Whole Exome Sequencing Identifies Novel De Novo Variants Interacting with Six Gene Networks in Autism Spectrum Disorder
Abstract
:1. Introduction
2. Methods
2.1. Subject Ascertainment and Ethnic Information
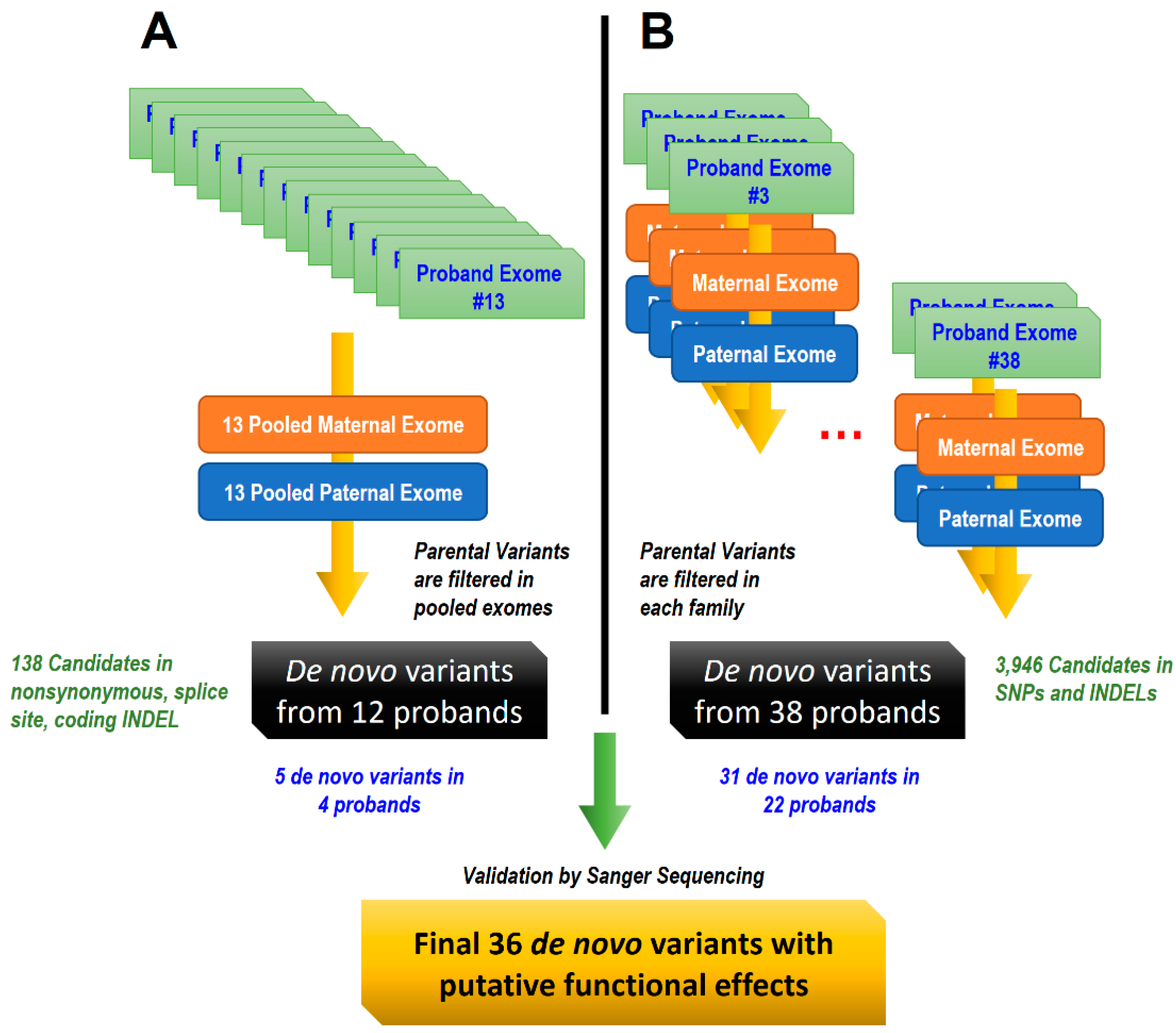
2.2. Study Design and Bioinformatics Analysis
2.3. Variant Discovery and Filtering
2.4. Gene Network Analysis
2.5. Genetic Validation using Sanger Sequencing
3. Results
3.1. Network A. DTX1, MTUS1, RASGRP1, and RUFY1
3.2. Network B. ADCY7, ATXN1, IWS1, KCTD9, MT-ND2, NFKB1, and PPP1R16B
3.3. Network C. AMIGO1, HAX1, MYH14, RNH1, SNF8, and SYNM
3.4. Network D. AKNA, GMIP, LARS2, NEK1, and TFPT
3.5. Network E. CELSR3, COL6A2, HMGXB4, PIEZO1, RBM27, SFMBT2, and UBAC1
3.6. Network F. ANKRD27, TMEM8A, and TTC21A
3.7. Evidence from Topologically Associating Domain (TAD) in TFPT and HAX1
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Yoo, H.J.; Kim, B.-N.; Kim, J.-W.; Shin, M.-S.; Park, T.-W.; Son, J.-W.; Chung, U.-S.; Park, M.; Kim, S.A. Family-based genetic association study of CNTNAP2 polymorphisms and sociality endophenotypes in Korean patients with autism spectrum disorders. Psychiatr. Genet. 2017, 27, 38–39. [Google Scholar] [CrossRef]
- Woodbury-Smith, M.; Scherer, S.W. Progress in the genetics of autism spectrum disorder. Dev. Med. Child Neurol. 2018, 60, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Ghirardi, L.; Brikell, I.; Kuja-Halkola, R.; Freitag, C.M.; Franke, B.; Asherson, P.; Lichtenstein, P.; Larsson, H. The familial co-aggregation of ASD and ADHD: A register-based cohort study. Mol. Psychiatry 2018, 23, 257–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, K.J. The genetics of neurodevelopmental disease. Curr. Opin. Neurobiol. 2011, 21, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Chahrour, M.H.; Yu, T.W.; Lim, E.T.; Ataman, B.; Coulter, M.E.; Hill, R.S.; Stevens, C.R.; Schubert, C.R.; Greenberg, M.E.; Gabriel, S.B.; et al. Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism. PLoS Genet. 2012, 8, e1002635. [Google Scholar] [CrossRef] [Green Version]
- Elia, J.; Gai, X.; Xie, H.M.; Perin, J.C.; Geiger, E.; Glessner, J.T.; Darcy, M.; Deberardinis, R.; Frackelton, E.; Kim, C.; et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 2009, 15, 637–646. [Google Scholar] [CrossRef]
- González, A.A.; Cabanas, M.C.; Rodriguez Fontenla, M.C.; Carracedo, A. Novel gene-based analysis of ASD GWAS: Insight into the biological role of associated genes. Front. Genet. 2019, 10, 733. [Google Scholar] [CrossRef] [Green Version]
- Robinson, E.B.; Pourcain, B.S.; Anttila, V.; Kosmicki, J.; Bulik-Sullivan, B.; Grove, J.; Maller, J.; Samocha, K.E.; Sanders, S.; Ripke, S.; et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet. 2016, 48, 552. [Google Scholar] [CrossRef] [Green Version]
