CDC-42 Interactions with Par Proteins Are Critical for Proper Patterning in Polarization
Abstract
:1. Introduction
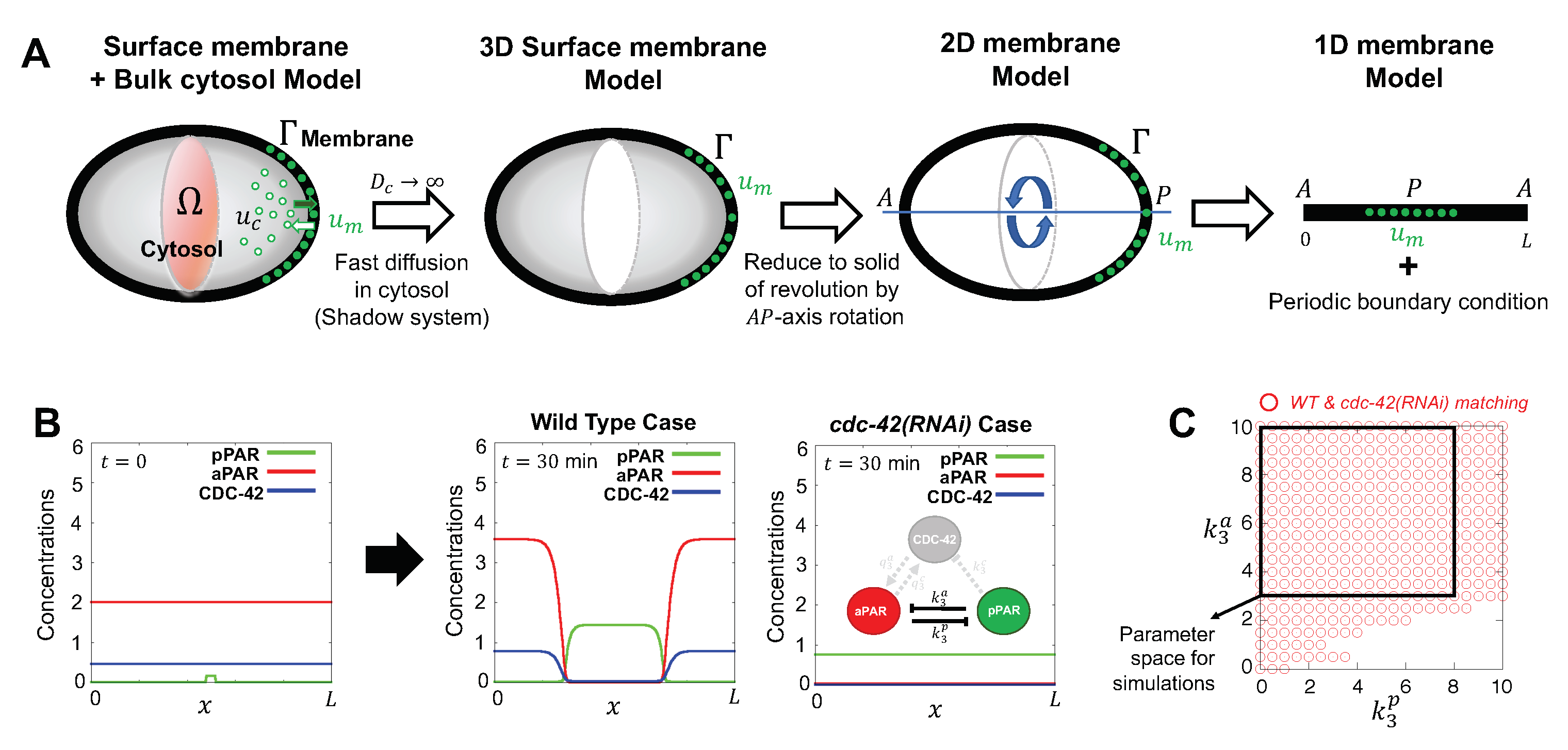
2. The Mathematical Model
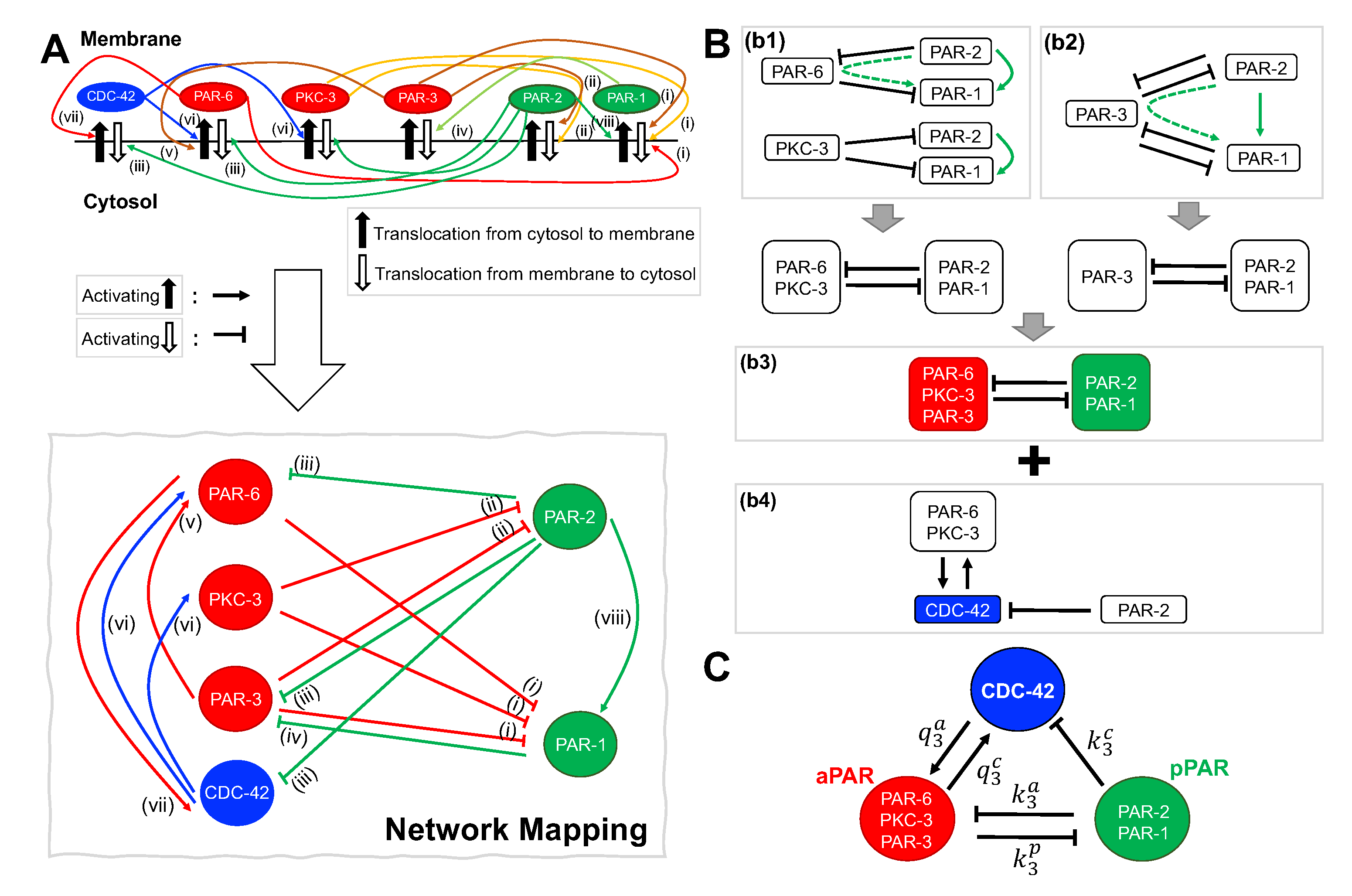
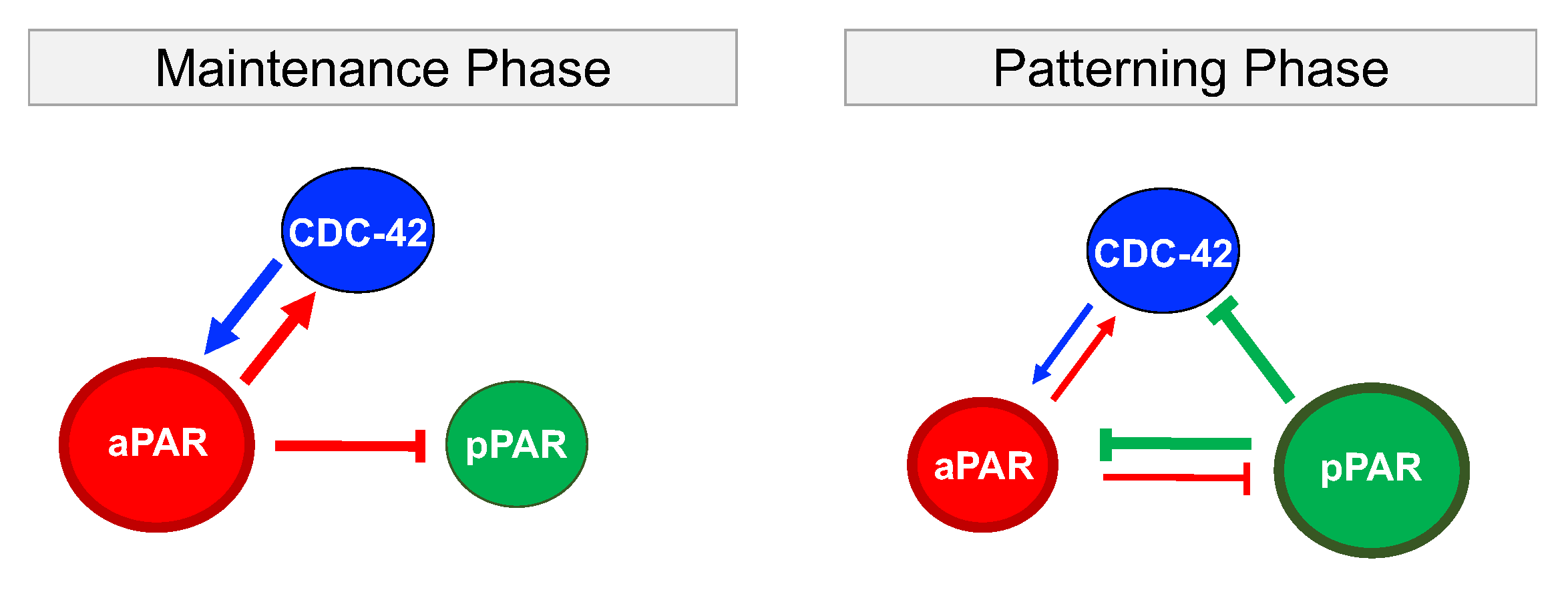
2.1. Network of Par Protein and Cdc-42 Interactions
2.2. Model Equations for Apar, Ppar and Cdc-42 Network
2.3. Parameter Values
2.4. Initial Conditions
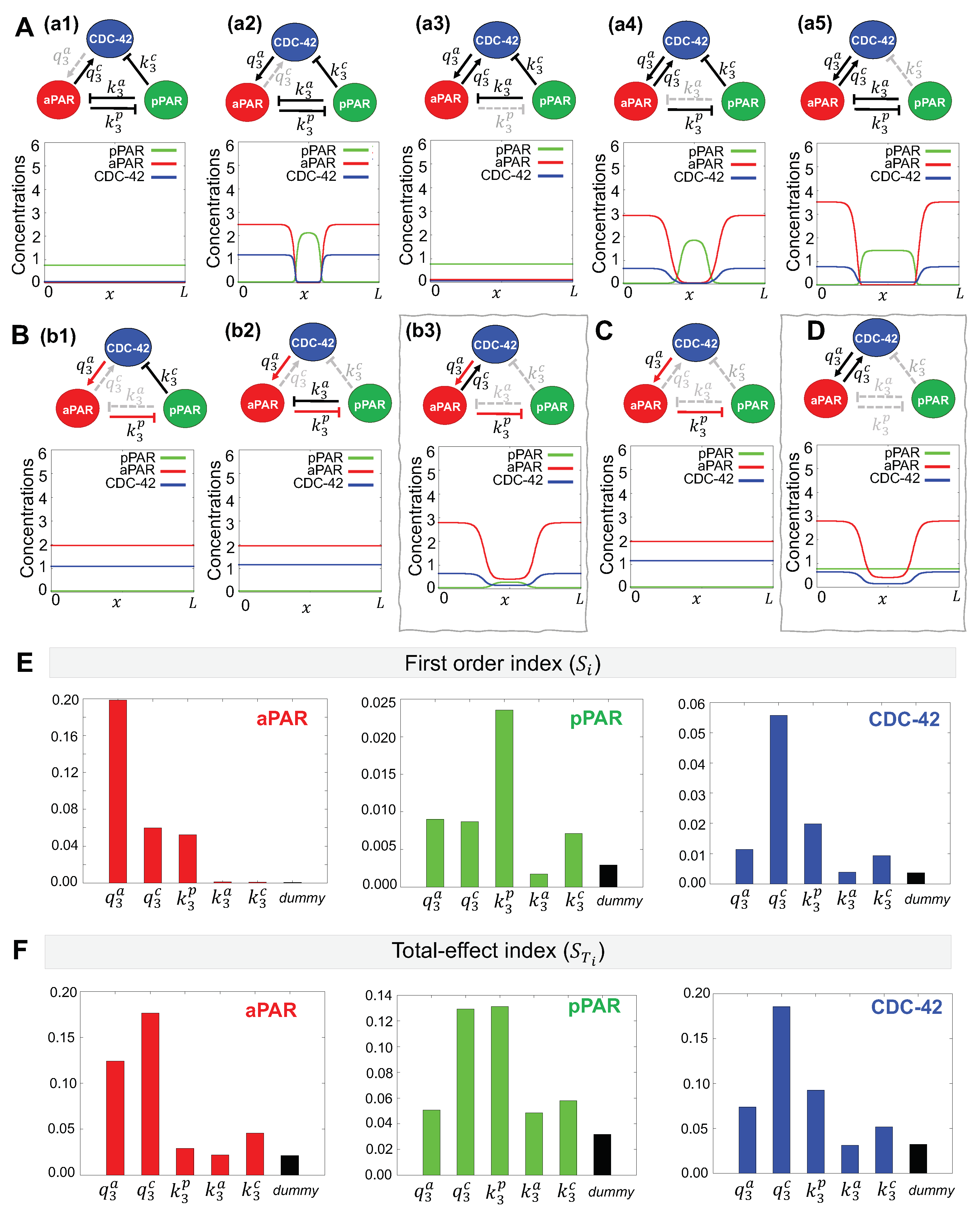
2.5. Minimal Network Analysis
2.6. eFAST Sensitivity Analysis
3. Results
3.1. Critical Network Interactions and Parameters for Maintenance of Par Protein Polarization
3.1.1. Minimal Network for Maintenance of Par Protein Polarization
3.1.2. Critical Parameters for Maintenance of Par Protein Polarization
3.2. Critical Network Interactions and Parameters for Generation of Par Protein Polarization
3.2.1. Minimal Network for Generation of Par Protein Polarization
3.2.2. Critical Parameters for Generation of Par Protein Polarization
4. Discussion
Author Contributions
Funding
Conflicts of Interest
Appendix A
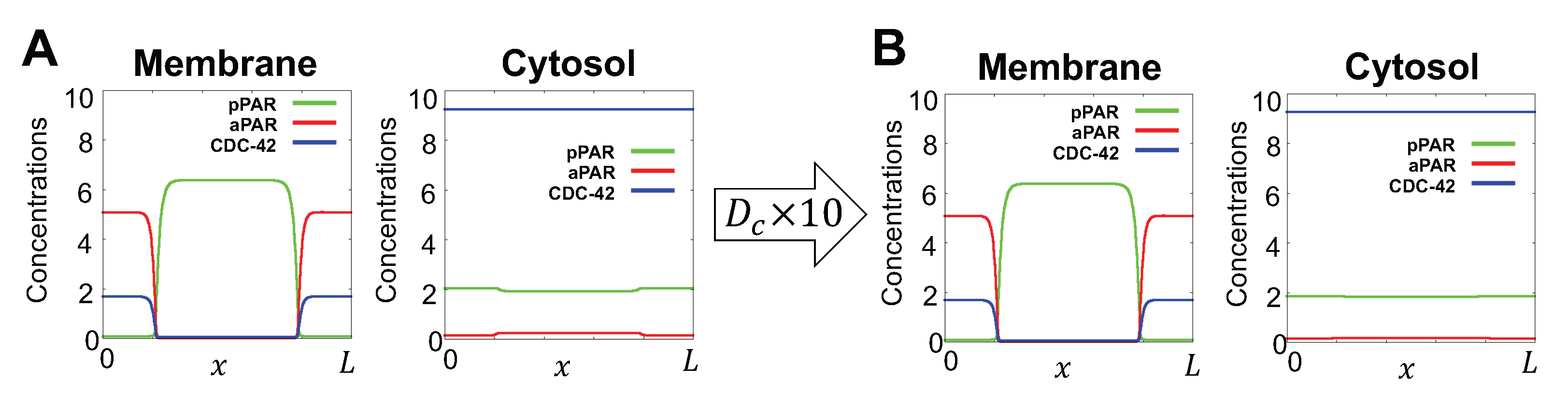
Appendix A.1. Model Reduction
Appendix A.2. Additional Simulations and Representative Parameter Set
| Parameter | Dimensional | Non-Dim. | Parameter | Dimensional | Non-Dim. |
| Value | Value | Value | Value | ||
| L | 142.75 m) | 1.0 | 1.0 | ||
| 0.28 m | 0.15 m | ||||
| 0.10 m | 14.0 m | ||||
| 7.50 m | 10.0 m | ||||
| 0.015 | 0.03 | 0.100 | 0.20 | ||
| 0.015 | 0.03 | 0.05 | 0.10 | ||
