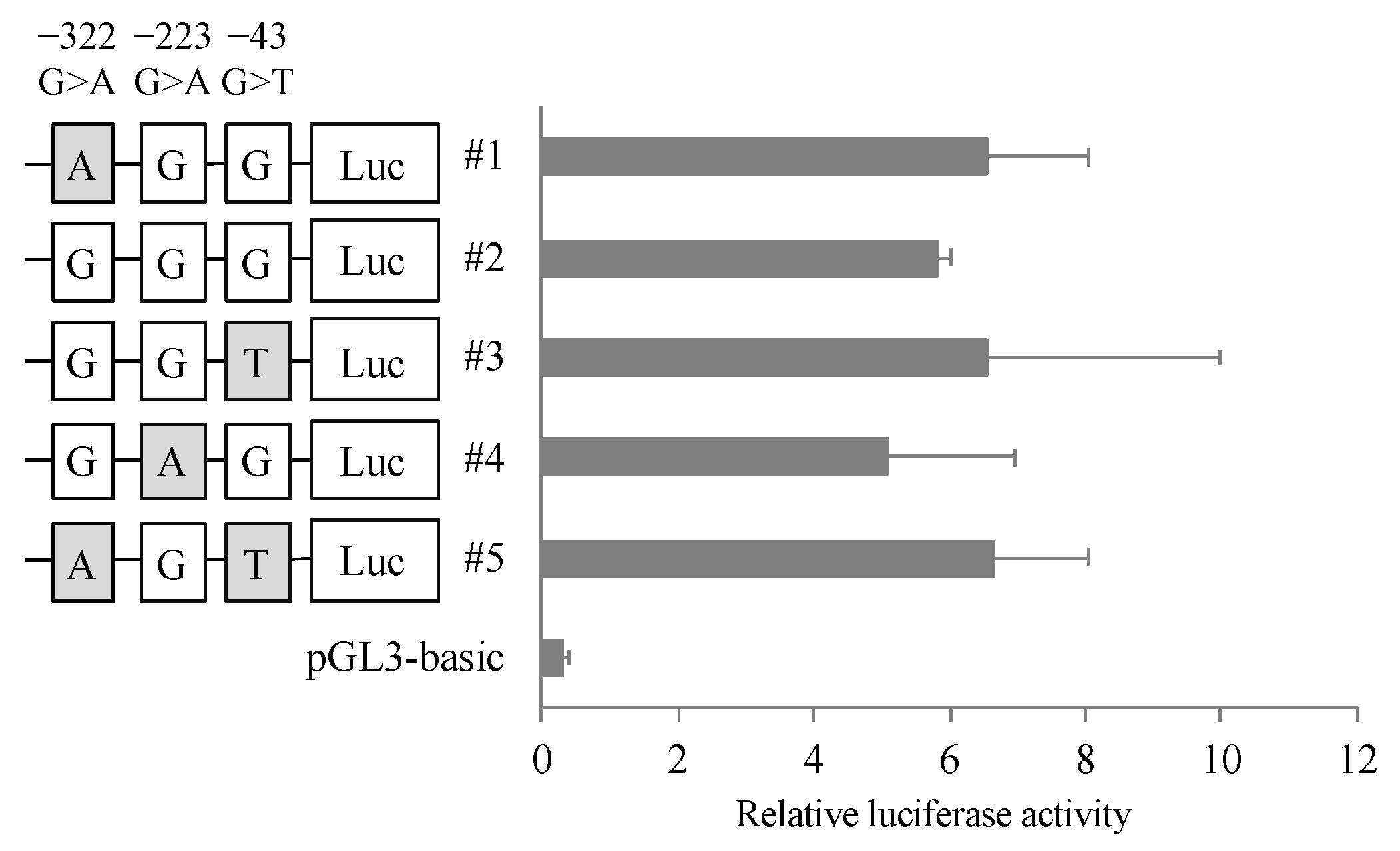
3.3. Effects of Polymorphisms of CatA Gene on Transcription Activity
3.4. Effects of Polymorphism of CatA Gene on Protein Expression
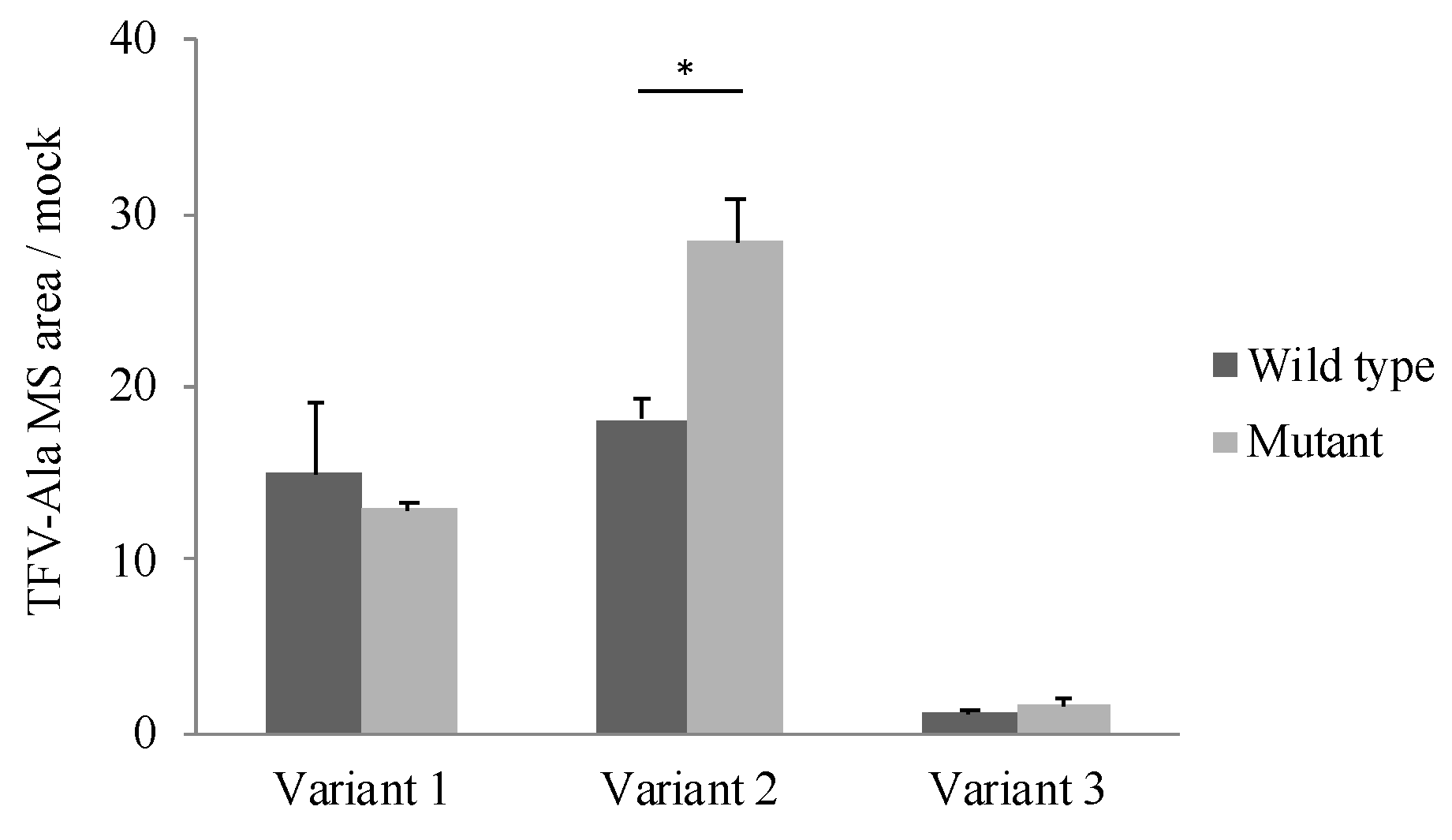
3.5. Effects of Polymorphism of CatA on Metabolic Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Najjar, A.; Karaman, R. The prodrug approach in the era of drug design. Expert Opin. Drug Deliv. 2019, 16, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Fukami, T.; Yokoi, T. The Emerging Role of Human Esterases. Drug Metab. Pharmacokinet. 2012, 27, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Paré, G.; Eriksson, N.; Lehr, T.; Connolly, S.; Eikelboom, J.; Ezekowitz, M.D.; Axelxxon, T.; Haertter, S.; Oldgren, J.; Reilly, P.; et al. Genetic determinants of dabigatran plasma levels and their relation to bleeding. Circulation 2013, 127, 1404–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crews, K.R.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Klein, T.E.; Caudle, K.E.; Haidar, C.E.; Shen, D.D.; Callaghan, J.T.; Sadhasivam, S.; et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 2014, 95, 376–382. [Google Scholar] [CrossRef] [Green Version]
- Chida, M.; Yokoi, T.; Kosaka, Y.; Chiba, K.; Nakamura, H.; Ishizaki, T.; Yokota, J.; Kinoshita, M.; Sato, K.; Inaba, M.; et al. Genetic polymorphism of CYP2D6 in the Japanese population. Pharmacogenetics 1999, 9, 601–605. [Google Scholar]
- Kobayashi, M.; Kajiwara, M.; Hasegawa, S. A Randomized Study of the Safety, Tolerability, Pharmacodynamics, and Pharmacokinetics of Clopidogrel in Three Different CYP2C19 Genotype Groups of Healthy Japanese Subjects. J. Atheroscler. Thromb. 2015, 22, 1186–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuta, T.; Sugimoto, M.; Shirai, N.; Ishizaki, T. CYP2C19 pharmacogenomics associated with therapy of Helicobacter pylori infection and gastro-esophageal reflux diseases with a proton pump inhibitor. Pharmacogenomics 2007, 8, 1199–1210. [Google Scholar] [CrossRef]