- Autism Spectrum Disorders Working Group of the Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism. 2017, 8, 21. [Google Scholar] [CrossRef]
- Sztainberg, Y.; Zoghbi, H.Y. Lessons learned from studying syndromic autism spectrum disorders. Nat. Neurosci. 2016, 19, 1408–1417. [Google Scholar] [CrossRef]
- Fernandez, B.A.; Scherer, S.W. Syndromic autism spectrum disorders: Moving from a clinically defined to a molecularly defined approach. Dialogues Clin. Neurosci. 2017, 19, 353. [Google Scholar] [PubMed]
- Nakanishi, M.; Nomura, J.; Ji, X.; Tamada, K.; Arai, T.; Takahashi, E.; Bućan, M.; Takumi, T. Functional significance of rare neuroligin 1 variants found in autism. PLoS Genet. 2017, 13, e1006940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafiq, M.A.; Leblond, C.S.; Saqib, M.A.N.; Vincent, A.K.; Ambalavanan, A.; Khan, F.S.; Ayaz, M.; Shaheen, N.; Spiegelman, D.; Ali, G.; et al. Novel VPS13B Mutations in Three Large Pakistani Cohen Syndrome Families Suggests a Baloch Variant with Autistic-Like Features. BMC Med. Genet. 2015, 16, 41. [Google Scholar] [CrossRef] [Green Version]
- Delahanty, R.J.; Kang, J.; Brune, C.W.; Kistner, E.O.; Courchesne, E.; Cox, N.J.; Cook, E.H., Jr.; Macdonald, R.L.; Sutcliffe, J.S. Maternal transmission of a rare GABRB3 signal peptide variant is associated with autism. Mol. Psychiatry 2011, 16, 86–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torrico, B.; Fernàndez-Castillo, N.; Hervás, A.; Milà, M.; Salgado, M.; Rueda, I.; Buitelaar, J.K.; Rommelse, N.; Oerlemans, A.M.; Bralten, J.; et al. Contribution of common and rare variants of the PTCHD1 gene to autism spectrum disorders and intellectual disability. Eur. J. Hum. Genet. 2015, 23, 1694–1701. [Google Scholar] [CrossRef] [Green Version]
- Schaaf, C.P.; Zoghbi, H.Y. Solving the Autism Puzzle a Few Pieces at a Time. Neuron 2011, 70, 806–808. [Google Scholar] [CrossRef] [Green Version]
- Weiner, D.J.; Wigdor, E.M.; Ripke, S.; Walters, R.K.; Kosmicki, J.A.; Grove, J.; Samocha, K.E.; Goldstein, J.I.; Okbay, A.; Bybjerg-Grauholm, J. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 2017, 49, 978–985. [Google Scholar] [CrossRef] [Green Version]
- Hazlett, H.C.; Gu, H.; Munsell, B.C.; Kim, S.H.; Styner, M.; Wolff, J.J.; Elison, J.T.; Swanson, M.R.; Zhu, H.; Botteron, K.N. Early brain development in infants at high risk for autism spectrum disorder. Nature 2017, 542, 348–351. [Google Scholar] [CrossRef]