| 0.03 | 0.06 | 0.05 | 0.10 | ||
| 0.20 L | 0.20 | 1.00 | 1.00 | ||
| 2.60 | 5.20 | 1.25 L | 1.25 | ||
| 1.00 | 1.00 | 2.60 | 5.20 | ||
| 1.25 L | 1.25 | 1.00 | 1.00 | ||
| 2.60 | 5.20 | 1.25 L | 1.25 | ||
| 1.00 | 1.00 | 2.60 | 5.20 | ||
| 1.00 L | 1.00 | 1.00 | 1.00 | ||
| 0.50 L | 1.00 | 1.00 | |||
| 2.00 | 5.00 | ||||
| Parameter range for sensitivity analysis | |||||
| Parameter | Dimensional | Non-Dim. | Parameter | Dimensional | Non-Dim. |
| Range | Range | Range | Range | ||
| [1.5, 5.0] | [3, 10] | [0.0, 4.0] | [0, 8] | ||
| [0.0, 5.0] | [0, 10] | [0.0, 5.0] | [0, 10] | ||
| [0.0, 5.0] L | [0, 10] | ||||
Appendix A.3. Variance-Based Sensitivity Analysis by Using Extended Fourier Amplitude Sensitivity Test (eFAST) Method
| Index | M | |||
|---|---|---|---|---|
| 4 | 2 | 2578 | ||
| 4 | 2 | 2050 |
Appendix A.4. First Order Index and Total Index for the Dummy Parameter
| Maintenance | |||
| aPAR | pPAR | CDC-42 | |
| 0.000869 | 0.002956 | 0.003730 | |
| 0.021139 | 0.031567 | 0.032265 | |
| Generation | |||
| aPAR | pPAR | CDC-42 | |
| 0.001711 | 0.004020 | 0.002282 | |
| 0.023502 | 0.160468 | 0.055702 |
References
- Hodge, R.G.; Ridley, A.J. Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 2016, 17, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Campanale, J.P.; Sun, T.Y.; Montell, D.J. Development and dynamics of cell polarity at a glance. J. Cell Sci. 2017, 130, 1201–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, A.; Ohno, S. The PAR-aPKC system: Lessons in polarity. J. Cell Sci. 2006, 119, 979–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, M.; Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef] [Green Version]