- Birkus, G.; Wang, R.; Liu, X.; Kutty, N.; MacArthur, H.; Cihlar, T.; Gibbs, C.; Swaminathan, S.; Lee, W.; McDermott, M.; et al. Cathepsin A is the major hydrolase catalyzing the intracellular hydrolysis of the antiretroviral nucleotide phosphonoamidate prodrugs GS-7340 and GS-9131. Antimicrob. Agents Chemother. 2007, 51, 543–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murakami, E.; Tolstykh, T.; Bao, H.; Niu, C.; Steuer, H.M.; Bao, D.; Chang, W.; Espiritu, C.; Bansal, S.; Lam, A.M.; et al. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 2010, 285, 34337–34347. [Google Scholar] [CrossRef] [Green Version]
- Kolli, N.; Garman, S.C. Proteolytic activation of human cathepsin A. J. Biol. Chem. 2014, 289, 11592–11600. [Google Scholar] [CrossRef] [Green Version]
- Satake, A.; Itoh, K.; Shimmoto, M.; Saido, T.C.; Sakuraba, H.; Suzuki, Y. Distribution of lysosomal protective protein in human tissues. Biochem. Biophys. Res. Commun. 1994, 205, 38–43. [Google Scholar] [CrossRef]
- Reich, M.; Spindler, K.D.; Burret, M.; Kalbacher, H.; Boehm, B.O.; Burster, T. Cathepsin A is expressed in primary human antigen-presenting cells. Immunol. Lett. 2010, 128, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, A.; Bonten, E.; d’Azzo, A. Transport of human lysosomal neuraminidase to mature lysosomes requires protective protein/cathepsin A. EMBO J. 1998, 17, 1588–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morreau, H.; Galjart, N.J.; Willemsen, R.; Gillemans, N.; Zhou, X.Y.; d’Azzo, A. Human lysosomal protective protein. Glycosylation, intracellular transport, and association with β-galactosidase in the endoplasmic reticulum. J. Biol. Chem. 1992, 267, 17949–17956. [Google Scholar] [CrossRef]
- Jackman, H.L.; Tan, F.L.; Tamei, C.; Beurling-Harbury, C.; Li, X.Y.; Skidgel, R.A.; Erdös, E.G. A peptidase in human Platelets that deamidates tachykinins. J. Biol. Chem. 1990, 265, 11265–11272. [Google Scholar] [CrossRef]
