Transcriptional Regulation of Differentiation and Functions of Effector T Regulatory Cells
Abstract
:1. Introduction
2. Transcription Factors in a Core eTreg Transcriptional Program
2.1. IRF4
2.2. BATF and JunB
2.3. Blimp1
2.4. Myb
2.5. NF-κB
2.6. Id2, Id3, E2A, and HEB
2.7. TCF1 and LEF1
2.8. Foxo1
3. Transcription Factors Regulating Differentiation of Distinct Subsets of eTreg Cells
3.1. T-Bet, GATA3, RORγt, Bcl6 and STAT5
3.2. c-Maf
3.3. STAT3
3.4. PPARγ and RORα
4. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sakaguchi, S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 2004, 22, 531–562. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.L.; Christie, J.; Ramsdell, F.; Brunkow, M.E.; Ferguson, P.J.; Whitesell, L.; Kelly, T.E.; Saulsbury, F.T.; Chance, P.F.; Ochs, H.D. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001, 27, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Brunkow, M.E.; Jeffery, E.W.; Hjerrild, K.A.; Paeper, B.; Clark, L.B.; Yasayko, S.A.; Wilkinson, J.E.; Galas, D.; Ziegler, S.F.; Ramsdell, F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001, 27, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Wildin, R.S.; Ramsdell, F.; Peake, J.; Faravelli, F.; Casanova, J.L.; Buist, N.; Levy-Lahad, E.; Mazzella, M.; Goulet, O.; Perroni, L.; et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001, 27, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Dooley, J.L.; Farr, A.G.; Rudensky, A.Y. Developmental regulation of Foxp3 expression during ontogeny. J. Exp. Med. 2005, 202, 901–906. [Google Scholar] [CrossRef] [Green Version]
- Abbas, A.K.; Benoist, C.; Bluestone, J.A.; Campbell, D.J.; Ghosh, S.; Hori, S.; Jiang, S.; Kuchroo, V.K.; Mathis, D.; Roncarolo, M.G.; et al. Regulatory T cells: Recommendations to simplify the nomenclature. Nat. Immunol. 2013, 14, 307–308. [Google Scholar] [CrossRef]
- Hsieh, C.S.; Lee, H.M.; Lio, C.W. Selection of regulatory T cells in the thymus. Nat. Rev. Immunol. 2012, 12, 157–167. [Google Scholar] [CrossRef]
- Klein, L.; Robey, E.A.; Hsieh, C.S. Central CD4(+) T cell tolerance: Deletion versus regulatory T cell differentiation. Nat. Rev. Immunol. 2019, 19, 7–18. [Google Scholar] [CrossRef]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003, 198, 1875–1886. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Louvet, C.; Davini, D.; Gardner, J.M.; Martinez-Llordella, M.; Bailey-Bucktrout, S.; Anthony, B.A.; Sverdrup, F.M.; Head, R.; Kuster, D.J.; et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 2012, 209, 1713–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, J.M.; Bilate, A.M.; Gobert, M.; Ding, Y.; Curotto de Lafaille, M.A.; Parkhurst, C.N.; Xiong, H.; Dolpady, J.; Frey, A.B.; Ruocco, M.G.; et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 2012, 209, 1723–1742. [Google Scholar] [CrossRef] [PubMed]
- Josefowicz, S.Z.; Niec, R.E.; Kim, H.Y.; Treuting, P.; Chinen, T.; Zheng, Y.; Umetsu, D.T.; Rudensky, A.Y. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 2012, 482, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Smigiel, K.S.; Richards, E.; Srivastava, S.; Thomas, K.R.; Dudda, J.C.; Klonowski, K.D.; Campbell, D.J. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 2014, 211, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.G.; Arvey, A.; Jin, W.; Rudensky, A.Y. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 2014, 15, 1070–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayatsu, N.; Miyao, T.; Tachibana, M.; Murakami, R.; Kimura, A.; Kato, T.; Kawakami, E.; Endo, T.A.; Setoguchi, R.; Watarai, H.; et al. Analyses of a Mutant Foxp3 Allele Reveal BATF as a Critical Transcription Factor in the Differentiation and Accumulation of Tissue Regulatory T Cells. Immunity 2017, 47, 268–283.e9. [Google Scholar] [CrossRef]