- Muthuswamy, S.K.; Xue, B. Cell polarity as a regulator of cancer cell behavior plasticity. Annu. Rev. Cell Dev. Biol. 2012, 28, 599–625. [Google Scholar] [CrossRef] [Green Version]
- Ridley, A.J. Rho GTPases and cell migration. J. Cell Sci. 2001, 114, 2713–2722. [Google Scholar]
- Johnson, D.I. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. MMBR 1999, 63, 54–105. [Google Scholar] [CrossRef] [Green Version]
- Amano, M.; Nakayama, M.; Kaibuchi, K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton 2010, 67, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Nobes, C.D.; Hall, A. Rho GTPases Control Polarity, Protrusion, and Adhesion during Cell Movement. J. Cell Biol. 1999, 144, 1235–1244. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, B.; Macara, I.G. The PAR proteins: Fundamental players in animal cell polarization. Dev. Cell 2007, 13, 609–622. [Google Scholar] [CrossRef] [Green Version]
- Macara, I.G. Parsing the Polarity Code. Nat. Rev. Mol. Cell Biol. 2004, 5, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Kemphues, K.J.; Priess, J.R.; Morton, D.G.; Cheng, N.S. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 1988, 52, 311–320. [Google Scholar] [CrossRef]
- Gérard, A.; Mertens, A.; van der Kammen, R.A.; Collard, J.G. The Par polarity complex regulates Rap1- and chemokine-induced T cell polarization. J. Cell Biol. 2007, 176, 863–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonegg, S.; Hyman, A. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Dev. Camb. Engl. 2006, 133, 3507–3516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumfer, K.T.; Cook, S.J.; Squirrell, J.M.; Eliceiri, K.W.; Peel, N.; O’Connell, K.F.; White, J.G. CGEF-1 and CHIN-1 Regulate CDC-42 Activity during Asymmetric Division in the Caenorhabditis elegans Embryo. Mol. Biol. Cell 2010, 21, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Cuenca, A.A.; Schetter, A.; Aceto, D.; Kemphues, K.; Seydoux, G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development 2002, 130, 1255–1265. [Google Scholar] [CrossRef] [Green Version]
- Munro, E.; Nance, J. Cortical Flows Powered by Asymmetrical Contraction Transport PAR Proteins to Establish and Maintain Anterior-Posterior Polarity in the Early C. elegans Embryo. Dev. Cell 2004, 7, 413–424. [Google Scholar] [CrossRef] [Green Version]
- Aceto, D.; Beers, M.; Kemphues, K.J. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev. Biol. 2006, 299, 386–397. [Google Scholar] [CrossRef] [Green Version]
- Gotta, M.; Abraham, M.C.; Ahringer, J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 2001, 11, 482–488. [Google Scholar] [CrossRef] [Green Version]
- Lang, C.F.; Munro, E. The PAR proteins: From molecular circuits to dynamic self-stabilizing cell polarity. Co. Biol. 2017, 144, 3405–3416. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, J.; Peglion, F.; Martin, J.; Hubatsch, L.; Reich, J.; Hirani, N.; Gubieda, A.G.; Roffey, J.; Fernandes, A.R.; Johnston, D.S.; et al. aPKC Cycles between Functionally Distinct PAR Protein Assemblies to Drive Cell Polarity. Dev. Cell 2017, 42, 400–415. [Google Scholar] [CrossRef] [Green Version]
- Small, L.E.; Dawes, A.T. PAR proteins regulate maintenance-phase myosin dynamics during Caenorhabditis elegans zygote polarization. Mol. Biol. Cell 2017, 28, 2220–2231. [Google Scholar] [CrossRef] [PubMed]
- Arata, Y.; Hiroshima, M.; Pack, C.G.; Ramanujam, R.; Motegi, F.; Nakazato, K.; Shindo, Y.; Wiseman, P.W.; Sawa, H.; Kobayashi, T.J.; et al. Cortical Polarity of the RING Protein PAR-2 Is Maintained by Exchange Rate Kinetics at the Cortical-Cytoplasmic Boundary. Cell Rep. 2016, 16, 2156–2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, A.J.; Hunter, C.P. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. CB 2001, 11, 474–481. [Google Scholar] [CrossRef] [Green Version]
- Beatty, A.; Morton, D.G.; Kemphues, K. PAR-2, LGL-1 and the CDC-42 GAP CHIN-1 act in distinct pathways to maintain polarity in the C. elegans embryo. Development 2013, 140, 2005–2014. [Google Scholar] [CrossRef] [Green Version]
- Boyd, L.; Guo, S.; Levitan, D.; Stinchcomb, D.T.; Kemphues, K.J. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 1996, 122, 3075–3084. [Google Scholar]
- Hao, Y.; Boyd, L.; Seydoux, G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell 2006, 10, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Seirin-Lee, S.; Shibata, T. Self-organization and advective transport in the cell polarity formation for asymmetric cell division. J. Theor. Biol. 2015, 382, 1–14. [Google Scholar] [CrossRef]