- Feliciano, P.; Zhou, X.; Astrovskaya, I.; Turner, T.N.; Wang, T.; Brueggeman, L.; Barnard, R.; Hsieh, A.; Snyder, L.G.; Muzny, D.M.; et al. Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom. Med. 2019, 4, 19. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Ha, S.; Lee, S.T.; Park, S.G.; Shin, S.; Choi, J.R.; Cheon, K.A. Next-Generation Sequencing in Korean Children With Autism Spectrum Disorder and Comorbid Epilepsy. Front. Pharmacol. 2020, 14, 585. [Google Scholar] [CrossRef]
- Stein, D.J.; Phillips, K.A. Patient advocacy and DSM-5. BMC Med. 2013, 11, 133. [Google Scholar] [CrossRef] [Green Version]
- Lord, C.; Rutter, M.; Le Couteur, A. Autism Diagnostic Interview-Revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994, 24, 659–685. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Risi, S.; Lambrecht, L.; Cook, E.H.; Leventhal, B.L.; DiLavore, P.C.; Pickles, A.; Rutter, M. The Autism Diagnostic Observation Schedule—Generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000, 30, 205–223. [Google Scholar] [CrossRef]
- Park, K.; Yoon, J.; Park, H.; Kwon, K. Korean Educational Developmental Institute-Wechsler Intelligence Scale for Children (KEDI-WISC); Korean Educational Development Institute: Seoul, Korea, 2002. [Google Scholar]
- Shin, M.; Cho, S. Korean Leiter International Performance Scale-Revised; Hakjisa: Seoul, Korea, 2010. [Google Scholar]
- Rutter, M.; Bailey, A.; Lord, C. The Social Communication Questionnaire: Manual; Western Psychological Services: Los Angeles, CA, USA, 2003. [Google Scholar]
- Constantino, J.N.; Gruber, C.P. Social Responsiveness Scale: SRS-2 Software Kit; Western Psychological Services: Torrane, CA, USA, 2012. [Google Scholar]
- Iossifov, I.; O’roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Liu, Z.; Li, X.; Li, F.; Hu, Y.; Chen, B.; Wang, Z.; Liu, Y. Whole-exome sequencing identifies a novel de novo mutation in DYNC1H1 in epileptic encephalopathies. Sci. Rep. 2017, 7, 258. [Google Scholar] [CrossRef] [Green Version]
- Seet, L.-F.; Hong, W. Endofin, an Endosomal FYVE Domain Protein. J. Biol. Chem. 2001, 276, 42445–42454. [Google Scholar] [CrossRef] [Green Version]
- Kishi, N.; MacDonald, J.L.; Ye, J.; Molyneaux, B.J.; Azim, E.; Macklis, J.D. Reduction of aberrant NF-κB signalling ameliorates Rett syndrome phenotypes in Mecp2-null mice. Nat. Commun. 2016, 7, 10520. [Google Scholar] [CrossRef] [Green Version]
- Shindler, A.E.; Hill-Yardin, E.L.; Petrovski, S.; Bishop, N.; Franks, A.E. Towards Identifying Genetic Biomarkers for Gastrointestinal Dysfunction in Autism. J. Autism Dev. Disord. 2019, 50, 76–86. [Google Scholar] [CrossRef]
- Li, H.; Wang, Z.; Ma, T.; Wei, G.; Ni, T. Alternative splicing in aging and age-related diseases. Transl. Med. Aging 2017, 1, 32–40. [Google Scholar] [CrossRef]
