Molecular Mechanisms and Biological Functions of Autophagy for Genetics of Hearing Impairment
Abstract
:1. Introduction
2. The Mechanisms and Biologic Functions of Autophagy
2.1. Autophagy Gene-Dependent Pathways for the Formation of Autophagosome
2.2. Autophagy Regulation by Lysosome through mTORC1 and v-ATPase
3. The Role of Autophagy in Auditory System (or Hearing)
3.1. Otic Epithelium
3.2. Hair Cells, Spiral Ganglion Cells, and Brain Stem Nuclei
3.3. Synapse Ribbon
3.4. Auditory Neurons
4. Autophagy- and Lysosomal-Function-Related Genes and Hearing Loss
4.1. Autophagy-Related Genes Essential for Autophagosome Formation
4.1.1. ATG5 Gene
4.1.2. miRNA 96 Gene
4.2. Lysosomal-Function-Related Genes Essential for the Autophagy–Lysosome Pathway
5. Congenital Disorder of Autophagy with Hearing Loss
β-Propeller Protein-Associated Neurodegeneration (BPAN): Mutations in the WDR45 Gene
6. The Effect of Autophagy for Genetics of Hearing Loss
6.1. Genetics of Sensorineural Hearing Loss (DFNA5 and DFNB59) and Autophagy
6.2. Presbycusis Accelerated by Connexin 26 Partial Loss and Autophagy through Nrf2/Keap1 Pathway
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fujimoto, C.; Yamasoba, T. Mitochondria-Targeted Antioxidants for Treatment of Hearing Loss: A Systematic Review. Antioxidants 2019, 8, 109. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.C.; Ryan, A.F. Mechanisms of sensorineural cell damage, death and survival in the cochlea. Front. Aging Neurosci. 2015, 7, 58. [Google Scholar] [CrossRef]
- Morrill, S.; He, D.Z.Z. Apoptosis in inner ear sensory hair cells. J. Otol. 2017, 12, 151–164. [Google Scholar] [CrossRef]
- Dirain, C.O.; Vasquez, T.K.; Antonelli, P.J. Prevention of Chlorhexidine Ototoxicity with Poloxamer in Rats. Otol. Neurotol. 2018, 39, e738–e742. [Google Scholar] [CrossRef]
- Jadidian, A.; Antonelli, P.J.; Ojano-Dirain, C.P. Evaluation of apoptotic markers in HEI-OC1 cells treated with gentamicin with and without the mitochondria-targeted antioxidant mitoquinone. Otol. Neurotol. 2015, 36, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Jankauskas, S.S.; Plotnikov, E.Y.; Morosanova, M.A.; Pevzner, I.B.; Zorova, L.D.; Skulachev, V.P.; Zorov, D.B. Mitochondria-targeted antioxidant SkQR1 ameliorates gentamycin-induced renal failure and hearing loss. Biochemistry 2012, 77, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.M.; Ryter, S.W.; Levine, B. Autophagy in human health and disease. N. Engl. J. Med. 2013, 368, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Tsuboyama, K.; Koyama-Honda, I.; Sakamaki, Y.; Koike, M.; Morishita, H.; Mizushima, N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 2016, 354, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef]