- Kuhn, T.; Ihalainen, T.O.; Hyvaluoma, J.; Dross, N.; Willman, S.F.; Langowski, J.; Vihinen-Ranta, M.; Timonen, J. Protein Diffusion in Mammalian Cell Cytoplasm. PLoS ONE 2011, 6, e22962. [Google Scholar] [CrossRef] [Green Version]
- Goehring, N.W.; Trong, P.K.; Bois, J.S.; Chowdhury, D.; Nicola, E.M.; Hyman, A.A.; Grill, S.W. Polarization of PAR Proteins by Advective Triggering of a Pattern-Forming System. Science 2011, 334, 1137–1141. [Google Scholar] [CrossRef] [Green Version]
- Saltelli, A.; Tarantola, S.; Campolongo, F.; Ratto, M. Sensitivity Analysis in Practice: A Guide to Assessing Scientific Models; Wiley: Hoboken, NJ, USA, 2004. [Google Scholar]
- Saltelli, A.; Tarantola, S.; Chan, K.P.S. A Quantitative Model-Independent Method for Global Sensitivity Analysis of Model Output. Technometrics 1999, 41, 39–56. [Google Scholar] [CrossRef]
- Gönczy, P. Asymmetric cell division and axis formation in the embryo. WormBook 2005. [Google Scholar] [CrossRef]
- Goehring, N.W.; Hoege, C.; Grill, S.W.; Hyman, A.A. PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. J. Cell Biol. 2011, 193, 583–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saltelli, A.; Tarantola, S.; Campolongo, F. Sensitivity Analysis as an Ingredient of Modeling. Stat. Sci. 2000, 15, 377–395. [Google Scholar]