- Luo, C.T.; Liao, W.; Dadi, S.; Toure, A.; Li, M.O. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature 2016, 529, 532–536. [Google Scholar] [CrossRef] [Green Version]
- Liston, A.; Gray, D.H. Homeostatic control of regulatory T cell diversity. Nat. Rev. Immunol. 2014, 14, 154–165. [Google Scholar] [CrossRef]
- Li, M.O.; Rudensky, A.Y. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 2016, 16, 220–233. [Google Scholar] [CrossRef] [Green Version]
- Vahl, J.C.; Drees, C.; Heger, K.; Heink, S.; Fischer, J.C.; Nedjic, J.; Ohkura, N.; Morikawa, H.; Poeck, H.; Schallenberg, S.; et al. Continuous T Cell Receptor Signals Maintain a Functional Regulatory T Cell Pool. Immunity 2014, 41, 722–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, S.; D’Amico, A.; Cretney, E.; Liao, Y.; Tellier, J.; Bruggeman, C.; Almeida, F.F.; Leahy, J.; Belz, G.T.; Smyth, G.K.; et al. Effector Regulatory T Cell Differentiation and Immune Homeostasis Depend on the Transcription Factor Myb. Immunity 2017, 46, 78–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koizumi, S.I.; Sasaki, D.; Hsieh, T.H.; Taira, N.; Arakaki, N.; Yamasaki, S.; Wang, K.; Sarkar, S.; Shirahata, H.; Miyagi, M.; et al. JunB regulates homeostasis and suppressive functions of effector regulatory T cells. Nat. Commun. 2018, 9, 5344. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, J.; Gu, Q.; Huang, M.; Zhang, W.; Guo, J.; Zhou, X. Reciprocal Expression of IL-35 and IL-10 Defines Two Distinct Effector Treg Subsets that Are Required for Maintenance of Immune Tolerance. Cell Rep. 2017, 21, 1853–1869. [Google Scholar] [CrossRef] [PubMed]
- Panduro, M.; Benoist, C.; Mathis, D. Tissue Tregs. Annu. Rev. Immunol. 2016, 34, 609–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, M.; Komai, K.; Nakamura, T.; Srirat, T.; Yoshimura, A. Tissue regulatory T cells and neural repair. Int. Immunol. 2019, 31, 361–369. [Google Scholar] [CrossRef]
- Arpaia, N.; Green, J.A.; Moltedo, B.; Arvey, A.; Hemmers, S.; Yuan, S.; Treuting, P.M.; Rudensky, A.Y. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell 2015, 162, 1078–1089. [Google Scholar] [CrossRef] [Green Version]
- Kuswanto, W.; Burzyn, D.; Panduro, M.; Wang, K.K.; Jang, Y.C.; Wagers, A.J.; Benoist, C.; Mathis, D. Poor Repair of Skeletal Muscle in Aging Mice Reflects a Defect in Local, Interleukin-33-Dependent Accumulation of Regulatory T Cells. Immunity 2016, 44, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Ali, N.; Zirak, B.; Rodriguez, R.S.; Pauli, M.L.; Truong, H.A.; Lai, K.; Ahn, R.; Corbin, K.; Lowe, M.M.; Scharschmidt, T.C.; et al. Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 2017, 169, 1119–1129.e11. [Google Scholar] [CrossRef] [Green Version]
- Ito, M.; Komai, K.; Mise-Omata, S.; Iizuka-Koga, M.; Noguchi, Y.; Kondo, T.; Sakai, R.; Matsuo, K.; Nakayama, T.; Yoshie, O.; et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 2019, 565, 246–250. [Google Scholar] [CrossRef]
- Mathur, A.N.; Zirak, B.; Boothby, I.C.; Tan, M.; Cohen, J.N.; Mauro, T.M.; Mehta, P.; Lowe, M.M.; Abbas, A.K.; Ali, N.; et al. Treg-Cell Control of a CXCL5-IL-17 Inflammatory Axis Promotes Hair-Follicle-Stem-Cell Differentiation During Skin-Barrier Repair. Immunity 2019, 50, 655–667.e4. [Google Scholar] [CrossRef] [Green Version]
- Feuerer, M.; Herrero, L.; Cipolletta, D.; Naaz, A.; Wong, J.; Nayer, A.; Lee, J.; Goldfine, A.B.; Benoist, C.; Shoelson, S.; et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009, 15, 930–939. [Google Scholar] [CrossRef]
- Cipolletta, D.; Feuerer, M.; Li, A.; Kamei, N.; Lee, J.; Shoelson, S.E.; Benoist, C.; Mathis, D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 2012, 486, 549–553. [Google Scholar] [CrossRef]