- Nakajima, H.; Ueno, M.; Adachi, K.; Nanba, E.; Naria, A.; Tsukimoto, J.; Itoh, K.; Kawakami, A. A new heterozygous compound mutation in the CTSA gene in galactosialidosis. Hum. Genome Var. 2019, 6, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Rottier, R.J.; d’Azzo, A. Identification of the Promoters for the Human and Murine Protective Protein/Cathepsin A Genes. DNA Cell Biol. 1997, 16, 599–610. [Google Scholar] [CrossRef]
- Galjart, N.J.; Gillemans, N.; Harris, A.; van der Horst, G.T.; Verheijen, F.W.; Galjaard, H.; d’Azzo, A. Expression of cDNA encoding the human “protective protein” associated with lysosomal β-galactosidase and neuraminidase: Homology to yeast proteases. Cell 1988, 54, 755–764. [Google Scholar] [CrossRef]
- Caciotti, A.; Catarzi, S.; Tonin, R.; Lugli, L.; Perez, C.R.; Michelakakis, H.; Mavridou, I.; Donati, M.A.; Guerrini, R.; d’Azzo, A.; et al. Galactosialidosis: Review and analysis of CTSA gene mutations. Orphanet J. Rare Dis. 2013, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Sardiello, M.; Palmieri, M.; di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A gene network regulating lysosomal biogenesis and function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef] [Green Version]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.H.; Szabady, R.L.; Bernstein, H.D. An unusual signal peptide extension inhibits the binding of bacterial presecretory proteins to the signal recognition particle, trigger factor, and the SecYEG complex. J. Biol. Chem. 2006, 281, 9038–9048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulemzin, S.; Chikaev, N.; Volkova, O.; Reshetnikova, E.; Taranin, A.; Najakshin, A.; Mechetina, L. Characterization of human FCRLA isoforms. Immunol. Lett. 2013, 152, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Riedl, E.; Koeppel, H.; Brinkkoetter, P.; Sternik, P.; Steinbeisser, H.; Sauerhoefer, S.; Janssen, B.; van Der Woude, F.J.; Yard, B.A. A CTG polymorphism in the CNDP1 gene determines the secretion of serum carnosinase in Cos-7-transfected cells. Diabetes 2007, 56, 2410–2413. [Google Scholar] [CrossRef] [Green Version]