- Kachur, T.M.; Audhya, A.; Pilgrim, D. UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev. Biol. 2008, 314, 287–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickinson, D.J.; Schwager, F.; Pintard, L.; Gotta, M.; Goldstein, B. A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization. Dev. Cell 2017, 42, 416–434.e11. [Google Scholar] [CrossRef]
- Mack, N.; Georgiou, M. The Interdependence of the Rho GTPases and Apicobasal Cell Polarity. Small GTPases 2014, 5, 10. [Google Scholar] [CrossRef]
- Cusseddu, D.; Edelstein-Keshet, L.; Mackenzie, J.A.; Portet, S.; Madzvamusea, A. A coupled bulk-surface model for cell polarisation. J. Theor. Biol. 2019, 481, 119–135. [Google Scholar] [CrossRef] [Green Version]
- Geßele, R.; Halatek, J.; Würthner, L.; Frey, E. Geometric cues stabilise long-axis polarisation of PAR protein patterns in C. elegans. Nat. Commun. 2020, 11, 539. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Jilkine, A.; Edelstein-Keshet, L. Wave-Pinning and Cell Polarity from a Bistable Reaction-Diffusion System. Biophys. J. 2008, 94, 3684–3697. [Google Scholar] [CrossRef] [Green Version]
- Morton, K.; Mayers, D. Numerical Solution of Partial Differential Equations; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]
- Woolley, T.; Gaffney, E.; Goriely, A. Membrane shrinkage and cortex remodelling are predicted to work in harmony to retract blebs. R. Soc. Open Sci. 2016, 2, 150184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, B.; Ishimoto, K.; Gaffney, E. A new basis for filament simulation in three dimensions. arXiv 2019, arXiv:1907.04823. [Google Scholar]