- Sun-Wada, G.H.; Wada, Y. Role of vacuolar-type proton ATPase in signal transduction. Biochim. Biophys. Acta 2015, 1847, 1166–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011, 334, 678–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Remaileh, M.; Wyant, G.A.; Kim, C.; Laqtom, N.N.; Abbasi, M.; Chan, S.H.; Freinkman, E.; Sabatini, D.M. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 2017, 358, 807–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aburto, M.R.; Sánchez-Calderón, H.; Hurlé, J.M.; Varela-Nieto, I.; Magariños, M. Early otic development depends on autophagy for apoptotic cell clearance and neural differentiation. Cell Death Dis. 2012, 3, e394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiuri, M.; Kroemer, G. Autophagy in stress and disease. Cell Death Differ. 2015, 22, 365–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nayagam, B.A.; Muniak, M.A.; Ryugo, D.K. The spiral ganglion: Connecting the peripheral and central auditory systems. Hear. Res. 2011, 278, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Reijntjes, D.O.J.; Pyott, S.J. The afferent signaling complex: Regulation of type I spiral ganglion neuron responses in the auditory periphery. Hear. Res. 2016, 336, 1–16. [Google Scholar] [CrossRef] [Green Version]
- MacLeod, K.M.; Carr, C.E. Beyond timing in the auditory brainstem: Intensity coding in the avian cochlear nucleus angularis. Prog. Brain Res. 2007, 165, 123–133. [Google Scholar]
- de Iriarte Rodríguez, R.; Pulido, S.; Rodríguez-de la Rosa, L.; Magariños, M.; Varela-Nieto, I. Age-regulated function of autophagy in the mouse inner ear. Hear. Res. 2015, 330, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, C.; Iwasaki, S.; Urata, S.; Morishita, H.; Sakamaki, Y.; Fujioka, M.; Kondo, K.; Mizushima, N.; Yamasoba, T. Autophagy is essential for hearing in mice. Cell Death Dis. 2017, 8, 2780. [Google Scholar] [CrossRef]
- Xiong, W.; Wei, W.; Qi, Y.; Du, Z.; Qu, T.; Liu, K.; Gong, S. Autophagy is Required for Remodeling in Postnatal Developing Ribbon Synapses of Cochlear Inner Hair Cells. Neuroscience 2020, 431, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Petitpré, C.; Wu, H.; Sharma, A.; Tokarska, A.; Fontanet, P.; Wang, Y.; Helmbacher, F.; Yackle, K.; Silberberg, G.; Hadjab, S.; et al. Neuronal heterogeneity and stereotyped connectivity in the auditory afferent system. Nat. Commun. 2018, 9, 3691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghavami, S.; Cunnington, R.H.; Gupta, S.; Yeganeh, B.; Filomeno, K.L.; Freed, D.H.; Chen, S.; Klonisch, T.; Halayko, A.J.; Ambrose, E.; et al. Autophagy is a regulator of TGF-β1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. 2015, 6, e1696. [Google Scholar] [CrossRef] [Green Version]