- Kolodin, D.; van Panhuys, N.; Li, C.; Magnuson, A.M.; Cipolletta, D.; Miller, C.M.; Wagers, A.; Germain, R.N.; Benoist, C.; Mathis, D. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 2015, 21, 543–557. [Google Scholar] [CrossRef]
- Ohnmacht, C.; Park, J.H.; Cording, S.; Wing, J.B.; Atarashi, K.; Obata, Y.; Gaboriau-Routhiau, V.; Marques, R.; Dulauroy, S.; Fedoseeva, M.; et al. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 2015, 349, 989–993. [Google Scholar] [CrossRef]
- Sefik, E.; Geva-Zatorsky, N.; Oh, S.; Konnikova, L.; Zemmour, D.; McGuire, A.M.; Burzyn, D.; Ortiz-Lopez, A.; Lobera, M.; Yang, J.; et al. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015, 349, 993–997. [Google Scholar] [CrossRef]
- Schiering, C.; Krausgruber, T.; Chomka, A.; Fröhlich, A.; Adelmann, K.; Wohlfert, E.A.; Pott, J.; Griseri, T.; Bollrath, J.; Hegazy, A.N.; et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 2014, 513, 564–568. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.; Tanaka, S.; Chu, F.; Nurieva, R.I.; Martinez, G.J.; Rawal, S.; Wang, Y.H.; Lim, H.; Reynolds, J.M.; Zhou, X.H.; et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 2011, 17, 983–988. [Google Scholar] [CrossRef]
- Linterman, M.A.; Pierson, W.; Lee, S.K.; Kallies, A.; Kawamoto, S.; Rayner, T.F.; Srivastava, M.; Divekar, D.P.; Beaton, L.; Hogan, J.J.; et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 2011, 17, 975–982. [Google Scholar] [CrossRef] [Green Version]
- Wollenberg, I.; Agua-Doce, A.; Hernandez, A.; Almeida, C.; Oliveira, V.G.; Faro, J.; Graca, L. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J. Immunol. 2011, 187, 4553–4560. [Google Scholar] [CrossRef]
- Fonseca, V.R.; Agua-Doce, A.; Maceiras, A.R.; Pierson, W.; Ribeiro, F.; Romao, V.C.; Pires, A.R.; da Silva, S.L.; Fonseca, J.E.; Sousa, A.E.; et al. Human blood Tfr cells are indicators of ongoing humoral activity not fully licensed with suppressive function. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef]
- Wing, J.B.; Kitagawa, Y.; Locci, M.; Hume, H.; Tay, C.; Morita, T.; Kidani, Y.; Matsuda, K.; Inoue, T.; Kurosaki, T.; et al. A distinct subpopulation of CD25(-) T-follicular regulatory cells localizes in the germinal centers. Proc. Natl. Acad. Sci. USA 2017, 114, E6400–E6409. [Google Scholar] [CrossRef]
- Maceiras, A.R.; Almeida, S.C.P.; Mariotti-Ferrandiz, E.; Chaara, W.; Jebbawi, F.; Six, A.; Hori, S.; Klatzmann, D.; Faro, J.; Graca, L. T follicular helper and T follicular regulatory cells have different TCR specificity. Nat. Commun. 2017, 8, 15067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; DiSpirito, J.R.; Zemmour, D.; Spallanzani, R.G.; Kuswanto, W.; Benoist, C.; Mathis, D. TCR Transgenic Mice Reveal Stepwise, Multi-site Acquisition of the Distinctive Fat-Treg Phenotype. Cell 2018, 174, 285–299.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, A.G.; Medoza, A.; Hemmers, S.; Moltedo, B.; Niec, R.E.; Schizas, M.; Hoyos, B.E.; Putintseva, E.V.; Chaudhry, A.; Dikiy, S.; et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature 2017, 546, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.A.; Tucker-Heard, G.; Perdue, N.R.; Killebrew, J.R.; Urdahl, K.B.; Campbell, D.J. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009, 10, 595–602. [Google Scholar] [CrossRef]
- Yu, F.; Sharma, S.; Edwards, J.; Feigenbaum, L.; Zhu, J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat. Immunol. 2015, 16, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chu, C.; Teng, F.; Bessman, N.J.; Goc, J.; Santosa, E.K.; Putzel, G.G.; Kabata, H.; Kelsen, J.R.; Baldassano, R.N.; et al. Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 2019, 568, 405–409. [Google Scholar] [CrossRef]