- Marée, A.F.M.; Jilkine, A.; Dawes, A.; Grieneisen, V.A.; Edelstein-Keshet, L. Polarization and Movement of Keratocytes: A Multiscale Modelling Approach. Bull. Math. Biol. 2006, 68, 1169–1211. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.; Hogue, I.B.; Ray, C.J.; Kirschner, D.E. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 2008, 254, 178–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
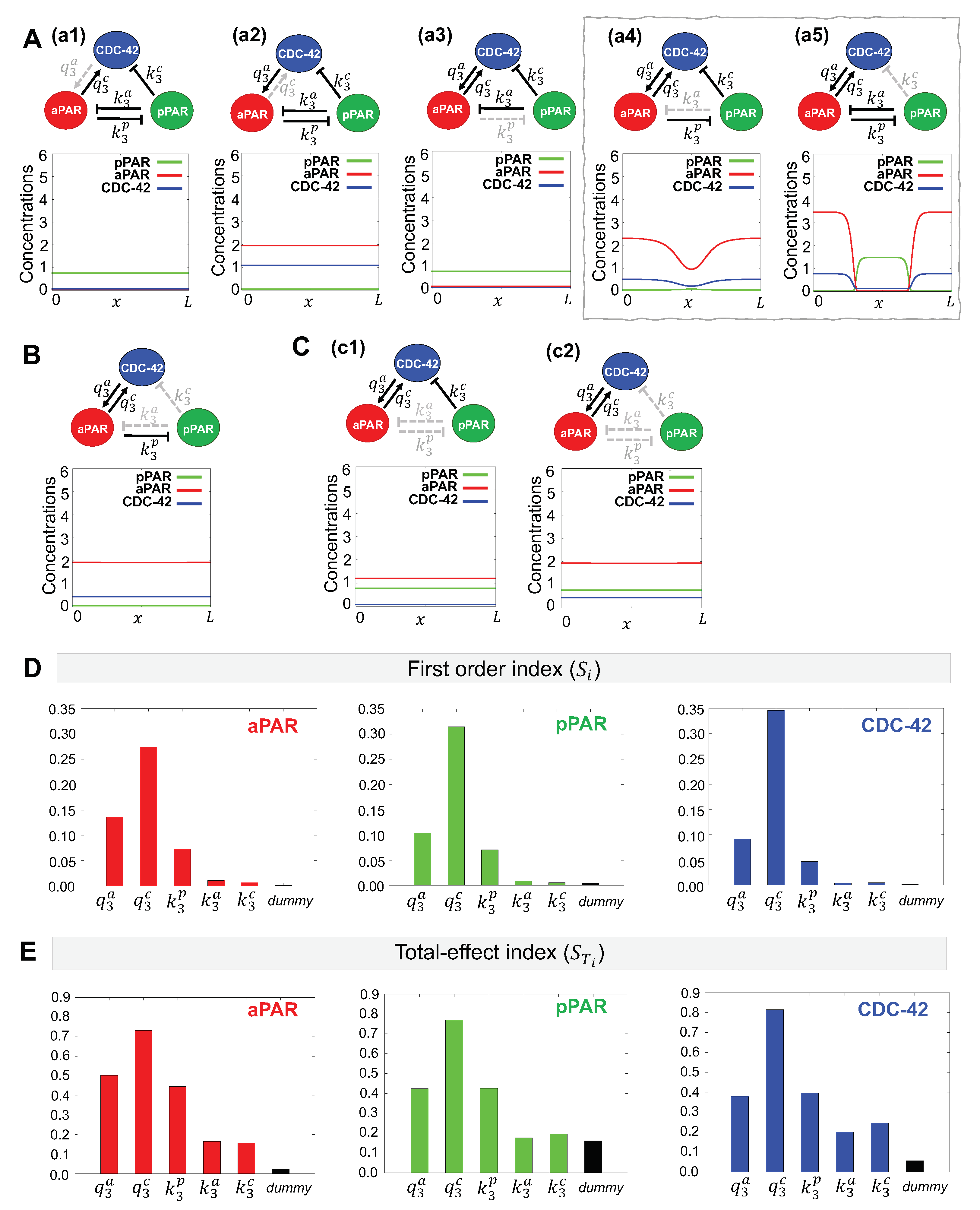
| Maintenance | Generation | ||||||
|---|---|---|---|---|---|---|---|
| Parameter/Interaction | MNA | MNA | |||||
| CDC-42→aPAR | ✓ | Apc | a | ✓ | apc | apc | |
| aPAR→CDC-42 | ✓ | apC | ApC | ✓ | APC | APC | |
| aPAR⊣pPAR | ✓ | aPc | Pc | ✓ | apc | apc | |
| pPAR⊣aPAR | p | ✓ | ac | ||||
| pPAR⊣CDC-42 | pc | apc | ✓ | ac | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seirin-Lee, S.; Gaffney, E.A.; Dawes, A.T. CDC-42 Interactions with Par Proteins Are Critical for Proper Patterning in Polarization. Cells 2020, 9, 2036. https://doi.org/10.3390/cells9092036
Seirin-Lee S, Gaffney EA, Dawes AT. CDC-42 Interactions with Par Proteins Are Critical for Proper Patterning in Polarization. Cells. 2020; 9(9):2036. https://doi.org/10.3390/cells9092036
Chicago/Turabian StyleSeirin-Lee, Sungrim, Eamonn A. Gaffney, and Adriana T. Dawes. 2020. "CDC-42 Interactions with Par Proteins Are Critical for Proper Patterning in Polarization" Cells 9, no. 9: 2036. https://doi.org/10.3390/cells9092036