- Pang, M.; Wang, H.; Rao, P.; Zhao, Y.; Xie, J.; Cao, Q.; Wang, Y.; Wang, Y.M.; Lee, V.W.; Alexander, S.I.; et al. Autophagy links β-catenin and Smad signaling to promote epithelial-mesenchymal transition via upregulation of integrin linked kinase. Int. J. Biochem. Cell Biol. 2016, 76, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Wang, Q.J.; Holstein, G.R.; Friedrich, V.L., Jr.; Iwata, J.; Kominami, E.; Chait, B.T.; Tanaka, K.; Yue, Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA 2007, 104, 14489–14494. [Google Scholar] [CrossRef] [Green Version]
- Ye, B.; Wang, Q.; Hu, H.; Shen, Y.; Fan, C.; Chen, P.; Ma, Y.; Wu, H.; Xiang, M. Restoring autophagic flux attenuates cochlear spiral ganglion neuron degeneration by promoting TFEB nuclear translocation via inhibiting MTOR. Autophagy 2019, 15, 998–1016. [Google Scholar] [CrossRef]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabeya, Y.; Suzuki, K.; Tokuhisa, T.; Ohsumi, Y.; Yoshimori, T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell. Biol. 2001, 152, 657–668. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Ohsumi, Y.; Yoshimori, T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 2002, 27, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.; Zhou, X.J.; Zhang, H. Exploring the Role of Autophagy-Related Gene 5 (ATG5) Yields Important Insights Into Autophagy in Autoimmune/Autoinflammatory Diseases. Front. Immunol. 2018, 9, 2334. [Google Scholar] [CrossRef]
- Mencía, A.; Modamio-Høybjør, S.; Redshaw, N.; Morín, M.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L.A.; del Castillo, I.; Steel, K.P.; Dalmay, T.; et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009, 41, 609–613. [Google Scholar]
- Soldà, G.; Robusto, M.; Primignani, P.; Castorina, P.; Benzoni, E.; Cesarani, A.; Ambrosetti, U.; Asselta, R.; Duga, S. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum. Mol. Genet. 2012, 21, 577–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Yang, H.Z.; Dong, B.J.; Zou, H.B.; Zhou, Y.; Kong, X.M.; Huang, Y.R. Biphasic regulation of autophagy by miR-96 in prostate cancer cells under hypoxia. Oncotarget 2014, 5, 9169–9182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Duve, C. The lysosome turns fifty. Nat. Cell. Biol. 2005, 7, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Klionsky, D.J. Autophagy in health and disease: A double-edged sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colacurcio, D.J.; Nixon, R.A. Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res. Rev. 2016, 32, 75–88. [Google Scholar] [CrossRef] [Green Version]
- Mindell, J.A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 2012, 74, 69–86. [Google Scholar] [CrossRef] [Green Version]