- Ingram, M.; Wozniak, E.A.; Duvick, L.; Yang, R.; Bergmann, P.; Carson, R.; O’Callaghan, B.; Zoghbi, H.Y.; Henzler, C.; Orr, H.T. Cerebellar Transcriptome Profiles of ATXN1 Transgenic Mice Reveal SCA1 Disease Progression and Protection Pathways. Neuron 2016, 89, 1194–1207. [Google Scholar] [CrossRef] [Green Version]
- Esteban, F.J.; Wall, D.P. Using game theory to detect genes involved in Autism Spectrum Disorder. Top 2011, 19, 121–129. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Devor, A.; Andreassen, O.A.; Wang, Y.; Mäki-Marttunen, T.; Smeland, O.B.; Fan, C.-C.; Schork, A.J.; Holland, D.; Thompson, W.K.; Witoelar, A.; et al. Genetic evidence for role of integration of fast and slow neurotransmission in schizophrenia. Mol. Psychiatry 2017, 22, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.C.; Archer, H.L.; Colley, J.P.; Ravn, K.; Nielsen, J.B.; Kerr, A.; Williams, E.; Christodoulou, J.; Gécz, J.; Jardine, P.E. Early onset seizures and Rett-like features associated with mutations in CDKL5. Eur. J. Hum. Genet. 2005, 13, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Mcbride, K.L.; Varga, E.A.; Pastore, M.T.; Prior, T.W.; Manickam, K.; Atkin, J.F.; Herman, G.E. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res. 2010, 3, 137–141. [Google Scholar] [CrossRef]
- Kolaitis, G.; Bouwkamp, C.G.; Papakonstantinou, A.; Otheiti, I.; Belivanaki, M.; Haritaki, S.; Korpa, T.; Albani, Z.; Terzioglou, E.; Apostola, P.; et al. A boy with conduct disorder (CD), attention deficit hyperactivity disorder (ADHD), borderline intellectual disability, and 47, XXY syndrome in combination with a 7q11.23 duplication, 11p15.5 deletion, and 20q13.33 deletion. Child Adolesc. Psychiatry Ment. Health 2016, 10, 33. [Google Scholar] [CrossRef] [Green Version]
- Shao, L.; Cui, L.; Lu, J.; Lang, Y.; Bottillo, I.; Zhao, X. A novel mutation in exon 9 of Cullin 3 gene contributes to aberrant splicing in pseudohypoaldosteronism type II. FEBS Open Bio 2018, 8, 461–469. [Google Scholar] [CrossRef] [Green Version]
- Roberts, J.L.; Hovanes, K.; Dasouki, M.; Manzardo, A.M.; Butler, M.G. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene 2014, 535, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Malenfant, P.; Reesor, C.; Lee, A.; Hudson, M.L.; Harvard, C.; Qiao, Y.; Persico, A.M.; Cohen, I.L.; Chudley, A.E.; et al. 2p15–p16.1 microdeletion syndrome: Molecular characterization and association of the OTX1 and XPO1 genes with autism spectrum disorders. Eur. J. Hum. Genet. 2011, 19, 1264–1270. [Google Scholar] [CrossRef]
- Wilkes, T.; Wang, E.; Perry, B. A neuro-developmentally sensitive and trauma informed service delivery approach for child and youth mental health and psychiatry. Eur. Psychiatry 2017, 41, S309. [Google Scholar] [CrossRef]