- Birkus, G.; Bam, R.A.; Willkom, M.; Frey, C.R.; Tsai, L.; Stray, K.M.; Yant, S.R.; Cihlar, T. Intracellular Activation of Tenofovir Alafenamide and the Effect of Viral and Host Protease Inhibitors. Antimicrob. Agents Chemother. 2015, 60, 316–322. [Google Scholar] [CrossRef] [Green Version]

| Location | Primer (5′ to 3′) | Primer Lengh (bp) | Product Length (bp) | |
|---|---|---|---|---|
| Total | Forward | CAGATTGCCGGCTTCGTGA | 19 | 116 |
| Reverse | AAGCGGGAGAACATGGTGAAG | 21 | ||
| Variant 1,2 | Forward | CCCTGGAGTACAACCCCTAT | 20 | 128 |
| Reverse | CTGGGCGACCTCAGTGTCAT | 20 | ||
| Variant 2 | Forward | CCCGGGATCGATGATCCGAG | 20 | 164 |
| Reverse | CGGAGTACTGGCGGAAAGACG | 21 | ||
| Varinat 3 | Forward | AGCATGGCCCCTTCCTGATT | 20 | 111 |
| Reverse | GGGCGACCTCAGTGTCATTA | 20 | ||
| GAPDH | Forward | ATCAAGAAGGTGGTGAAGCAG | 21 | 95 |
| Reverse | TCGCTGTTGAAGTCAGAGGAG | 21 | ||
| LC | LC: | Nexera (Shimadzu) | ||||
| Column: | Mastro C18, 3 μm, 2.1 × 150 mm (Shimadzu GLC) | |||||
| Column temp.: | 35 °C | |||||
| Auto sampler temp: | 4 °C | |||||
| Mobile phase A: | 10 mM Ammonium bicarbonate | |||||
| Mobile phase B: | Acetonitrile (MeCN) | |||||
| Weak wash: | 5% MeCN | |||||
| Strong wash: | MeCN/MeOH/IPA/H2O = 1/1/1/1 (v/v), 0.2% formic acid | |||||
| Gradient | ||||||
| Time (min) | Flow rate (mL/min) | A (%) | B(%) | |||
| 0 | 0.4 | 98 | 2 | |||
| 1.5 | 0.4 | 98 | 2 | |||
| 5 | 0.4 | 5 | 95 | |||
| 6.5 | 0.4 | 5 | 95 | |||
| 6.6 | 0.4 | 98 | 2 | |||
| 8 | 0.4 | 100 | stop | |||
| MS | Mass spectrometer: | LCMS8050 | ||||
| Scan type: | MRM | |||||
| Ionization: | ESI | |||||
| Polarity: | Positive | |||||
| Interface voltage: | 4 kV | |||||
| Nebulizing gas: | 3 L/min | |||||
| Heating gas: | 10 L/min | |||||
| DL temperature: | 250 °C | |||||
| Heat block temperature: | 400 °C | |||||
| Drying gas: | 10 L/min | |||||
| MRM condition | ||||||
| Analyte | Q1 m/z | Q3 m/z | Q1 pre bias | CE | Q3 pre bias | |