- Escobar, L.I.; Simian, C.; Treard, C.; Hayek, D.; Salvador, C.; Guerra, N.; Matos, M.; Medeiros, M.; Enciso, S.; Camargo, M.D.; et al. Mutations in ATP6V1B1 and ATP6V0A4 genes cause recessive distal renal tubular acidosis in Mexican families. Mol. Genet. Genom. Med. 2016, 4, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Menendez, I.; Carranza, C.; Herrera, M.; Marroquin, N.; Foster, J., 2nd; Cengiz, F.B.; Bademci, G.; Tekin, M. Dominant deafness-onychodystrophy syndrome caused by an ATP6V1B2mutation. Clin. Case Rep. 2017, 5, 376–379. [Google Scholar] [CrossRef]
- Zhao, W.; Gao, X.; Qiu, S.; Gao, B.; Gao, S.; Zhang, X.; Kang, D.; Han, W.; Dai, P.; Yuan, Y. A subunit of V-ATPases, ATP6V1B2, underlies the pathology of intellectual disability. Ebiomedicine 2019, 45, 408–421. [Google Scholar] [CrossRef] [Green Version]
- Palazzo, V.; Provenzano, A.; Becherucci, F.; Sansavini, G.; Mazzinghi, B.; Orlandini, V.; Giunti, L.; Roperto, R.M.; Pantaleo, M.; Artuso, R.; et al. The genetic and clinical spectrum of a large cohort of patients with distalrenal tubular acidosis. Kidney Int. 2017, 91, 1243–1255. [Google Scholar] [CrossRef]
- Gonda, X.; Eszlari, N.; Anderson, I.M.; Deakin, J.F.; Bagdy, G.; Juhasz, G. Association of ATP6V1B2 rs1106634 with lifetime risk of depression and hippocampal neurocognitive deficits: Possible novel mechanisms in the etiopathology of depression. Transl. Psychiatry 2016, 6, e945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, A.L.; Gottschling, D.E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 2012, 492, 261–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molin, M.; Demir, A.B. Linking Peroxiredoxin and Vacuolar-ATPase Functions in Calorie Restriction-Mediated Life Span Extension. Int. J. Cell Biol. 2014, 2014, 913071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, A.; Castells-Nobau, A.; Meziane, H.; Oortveld, M.A.; Houbaert, X.; Iacono, G.; Martin, C.; Mittelhaeuser, C.; Lalanne, V.; Kramer, J.M.; et al. Conditional depletion of intellectual disability and Parkinsonism candidate gene ATP6AP2 in fly and mouse induces cognitive impairment and neurodegeneration. Mol. Genet. 2015, 24, 6736–6755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, C.R.; Gahl, W.A. Lysosomal storage diseases. Tranlational Sci. Rare Dis. 2017, 2, 1–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bamiou, D.E.; Campbell, P.; Liasis, A.; Page, J.; Sirimanna, T.; Boyd, S.; Vellodi, A.; Harris, C. Audiometric abnormalities in children with Gaucher disease type 3. Neuropediatry 2001, 32, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Keilmann, A.; Hajioff, D.; Ramaswami, U. FOS Investigators. Ear symptoms in children with Fabry disease: Data from the Fabry Outcome Survey. J. Inherit. Metab Dis. 2009, 32, 739. [Google Scholar] [CrossRef] [PubMed]