- Rosenblum, M.D.; Gratz, I.K.; Paw, J.S.; Lee, K.; Marshak-Rothstein, A.; Abbas, A.K. Response to self antigen imprints regulatory memory in tissues. Nature 2011, 480, 538–542. [Google Scholar] [CrossRef]
- Rosenblum, M.D.; Way, S.S.; Abbas, A.K. Regulatory T cell memory. Nat. Rev. Immunol. 2016, 16, 90–101. [Google Scholar] [CrossRef]
- Zemmour, D.; Zilionis, R.; Kiner, E.; Klein, A.M.; Mathis, D.; Benoist, C. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol. 2018, 19, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Miragaia, R.J.; Gomes, T.; Chomka, A.; Jardine, L.; Riedel, A.; Hegazy, A.N.; Whibley, N.; Tucci, A.; Chen, X.; Lindeman, I.; et al. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity 2019, 50, 493–504.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Chaudhry, A.; Kas, A.; deRoos, P.; Kim, J.M.; Chu, T.T.; Corcoran, L.; Treuting, P.; Klein, U.; Rudensky, A.Y. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature 2009, 458, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Vasanthakumar, A.; Moro, K.; Xin, A.; Liao, Y.; Gloury, R.; Kawamoto, S.; Fagarasan, S.; Mielke, L.A.; Afshar-Sterle, S.; Masters, S.L.; et al. The transcriptional regulators IRF4, BATF and IL-33 orchestrate development and maintenance of adipose tissue-resident regulatory T cells. Nat. Immunol. 2015, 16, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Delacher, M.; Imbusch, C.D.; Weichenhan, D.; Breiling, A.; Hotz-Wagenblatt, A.; Trager, U.; Hofer, A.C.; Kagebein, D.; Wang, Q.; Frauhammer, F.; et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat. Immunol. 2017, 18, 1160–1172. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ma, S.; Hotz-Wagenblatt, A.; Angel, P.; Mohr, K.; Schlimbach, T.; Schmitt, M.; Cui, G. Regulatory T cells sense effector T-cell activation through synchronized JunB expression. FEBS Lett. 2019, 593, 1020–1029. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, T.; Yamazaki, S.; Fukui, Y.; Aoki, K.; Yagita, H.; Nishina, T.; Mikami, T.; Katagiri, S.; Shiraishi, A.; Kimura, S.; et al. JunB plays a crucial role in development of regulatory T cells by promoting IL-2 signaling. Mucosal Immunol. 2019. [Google Scholar] [CrossRef]
- Cretney, E.; Xin, A.; Shi, W.; Minnich, M.; Masson, F.; Miasari, M.; Belz, G.T.; Smyth, G.K.; Busslinger, M.; Nutt, S.L.; et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011, 12, 304–311. [Google Scholar] [CrossRef]
- Oh, H.; Grinberg-Bleyer, Y.; Liao, W.; Maloney, D.; Wang, P.; Wu, Z.; Wang, J.; Bhatt, D.M.; Heise, N.; Schmid, R.M.; et al. An NF-κB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T Cell Identity and Function. Immunity 2017, 47, 450–465.e5. [Google Scholar] [CrossRef]
- Vasanthakumar, A.; Liao, Y.; Teh, P.; Pascutti, M.F.; Oja, A.E.; Garnham, A.L.; Gloury, R.; Tempany, J.C.; Sidwell, T.; Cuadrado, E.; et al. The TNF Receptor Superfamily-NF-kappaB Axis Is Critical to Maintain Effector Regulatory T Cells in Lymphoid and Non-lymphoid Tissues. Cell Rep. 2017, 20, 2906–2920. [Google Scholar] [CrossRef]
- Grinberg-Bleyer, Y.; Oh, H.; Desrichard, A.; Bhatt, D.M.; Caron, R.; Chan, T.A.; Schmid, R.M.; Klein, U.; Hayden, M.S.; Ghosh, S. NF-κB c-Rel Is Crucial for the Regulatory T Cell Immune Checkpoint in Cancer. Cell 2017, 170, 1096–1108.e13. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, M.; Miyazaki, K.; Chen, S.; Itoi, M.; Miller, M.; Lu, L.F.; Varki, N.; Chang, A.N.; Broide, D.H.; Murre, C. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat. Immunol. 2014, 15, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.M.; Höllbacher, B.; Campbell, D.J. Cutting Edge: Dynamic Expression of Id3 Defines the Stepwise Differentiation of Tissue-Resident Regulatory T Cells. J. Immunol. 