- Yoo, H.; Kim, S.; Park, M.; Kim, J.; Lim, W.; Noh, D.; Han, D.; Shin, C.; Kim, N. Family-based Whole Exome Sequencing of Autism Spectrum Disorder Reveals Novel De Novo Variants in Korean Population. Eur. Psychiatry 2017, 41, S309. [Google Scholar] [CrossRef]
- Kim, J.-H.; Yoon, K.-O.; Kim, J.-K.; Kim, J.-W.; Lee, S.-K.; Kong, S.-Y.; Seo, J.-M. Novel mutations of RET gene in Korean patients with sporadic Hirschsprungs disease. J. Pediatr. Surg. 2006, 41, 1250–1254. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.K.; Merico, D.; Bookman, M.; Howe, J.L.; Thiruvahindrapuram, B.; Patel, R.V.; Whitney, J.; Deflaux, N.; Bingham, J.; Wang, Z. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017, 20, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Guo, H.; Xiong, B.; Stessman, H.; Xia, K.; Eichler, E. De Novo Genic Mutations Among A Chinese Autism Spectrum Disorder Cohort. Nat. Commun. 2016, 7, 13316. [Google Scholar] [CrossRef] [PubMed]
- Stessman, H.A.F.; Xiong, B.; Coe, B.P.; Wang, T.; Hoekzema, K.; Fenckova, M.; Kvarnung, M.; Gerdts, J.; Trinh, S.; Cosemans, N.; et al. argeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017, 49, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, S.; Choi, E.Y.; Kim, S.; Kwak, H.J.; Yoo, B.C.; Yoo, H.; Lee, S.; Kim, D.; Park, J.B.; et al. PTEN modulates miR-21 processing via RNA-regulatory protein RNH1. PLoS ONE 2011, 6, e28308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjorklund, G.; Saad, K.; Chirumbolo, S.; Kern, J.K.; Geier, D.A.; Geier, M.R.; Urbina, M.A. Immune dysfunction and neuroinflammation in autism spectrum disorder. Acta Neurobiol. Exp. 2016, 76, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Karin, M. Is NF-κB the sensor of oxidative stress? FASEB J. 1999, 13, 1137–1143. [Google Scholar] [CrossRef]
- Gutierrez, H.; Davies, A.M. Regulation of neural process growth, elaboration and structural plasticity by NF-κB. Trends Neurosci. 2011, 34, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hu, W. NFκB signaling regulates embryonic and adult neurogenesis. Front. Biol. 2012, 7, 277–291. [Google Scholar] [CrossRef]
- Roussos, P.; Katsel, P.; Davis, K.L.; Giakoumaki, S.G.; Siever, L.J.; Bitsios, P.; Haroutunian, V. Convergent Findings for Abnormalities of the NF-κB Signaling Pathway in Schizophrenia. Neuropsychopharmacology 2012, 38, 533–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Riordan, K.J.; Huang, I.C.; Pizzi, M.; Spano, P.; Boroni, F.; Egli, R.; Desai, P.; Fitch, O.; Malone, L.; Ahn, H.J.; et al. Regulation of Nuclear Factor B in the Hippocampus by Group I Metabotropic Glutamate Receptors. J. Neurosci. 2006, 26, 4870–4879. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M. Transcription Factors in Long-Term Memory and Synaptic Plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.I.; Kern, J.K. Evidence of microglial activation in autism and its possible role in brain underconnectivity. Neuron Glia Biol. 2011, 7, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Dodurga, Y.; Gundogdu, G.; Tekin, V.; Koc, T.; Satiroglu-Tufan, N.L.; Bagci, G.; Kucukatay, V. Valproic acid inhibits the proliferation of SHSY5Y neuroblastoma cancer cells by downregulating URG4/URGCP and CCND1 gene expression. Mol. Biol. Rep. 2014, 41, 4595–4599. [Google Scholar] [CrossRef]
- Go, H.; Seo, J.; Kim, K.; Han, S.; Kim, P.; Kang, Y.; Han, S.; Shin, C.; Ko, K. Valproic acid inhibits neural progenitor cell death by activation of NF-κB signaling pathway and up-regulation of Bcl-XL. J. Biomed. Sci. 2011, 18, 48. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Salam, O.M.E.; Youness, E.R.; Mohammed, N.A.; Elhamed, W.A.A. Nuclear Factor-Kappa B and Other Oxidative Stress Biomarkers in Serum of Autistic Children. Open J. Mol. Integr. Physiol. 2015, 5, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Young, A.M.H.; Campbell, E.; Lynch, S.; Suckling, J.; Powis, S.J. Aberrant NF-KappaB Expression in Autism Spectrum Condition: A Mechanism for Neuroinflammation. Front. Psychiatry 2011, 2, 27. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, A.; Ahmad, S.F.; Attia, S.M.; Bakheet, S.A.; Al-Harbi, N.O.; AL-Ayadhi, L.Y. Activation of IL-17 receptor leads to increased oxidative inflammation in peripheral monocytes of autistic children. Brain Behav. Immun. 2018, 67, 335–344. [Google Scholar] [CrossRef]
- Baranova, J.; Dragunas, G.; Botellho, M.C.S.; Ayub, A.L.P.; Bueno‑Alves, R.; Alencar, R.R.; Papaiz, D.D.; Sogayar, M.C.; Ulrich, H.; Correa, R.G. Autism Spectrum Disorder: Signaling Pathways and Prospective Therapeutic Targets. Cell. Mol. Neurobiol. 2020, 1–31. [Google Scholar] [CrossRef]
- Liao, X.; Li, Y. Nuclear factor kappa B in autism spectrum disorder: A systematic review. Pharmacol. Res. 2020, 159, 104918. [Google Scholar] [CrossRef] [PubMed]
- Strenn, N.; Hovey, D.; Jonsson, L.; Anckarster, H.; Anckarsäter, H.; Lundström, S.; Lichtenstein, P.; Ekman, A. Associations between autistic-like traits and polymorphisms in NFKBIL1. Acta Neuropsychiatr. 2019, 31, 220–229. [Google Scholar] [CrossRef] [PubMed]


| Clinical Characteristics | First Stage (n = 13) | Second Stage (n = 38) | |||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| Age (months) | 59.50 | 12.41 | 73.53 | 29.94 | |
| Sex (number of females) | 0 | 4 | |||
| ADI-R | social interaction | 24.33 | 3.65 | 22.05 | 5.67 |
| communication: verbal | 18.43 | 5.68 | 18.58 | 4.50 | |