- van Capelle, C.I.; Goedegebure, A.; Homans, N.C.; Hoeve, H.L.; Reuser, A.J.; van der Ploeg, A.T. Hearing loss in Pompe disease revisited: Results from a study of 24 children. J. Inherit. Metab. Dis. 2010, 33, 597–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, K.A.; Gordon-Salant, S.; Yanjanin, N.; Zalewski, C.; Houser, A.; Porter, F.D.; Brewer, C.C. Auditory phenotype of Niemann-Pick disease, type C1. Ear Hear. 2014, 35, 110–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schachern, P.A.; Cureoglu, S.; Tsuprun, V.; Paparella, M.M.; Whitley, C.B. Age-related functional and histopathological changes of the ear in the MPS I mouse. Int. J. Pediatr. Otorhinolaryngol. 2007, 71, 197–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skowyra, M.L.; Schlesinger, P.H.; Naismith, T.V.; Hanson, P.I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 2018, 360, 6384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, A.R.A.; Saftig, P. Lysosomal storage disorders—Challenges, concepts and avenues for therapy: Beyond rare diseases. J. Cell Sci. 2019, 132, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi-Fakhari, D.; Saffari, A.; Wahlster, L.; Lu, J.; Byrne, S.; Hoffmann, G.F.; Jungbluth, H.; Sahin, M. Congenital disorders of autophagy: An emerging novel class of inborn errors of neuro-metabolism. Brain 2016, 139, 317–337. [Google Scholar] [CrossRef] [PubMed]
- Haack, T.B.; Hogarth, P.; Kruer, M.C.; Gregory, A.; Wieland, T.; Schwarzmayr, T. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 2012, 91, 1144–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proikas-Cezanne, T.; Waddell, S.; Gaugel, A.; Frickey, T.; Lupas, A.; Nordheim, A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 2004, 23, 9314–9325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stirnimann, C.U.; Petsalaki, E.; Russell, R.B.; Muller, C.W. WD40 proteins propel cellular networks. Trends Biochem. Sci. 2010, 35, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network organization of the human autophagy system. Nature 2010, 466, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Yang, P.; Huang, X.; Hu, W.; Guo, B.; Wu, F. TheWD40 repeat PtdIns (3) P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell 2011, 21, 343–357. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.G.; Sun, L.; Miao, G.; Ji, C.; Zhao, H.; Sun, H.; Miao, L.; Yoshii, S.R.; Mizushima, N.; Wang, X.; et al. The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy 2015, 11, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Asano, T.; Komatsu, M.; Yamaguchi-Iwai, Y.; Ishikawa, F.; Mizushima, N.; Iwai, K. Distinct mechanisms of ferritin delivery to lysosomes in iron- depleted and iron-replete cells. Mol. Cell Biol. 2011, 31, 2040–2052. [Google Scholar] [CrossRef] [Green Version]