| TFV-Ala | 359.0 | 270.0 | −26.0 | −20.0 | −20.0 | |
| Daidzein (IS) | 255.1 | 199.1 | −17.0 | −25.0 | −22.0 | |
| Location | CDS Position * | Reference Number | V/R Allele | Amino Acid Substitution | n | Number of | Frequency of Variant Allele (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| R/R | V/R | V/V | |||||||
| Exon 1 | −322 | rs2868362 | G>A | - | 76 | 25 | 33 | 18 | 42.11 |
| −223 | rs117529875 | G>A | - | 76 | 74 | 2 | 0 | 1.32 | |
| −43 | rs116893852 | G>T | - | 76 | 54 | 19 | 3 | 16.45 | |
| Exon 2 | 85_87 | rs72555383 | CTG>- | 29 Leu >- | 48 | 2 | 17 | 29 | 78.13 |
| 108 | rs181943893 | G>C | Synonymous | 48 | 39 | 6 | 3 | 12.5 | |
| Exon 3 | 273 | rs742035 | C>G | Synonymous | 48 | 39 | 9 | 0 | 9.38 |
| Intron 9 | 924-19 | rs3215446 | C>- | - | 48 | 14 | 19 | 15 | 51.04 |
| Intron 10 | 1002 + 7 | rs2075961 | G>A | - | 48 | 0 | 13 | 35 | 86.46 |
| Intron 11 | 1142 + 10 | rs4608591 | C>T | - | 48 | 0 | 13 | 35 | 86.46 |
| Haplotype Pattern | Genetic Variations | Estimated Frequency (%) | ||
|---|---|---|---|---|
| −322G>A | −223G>A | −43G>T | ||
| #1 | A | G | G | 45.4 |
| #2 | G | G | G | 37.0 |
| #3 | G | G | T | 16.3 |
| #4 | G | A | G | 1.18 |
| #5 | A | G | T | NE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ito, S.; Hirota, T.; Yanai, M.; Muto, M.; Watanabe, E.; Taya, Y.; Ieiri, I. Effects of Genetic Polymorphisms of Cathepsin A on Metabolism of Tenofovir Alafenamide. Genes 2021, 12, 2026. https://doi.org/10.3390/genes12122026
Ito S, Hirota T, Yanai M, Muto M, Watanabe E, Taya Y, Ieiri I. Effects of Genetic Polymorphisms of Cathepsin A on Metabolism of Tenofovir Alafenamide. Genes. 2021; 12(12):2026. https://doi.org/10.3390/genes12122026
Chicago/Turabian StyleIto, Soichiro, Takeshi Hirota, Miyu Yanai, Mai Muto, Eri Watanabe, Yuki Taya, and Ichiro Ieiri. 2021. "Effects of Genetic Polymorphisms of Cathepsin A on Metabolism of Tenofovir Alafenamide" Genes 12, no. 12: 2026. https://doi.org/10.3390/genes12122026