| communication: nonverbal | 12.00 | 2.73 | 11.71 | 2.34 | |
| repetitive behavior | 6.50 | 2.71 | 5.74 | 2.58 | |
| ADOS | Communication | 6.67 | 1.23 | 6.05 | 1.23 |
| social interaction | 10.25 | 2.34 | 10.16 | 2.15 | |
| Play | 2.92 | 1.24 | 2.34 | 1.17 | |
| repetitive behavior | 3.00 | 1.65 | 2.50 | 1.39 | |
| KEDI-WISC-R | 43.00 | 33.41 | 57.47 | 15.88 | |
| K-Leiter-R | 78.86 | 25.60 | 62.56 | 25.64 | |
| SCQ | 23.75 | 5.55 | 19.97 | 7.71 | |
| SRS | 109.08 | 27.62 | 95.08 | 31.05 | |
| Genomic Position | Gene Symbol | dbSNP RS ID | REF>ALT Allele | Sequence Variant Nomenclature (2) | Variant Type | Probands (:occur.) |
|---|---|---|---|---|---|---|
| (GRCh38/hg38) (1) | Build 154 | |||||
| chr1:109507558 | AMIGO1 | C>A | ENST00000369864.5 c.1355G>T/p.Gly452Val | Missense | B34 | |
| chr1:154274980 | HAX1 | C>A | ENST00000328703.11|ENST00000457918.6 c.535C>A|c.391C>A/p.Pro179Thr|p.Pro131Thr | Missense | B15 | |
| chr2:127493330 | IWS1 | C>G | ENST00000295321.9|ENST00000637187.1 c.1880G>C|c.47G>C/p.Ser627Thr|p.Ser16Thr | Missense | B5 :1st | |
| chr3:39112529 | TTC21A | C>A | ENST00000431162.6 c.507C>A/p.Tyr169 * | Nonsense | B16 | |
| chr3:45513187 | LARS2 | rs147745374 | C>T | ENST00000415258.6 c.1813C>T/p.Arg605Cys | Missense | B1 |
| chr3:48645607 | CELSR3 | rs756781285 | C>T | ENST00000164024.5 c.7633G>A/p.Val2545Met | Missense | A12 :1st |
| chr4:990079 | SLC26A1 | rs748023701 | C>T | ENST00000361661.6 c.860G>A/p.Arg287His | Missense | B27 |
| chr4:102607251 | NFKB1 | rs934744465 | G>A | ENST00000394820.8 c.2053G>A/p.Gly685Arg | Missense | B25 :1st |
| chr4:169602542 | NEK1 | rs200825809 | T>C | ENST00000507142.5 c.89A>G/p.Tyr30Cys | Missense | B11 |
| chr5:146233644 | RBM27 | C>A | ENST00000265271.7 c.1045C>A/p.Pro349Thr | Missense | B26 :1st | |
| chr5:179609380 | RUFY1 | A>G | ENST00000393438.6 c.1664A>G/p.His555Arg | Missense | B29 | |
| chr6:16306637 | ATXN1 | C>T | ENST00000244769.8 c.2140G>A/p.Asp714Asn | Missense | B26 :2nd | |
| chr6:44282283 | TCTE1 | rs746423492 | C>T | ENST00000371505.5 c.1123G>A/p.Glu375Lys | Missense | A4 |
| chr8:17697396 | MTUS1 | rs377560516 | G>A | ENST00000297488.10 c.-19C>T/5’UTR | 5’UTR | B26 :3rd |
| chr8:25436242 | KCTD9 | rs1448423271 | C>T | ENST00000221200.9 c.656G>A/p.Arg219Gln | Missense | A9 |
| chr9:114342063 | AKNA | A>T | ENST00000307564.8 c.3820T>A/p.Cys1274Ser | Missense | A12 :2nd | |
| chr9:135939739 | UBAC1 | C>A | ENST00000371756.3 c.897G>T/p.Glu299Asp | Missense | B10 :1st | |
| chr10:3141725 | PITRM1-AS1 | G>A | ENST00000430356.3 (PITRM1-AS1, Antisense) ENST00000451454.5 (PITRM1, Intronic) | Antisense/Intronic | B25 :2nd | |