- Mancias, J.D.; Wang, X.; Gygi, S.P.; Harper, J.W.; Kimmelman, A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014, 509, 105–109. [Google Scholar] [CrossRef] [PubMed]
- De Beeck, K.O.; Van Camp, G.; Thys, S.; Cools, N.; Callebaut, I.; Vrijens, K.; Van Laer, L. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. Eur. J. Hum. Genet. 2011, 19, 965–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Gao, W.; Shi, X.; Ding, J.; Liu, W.; He, H.; Wang, K.; Shao, F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017, 547, 99–103. [Google Scholar] [CrossRef]
- Van Laer, L.; Huizing, E.H.; Verstreken, M.; van Zuijlen, D.; Wauters, J.G.; Bossuyt, P.J.; Van de Heyning, P.; McGuirt, W.T.; Smith, R.J.; Willems, P.J.; et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat. Genet. 1998, 20, 194–197. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, L.; Liu, L.; Gao, P.; Tian, W.; Wang, X.; Jin, H.; Xu, H.; Chen, Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 2010, 1, 468–477. [Google Scholar] [CrossRef] [Green Version]
- Rogers, C.; Fernandes-Alnemri, T.; Mayes, L.; Alnemri, D.; Cingolani, G.; Alnemri, E.S. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat. Commun. 2017, 8, 14128. [Google Scholar] [CrossRef] [Green Version]
- Collin, R.W.; Kalay, E.; Oostrik, J.; Caylan, R.; Wollnik, B.; Arslan, S.; den Hollander, A.I.; Birinci, Y.; Lichtner, P.; Strom, T.M.; et al. Involvement of DFNB59 mutations in autosomal recessive nonsyndromic hearing impairment. Hum. Mutat. 2007, 28, 718–723. [Google Scholar] [CrossRef]
- Mujtaba, G.; Bukhari, I.; Fatima, A.; Naz, S. A p.C343S missense mutation in PJVK causes progressive hearing loss. Gene 2012, 504, 98–101. [Google Scholar] [CrossRef] [Green Version]
- Delmaghani, S.; del Castillo, F.J.; Michel, V.; Leibovici, M.; Aghaie, A.; Ron, U.; Van Laer, L.; Ben-Tal, N.; Van Camp, G.; Weil, D.; et al. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat. Genet. 2006, 38, 770–778. [Google Scholar] [CrossRef]
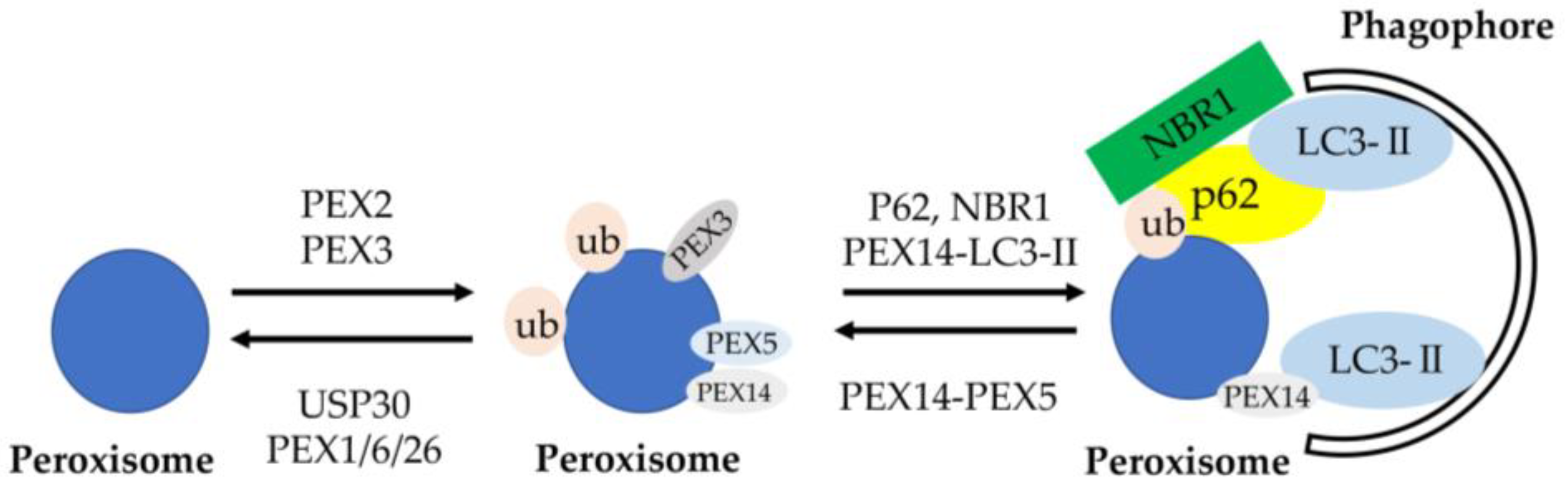
- Defourny, J.; Aghaie, A.; Perfettini, I.; Avan, P.; Delmaghani, S.; Petit, C. Pejvakin-mediated pexophagy protects auditory hair cells against noise-induced damage. Proc. Natl. Acad. Sci. USA 2019, 116, 8010–8017. [Google Scholar] [CrossRef] [Green Version]
- Germain, K.; Kim, P.K. Pexophagy: A Model for Selective Autophagy. Int. J. Mol. Sci. 2020, 21, 578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, M.; Minns, M.; Greenberg, E.N.; Diaz-Aponte, J.; Pestonjamasp, K.; Johnson, J.L.; Rathkey, J.K.; Abbott, D.W.; Wang, K.; Shao, F.; et al. N-GSDMD trafficking to neutrophil organelles facilitates IL-1β release independently of plasma membrane pores and pyroptosis. Nat. Commun. 2020, 11, 2212. [Google Scholar] [CrossRef]