2019, 202, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.M.; Sharma, G.; Verma, R.; Byun, S.; Rudra, D.; Im, S.H. Inflammation-induced Id2 promotes plasticity in regulatory T cells. Nat. Commun. 2018, 9, 4736. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Huang, H.; Gao, P.; Zhang, Q.; Liu, X.; Jia, B.; Strober, W.; Hou, B.; Zhou, X.; Gao, G.F.; et al. E-protein regulatory network links TCR signaling to effector Treg cell differentiation. Proc. Natl. Acad. Sci. USA 2019. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.H.; Wang, K.; Wan, S.; Liang, Y.; Yuan, X.; Dong, Y.; Cho, S.; Xu, W.; Jepsen, K.; Feng, G.S.; et al. TCF1 and LEF1 Control Treg Competitive Survival and Tfr Development to Prevent Autoimmune Diseases. Cell Rep. 2019, 27, 3629–3645.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, S.; Gai, K.; Li, X.; Shao, P.; Zeng, Z.; Zhao, X.; Zhao, X.; Chen, X.; Paradee, W.J.; Meyerholz, D.K.; et al. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J. Exp. Med. 2019, 216, 847–866. [Google Scholar] [CrossRef]
- Ouyang, W.; Liao, W.; Luo, C.T.; Yin, N.; Huse, M.; Kim, M.V.; Peng, M.; Chan, P.; Ma, Q.; Mo, Y.; et al. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature 2012, 491, 554–559. [Google Scholar] [CrossRef]
- Koch, M.A.; Thomas, K.R.; Perdue, N.R.; Smigiel, K.S.; Srivastava, S.; Campbell, D.J. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity 2012, 37, 501–510. [Google Scholar] [CrossRef]
- Harrison, O.J.; Linehan, J.L.; Shih, H.-Y.; Bouladoux, N.; Han, S.-J.; Smelkinson, M.; Sen, S.K.; Byrd, A.L.; Enamorado, M.; Yao, C.; et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 2019, 363, eaat6280. [Google Scholar] [CrossRef]
- Wohlfert, E.A.; Grainger, J.R.; Bouladoux, N.; Konkel, J.E.; Oldenhove, G.; Ribeiro, C.H.; Hall, J.A.; Yagi, R.; Naik, S.; Bhairavabhotla, R.; et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J. Clin. Invest. 2011, 121, 4503–4515. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Hong, S.W.; Han, D.; Yi, J.; Jung, J.; Yang, B.G.; Lee, J.Y.; Lee, M.; Surh, C.D. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 2016, 351, 858–863. [Google Scholar] [CrossRef] [PubMed]
- Botta, D.; Fuller, M.J.; Marquez-Lago, T.T.; Bachus, H.; Bradley, J.E.; Weinmann, A.S.; Zajac, A.J.; Randall, T.D.; Lund, F.E.; Leon, B.; et al. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat. Immunol. 2017, 18, 1249–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Xie, M.M.; Liu, H.; Dent, A.L. Stat3 Is Important for Follicular Regulatory T Cell Differentiation. PLoS ONE 2016, 11, e0155040. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Pokrovskii, M.; Ding, Y.; Yi, R.; Au, C.; Harrison, O.J.; Galan, C.; Belkaid, Y.; Bonneau, R.; Littman, D.R. C-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018, 554, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Neumann, C.; Blume, J.; Roy, U.; Teh, P.P.; Vasanthakumar, A.; Beller, A.; Liao, Y.; Heinrich, F.; Arenzana, T.L.; Hackney, J.A.; et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat. Immunol. 2019, 20, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Wheaton, J.D.; Yeh, C.H.; Ciofani, M. Cutting Edge: C-Maf Is Required for Regulatory T Cells To Adopt RORgammat(+) and Follicular Phenotypes. J. Immunol. 2017, 199, 3931–3936. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, A.; Rudra, D.; Treuting, P.; Samstein, R.M.; Liang, Y.; Kas, A.; Rudensky, A.Y. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 2009, 326, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, Q.; Wang, H.; Hao, Y.; Bai, Q.; Hu, J.; Li, Y.; Wang, P.; Chen, X.; He, R.; et al. The Kinase mTORC1 Promotes the Generation and Suppressive Function of Follicular Regulatory T Cells. Immunity 2017, 47, 538–551.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malhotra, N.; Leyva-Castillo, J.M.; Jadhav, U.; Barreiro, O.; Kam, C.; O’Neill, N.K.; Meylan, F.; Chambon, P.; von Andrian, U.H.; Siegel, R.M.; et al. RORalpha-expressing T regulatory cells restrain allergic skin inflammation. Sci. Immunol. 2018, 3. [Google Scholar] [CrossRef]