| chr10:7370280 | SFMBT2 | rs1373335289 | C>T | ENST00000361972.8 c.195+1G>A/Splice Donor Variant | Splice Site | B5 :2nd |
| chr11:499976 | RNH1 | C>T | ENST00000534797.5 c.296G>A/p.Gly99Glu | Missense | B33 | |
| chr12:113094062 | DTX1 | rs1232450939 | G>A | ENST00000257600.3 c.1190G>A/p.Arg397Gln | Missense | B6 |
| chr15:38516208 | RASGRP1 | rs1595848108 | G>A | ENST00000310803.10 c.664C>T/p.Arg222Trp | Missense | B12 |
| chr15:99131660 | SYNM | rs782771735 | G>A | ENST00000336292.10 c.3300G>A/p.Ser1100Ser | Silent | B18 |
| chr16:377706 | TMEM8A | C>G | ENST00000431232.7 c.264G>C/p.Glu88Asp | Missense | B25 :3rd | |
| chr16:50293480 | ADCY7 | rs199730202 | G>A | ENST00000254235.7 c.814G>A/p.Val272Ile | Missense | B10 :2nd |
| chr16:88738626 | PIEZO1 | rs1475582880 | GGCCGTGACTCGGAAACGAGCGGCCA>G | ENST00000301015.14 c.551_575delTGGCCGCTCGTTTCCGAGTCACGGC/p.Leu184fs | Frameshift Deletion | B3 |
| chr17:48937128 | SNF8 | C>T | ENST00000502492.5 c.245-4G>A/Spice Region Variant | Splice Region | B25 :4th | |
| chr19:19637386 | GMIP | T>A | ENST00000203556.9 c.1103A>T/p.Glu368Val | Missense | B21 :1st | |
| chr19:32628120 | ANKRD27 | G>C | ENST00000306065.9 c.1383C>G/p.Asp461Glu | Missense | B21 :2nd | |
| chr19:50257343 | MYH14 | rs778416774 | G>A | ENST00000599920.5 c.1990G>A/p.Gly664Ser | Missense | A3 |
| chr19:54107093 | TFPT | TTGTC>T | ENST00000391759.5 c.715_718delGACA/p.Asp239fs | Frameshift Deletion | B4 | |
| chr20:32453760 | NOL4L | rs749152593 | G>C | ENST00000359676.9 c.389C>G/p.Ser130Cys | Missense | B9 |
| chr20:38902679 | PPP1R16B | G>A | ENST00000299824.6 c.583G>A/p.Glu195Lys | Missense | B7 | |
| chr21:46121590 | COL6A2 | rs267606749 | G>A | ENST00000300527.8 c.1493G>A/p.Arg498His | Missense | B37 |
| chr22:35265292 | HMGXB4 | C>G | ENST00000455359.5 c.577C>G/p.Leu193Val | Missense | B32 :1st | |
| chrM:4943 | MT-ND2 | rs1603219681 | A>G | ENST00000361453.3 c.474A>G/p.Ser158Ser | Silent | B32 :2nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Kim, K.H.; Lim, W.-J.; Kim, J.; Kim, S.A.; Yoo, H.J. Whole Exome Sequencing Identifies Novel De Novo Variants Interacting with Six Gene Networks in Autism Spectrum Disorder. Genes 2021, 12, 1. https://doi.org/10.3390/genes12010001
Kim N, Kim KH, Lim W-J, Kim J, Kim SA, Yoo HJ. Whole Exome Sequencing Identifies Novel De Novo Variants Interacting with Six Gene Networks in Autism Spectrum Disorder. Genes. 2021; 12(1):1. https://doi.org/10.3390/genes12010001
Chicago/Turabian StyleKim, Namshin, Kyoung Hyoun Kim, Won-Jun Lim, Jiwoong Kim, Soon Ae Kim, and Hee Jeong Yoo. 2021. "Whole Exome Sequencing Identifies Novel De Novo Variants Interacting with Six Gene Networks in Autism Spectrum Disorder" Genes 12, no. 1: 1. https://doi.org/10.3390/genes12010001