- Kelsell, D.P.; Dunlop, J.; Stevens, H.P.; Lench, N.J.; Liang, J.N.; Parry, G.; Mueller, R.F.; Leigh, I.M. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 1997, 387, 80–88. [Google Scholar] [CrossRef]
- Kenna, M.A.; Feldman, H.A.; Neault, M.W.; Frangulov, A.; Wu, B.L.; Fligor, B.; Rehm, H.L. Audiologic phenotype and progression in GJB2 (Connexin 26) hearing loss. Arch. Otolaryngol.-Head Neck Surg. 2010, 136, 81–87. [Google Scholar] [CrossRef] [Green Version]
- Snoeckx, R.L.; Huygen, P.L.; Feldmann, D.; Marlin, S.; Denoyelle, F.; Waligora, J.; Mueller-Malesinska, M.; Pollak, A.; Ploski, R.; Murgia, A.; et al. GJB2 mutations and degree of hearing loss: A multicenter study. Am. J. Hum. Genet. 2005, 77, 945–957. [Google Scholar] [CrossRef] [Green Version]
- Font, M.A.; Feliubadaló, L.; Estivill, X.; Nunes, V.; Golomb, E.; Kreiss, Y.; Pras, E.; Bisceglia, L.; d’Adamo, A.P.; Zelante, L.; et al. International Cystinuria Consortium. Functional analysis of mutations in SLC7A9, and genotype-phenotype correlation in non-Type I cystinuria. Hum. Mol. Genet. 2001, 10, 305–316. [Google Scholar] [CrossRef] [Green Version]
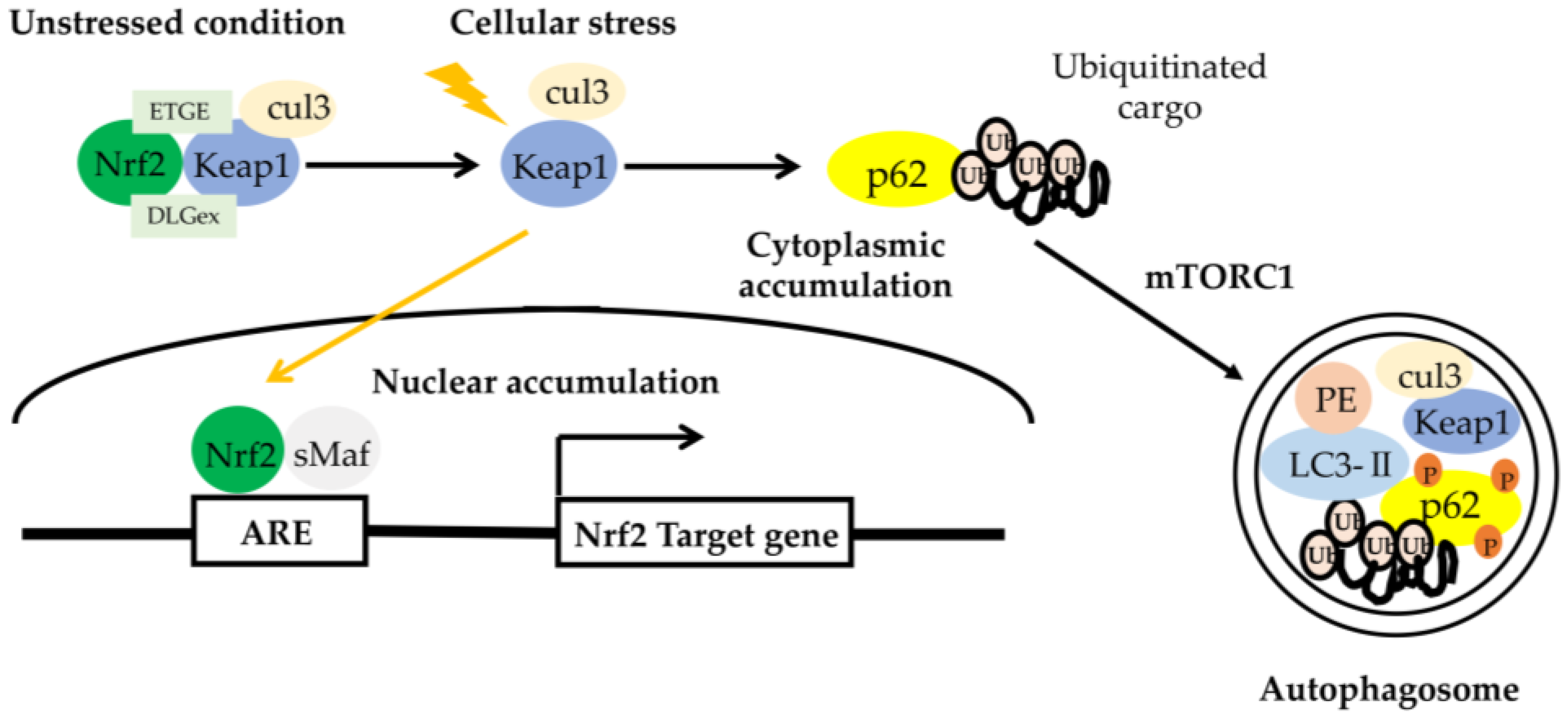
- Fetoni, A.R.; Zorzi, V.; Paciello, F.; Ziraldo, G.; Peres, C.; Raspa, M.; Scavizzi, F.; Salvatore, A.M.; Crispino, G.; Tognola, G.; et al. Cx26 partial loss causes accelerated presbycusis by redox imbalance and dysregulation of Nfr2 pathway. Redox Biol. 2018, 19, 301–317. [Google Scholar] [CrossRef]
- Johnson, S.L.; Ceriani, F.; Houston, O.; Polishchuk, R.; Polishchuk, E.; Crispino, G.; Zorzi, V.; Mammano, F.; Marcotti, W. Connexin-Mediated Signaling in Nonsensory Cells Is Crucial for the Development of Sensory Inner Hair Cells in the Mouse Cochlea. Neuroscience 2017, 37, 258–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsuishi, Y.; Motohashi, H.; Yamamoto, M. The Keap1-Nrf2 system in cancers: Stress response and anabolic metabolism. Front. Oncol. 2012, 2, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, K.; Dan, K.; Goto, F.; Tsuchihashi, N.; Nomura, Y.; Fujioka, M.; Kanzaki, S.; Ogawa, K. The autophagy pathway maintained signaling crosstalk with the Keap1-Nrf2 system through p62 in auditory cells under oxidative stress. Cell Signal. 2015, 27, 382–393. [Google Scholar] [CrossRef]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768. [Google Scholar] [CrossRef] [Green Version]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, S.; Saito, T.; Obata, M.; Koide, R.H.; Ichimura, Y.; Komatsu, M. Negative Regulation of the Keap1-Nrf2 Pathway by a p62/Sqstm1 Splicing Variant. Mol. Cell Biol. 2018, 38, e00642-17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.D.; Hannink, M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell Biol. 2003, 23, 8137–8151. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, M.; Li, L.; Iwamoto, N.; Nakajima-Takagi, Y.; Kaneko, H.; Nakayama, Y.; Eguchi, M.; Wada, Y.; Kumagai, Y.; Yamamoto, M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell Biol. 2009, 29, 493–502. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Kominami, E.; Tanaka, K.; Komatsu, M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 2008, 4, 1063–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene | Gene Locus | Encoding | Genetic Defects | Related Disease | Affected Process of Autophagy |
|---|---|---|---|---|---|
| Atg5 | 6q21 | ATG protein | Deletion | Autoinflammatory disease Autoimmune disease | Autophagosome formation |
| miRNA96 | 7q32.2 | DFNA50 (OMIM #613074) | Point mutations | Sensorineural hearing loss | Autophagosome formation |
| WDR45 | Xp11.23 | WD repeat protein | Uncovered mutations | BPAN | Autophagosome formation |
| GBA | 1q21 | (Lyso)glucosylceramide | Missense mutations Point mutations Deletions Insertions Splicing aberrations Various rearrangements | Gaucher disease Type 1 (GD1) Type 2 (GD2) Type 3 (GD3) | Lysosome biogenesis |
| GLA | Xq22.1 | lysosomal α-galactosidase A | Missense mutations Nonsense mutations Splicing mutations Deletions Insertions | Fabry disease | Lysosome biogenesis |
| GAA | 17q25.3 | lysosomal α-glucosidase | Nonsense mutations Multiple exon deletion | Pompe disease | Lysosome biogenesis |
| NPC1 | 18q11.2 | NPC protein | Missense mutations Point mutation Duplication mutation Splicing mutation Frame deletion | Niemann–Pick type C | Lysosome biogenesis |
| NPC2 | 14q24.3 | NPC protein | Missense mutations of homozygous state | Niemann–Pick type C | Lysosome biogenesis |
| IDUA | 4p16.3 | alpha-L-iduronidase | Missense mutations Nonsense mutation Deletion | Mucopolysaccharidoses | Lysosome biogenesis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayashi, K.; Suzuki, Y.; Fujimoto, C.; Kanzaki, S. Molecular Mechanisms and Biological Functions of Autophagy for Genetics of Hearing Impairment. Genes 2020, 11, 1331. https://doi.org/10.3390/genes11111331
Hayashi K, Suzuki Y, Fujimoto C, Kanzaki S. Molecular Mechanisms and Biological Functions of Autophagy for Genetics of Hearing Impairment. Genes. 2020; 11(11):1331. https://doi.org/10.3390/genes11111331
Chicago/Turabian StyleHayashi, Ken, Yuna Suzuki, Chisato Fujimoto, and Sho Kanzaki. 2020. "Molecular Mechanisms and Biological Functions of Autophagy for Genetics of Hearing Impairment" Genes 11, no. 11: 1331. https://doi.org/10.3390/genes11111331