- Tamura, T.; Yanai, H.; Savitsky, D.; Taniguchi, T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008, 26, 535–584. [Google Scholar] [CrossRef] [PubMed]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.L.; Tussiwand, R.; Murphy, K.M. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat. Rev. Immunol. 2013, 13, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Hasan, Z.; Koizumi, S.I.; Sasaki, D.; Yamada, H.; Arakaki, N.; Fujihara, Y.; Okitsu, S.; Shirahata, H.; Ishikawa, H. JunB is essential for IL-23-dependent pathogenicity of Th17 cells. Nat. Commun. 2017, 8, 15628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, T.M.; Wheaton, J.D.; Houtz, G.M.; Ciofani, M. JunB promotes Th17 cell identity and restrains alternative CD4(+) T-cell programs during inflammation. Nat. Commun. 2017, 8, 301. [Google Scholar] [CrossRef] [PubMed]
- Kurachi, M.; Barnitz, R.A.; Yosef, N.; Odorizzi, P.M.; DiIorio, M.A.; Lemieux, M.E.; Yates, K.; Godec, J.; Klatt, M.G.; Regev, A.; et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 2014, 15, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Brustle, A.; Heink, S.; Huber, M.; Rosenplanter, C.; Stadelmann, C.; Yu, P.; Arpaia, E.; Mak, T.W.; Kamradt, T.; Lohoff, M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol. 2007, 8, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Schraml, B.U.; Hildner, K.; Ise, W.; Lee, W.L.; Smith, W.A.; Solomon, B.; Sahota, G.; Sim, J.; Mukasa, R.; Cemerski, S.; et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 2009, 460, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Buzo, B.F.; Pham, D.; Jiang, L.; Taparowsky, E.J.; Kaplan, M.H.; Sun, J. Interferon regulatory factor 4 sustains CD8(+) T cell expansion and effector differentiation. Immunity 2013, 39, 833–845. [Google Scholar] [CrossRef]
- Man, K.; Miasari, M.; Shi, W.; Xin, A.; Henstridge, D.C.; Preston, S.; Pellegrini, M.; Belz, G.T.; Smyth, G.K.; Febbraio, M.A.; et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 2013, 14, 1155–1165. [Google Scholar] [CrossRef]
- Ise, W.; Kohyama, M.; Schraml, B.U.; Zhang, T.; Schwer, B.; Basu, U.; Alt, F.W.; Tang, J.; Oltz, E.M.; Murphy, T.L.; et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 2011, 12, 536–543. [Google Scholar] [CrossRef]
- Wang, C.; Thangamani, S.; Kim, M.; Gu, B.H.; Lee, J.H.; Taparowsky, E.J.; Kim, C.H. BATF is required for normal expression of gut-homing receptors by T helper cells in response to retinoic acid. J. Exp. Med. 2013, 210, 475–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cretney, E.; Leung, P.S.; Trezise, S.; Newman, D.M.; Rankin, L.C.; Teh, C.E.; Putoczki, T.L.; Gray, D.H.; Belz, G.T.; Mielke, L.A.; et al. Characterization of Blimp-1 function in effector regulatory T cells. J. Autoimmun. 2018, 91, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Bankoti, R.; Ogawa, C.; Nguyen, T.; Emadi, L.; Couse, M.; Salehi, S.; Fan, X.; Dhall, D.; Wang, Y.; Brown, J.; et al. Differential regulation of Effector and Regulatory T cell function by Blimp1. Sci Rep. 2017, 7, 12078. [Google Scholar] [CrossRef] [PubMed]
- Garg, G.; Muschaweckh, A.; Moreno, H.; Vasanthakumar, A.; Floess, S.; Lepennetier, G.; Oellinger, R.; Zhan, Y.; Regen, T.; Hiltensperger, M.; et al. Blimp1 Prevents Methylation of Foxp3 and Loss of Regulatory T Cell Identity at Sites of Inflammation. Cell Rep. 2019, 26, 1854–1868.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, C.; Bankoti, R.; Nguyen, T.; Hassanzadeh-Kiabi, N.; Nadeau, S.; Porritt, R.A.; Couse, M.; Fan, X.; Dhall, D.; Eberl, G.; et al. Blimp-1 Functions as a Molecular Switch to Prevent Inflammatory Activity in Foxp3(+)RORgammat(+) Regulatory T Cells. Cell Rep. 2018, 25, 19–28.e5. [Google Scholar] [CrossRef] [PubMed]
- Maurice, D.; Hooper, J.; Lang, G.; Weston, K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007, 26, 3629–3640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakata, Y.; Brignier, A.C.; Jin, S.; Shen, Y.; Rudnick, S.I.; Sugita, M.; Gewirtz, A.M. c-Myb, Menin, GATA-3, and MLL form a dynamic transcription complex that plays a pivotal role in human T helper type 2 cell development. Blood 2010, 116, 1280–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, K.; Karin, M. NF-kappaB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Gerondakis, S.; Fulford, T.S.; Messina, N.L.; Grumont, R.J. NF-kappaB control of T cell development. Nat. Immunol. 2014, 15, 15–25. [Google Scholar] [CrossRef]
- Long, M.X.; Park, S.G.; Strickland, I.; Hayden, M.S.; Ghosh, S. Nuclear Factor-kappa B Modulates Regulatory T Cell Development by Directly Regulating Expression of Foxp3 Transcription Factor. Immunity 2009, 31, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Kameswaran, V.; Tone, Y.; Li, L.; Liou, H.C.; Greene, M.I.; Tone, M.; Chen, Y.H. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity 2009, 31, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Messina, N.; Fulford, T.; O’Reilly, L.; Loh, W.X.; Motyer, J.M.; Ellis, D.; McLean, C.; Naeem, H.; Lin, A.; Gugasyan, R.; et al. The NF-kappaB transcription factor RelA is required for the tolerogenic function of Foxp3(+) regulatory T cells. J. Autoimmun. 2016, 70, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Kee, B.L. E and ID proteins branch out. Nat. Rev. Immunol. 2009, 9, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Steinke, F.C.; Xue, H.H. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol. Res. 2014, 59, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, B.; Omenetti, S. The dichotomous nature of T helper 17 cells. Nat. Rev. Immunol. 2017, 17, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Vinuesa, C.G.; Linterman, M.A.; Yu, D.; MacLennan, I.C. Follicular Helper T Cells. Annu. Rev. Immunol. 2016, 34, 335–368. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, G.H.; Kedl, R.M.; Hunter, C.A. The evolving role of T-bet in resistance to infection. Nat. Rev. Immunol. 2019, 19, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Tindemans, I.; Serafini, N.; DiSanto, J.P.; Hendriks, R.W. GATA-3 function in innate and adaptive immunity. Immunity 2014, 41, 191–206. [Google Scholar] [CrossRef]
- Wang, Y.; Su, M.A.; Wan, Y.Y. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 2011, 35, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Eberl, G. RORgammat, a multitask nuclear receptor at mucosal surfaces. Mucosal Immunol. 2017, 10, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Stewart, K.M.; Wang, X.; Liu, K.; Xie, M.; Ryu, J.K.; Li, K.; Ma, T.; Wang, H.; Ni, L.; et al. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature 2017, 548, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Liu, X.; Lin, X.; Feng, H.; Sun, L.; Li, S.; Chen, H.; Tang, H.; Lu, L.; Jin, W.; et al. Deficiency in T follicular regulatory cells promotes autoimmunity. J. Exp. Med. 2018, 215, 815–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatzi, K.; Nance, J.P.; Kroenke, M.A.; Bothwell, M.; Haddad, E.K.; Melnick, A.; Crotty, S. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 2015, 212, 539–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oestreich, K.J.; Mohn, S.E.; Weinmann, A.S. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 2012, 13, 405–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, R.J.; Choi, Y.S.; Diamond, J.A.; Yang, J.A.; Crotty, S. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 2012, 209, 243–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, P.W.; Read, K.A.; Baker, C.E.; Anderson, A.E.; Powell, M.D.; Ballesteros-Tato, A.; Oestreich, K.J. IL-7 signalling represses Bcl-6 and the TFH gene program. Nat. Commun. 2016, 7, 10285. [Google Scholar] [CrossRef]
- Fan, M.Y.; Low, J.S.; Tanimine, N.; Finn, K.K.; Priyadharshini, B.; Germana, S.K.; Kaech, S.M.; Turka, L.A. Differential Roles of IL-2 Signaling in Developing versus Mature Tregs. Cell Rep. 2018, 25, 1204–1213.e4. [Google Scholar] [CrossRef] [Green Version]
- Gabrysova, L.; Alvarez-Martinez, M.; Luisier, R.; Cox, L.S.; Sodenkamp, J.; Hosking, C.; Perez-Mazliah, D.; Whicher, C.; Kannan, Y.; Potempa, K.; et al. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4(+) T cells. Nat. Immunol. 2018, 19, 497–507. [Google Scholar] [CrossRef]
- Gratz, I.K.; Truong, H.A.; Yang, S.H.; Maurano, M.M.; Lee, K.; Abbas, A.K.; Rosenblum, M.D. Cutting Edge: Memory regulatory t cells require IL-7 and not IL-2 for their maintenance in peripheral tissues. J. Immunol. 2013, 190, 4483–4487. [Google Scholar] [CrossRef] [PubMed]
- DuPage, M.; Chopra, G.; Quiros, J.; Rosenthal, W.L.; Morar, M.M.; Holohan, D.; Zhang, R.; Turka, L.; Marson, A.; Bluestone, J.A. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity 2015, 42, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Quiros, J.; Mahuron, K.; Pai, C.C.; Ranzani, V.; Young, A.; Silveria, S.; Harwin, T.; Abnousian, A.; Pagani, M.; et al. Targeting EZH2 Reprograms Intratumoral Regulatory T Cells to Enhance Cancer Immunity. Cell Rep. 2018, 23, 3262–3274. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Pfleger, C.; Lai, J.F.; Roan, F.; Sun, S.C.; Ziegler, S.F. KAP1 Regulates Regulatory T Cell Function and Proliferation in Both Foxp3-Dependent and -Independent Manners. Cell Rep. 2018, 23, 796–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.; Clement, R.L.; Diallo, A.; Blazar, B.R.; Rudensky, A.Y.; Sharpe, A.H.; Sage, P.T. FoxP3 and Ezh2 regulate Tfr cell suppressive function and transcriptional program. J. Exp. Med. 2019, 216, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Lio, C.J.; Samaniego-Castruita, D.; Li, X.; Rao, A. Loss of TET2 and TET3 in regulatory T cells unleashes effector function. Nat. Commun. 2019, 10, 2011. [Google Scholar] [CrossRef] [PubMed]
- Nakatsukasa, H.; Oda, M.; Yin, J.; Chikuma, S.; Ito, M.; Koga-Iizuka, M.; Someya, K.; Kitagawa, Y.; Ohkura, N.; Sakaguchi, S.; et al. Loss of TET proteins in regulatory T cells promotes abnormal proliferation, Foxp3 destabilization and IL-17 expression. Int. Immunol. 2019, 31, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Obata, Y.; Furusawa, Y.; Endo, T.A.; Sharif, J.; Takahashi, D.; Atarashi, K.; Nakayama, M.; Onawa, S.; Fujimura, Y.; Takahashi, M.; et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat. Immunol. 2014, 15, 571–579. [Google Scholar] [CrossRef]
- Zeng, H.; Yang, K.; Cloer, C.; Neale, G.; Vogel, P.; Chi, H. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature 2013, 499, 485–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, N.M.; Zeng, H.; Nguyen, T.M.; Wang, Y.; Vogel, P.; Dhungana, Y.; Liu, X.; Neale, G.; Locasale, J.W.; Chi, H. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat. Commun. 2018, 9, 2095. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Yin, N.; Chhangawala, S.; Xu, K.; Leslie, C.S.; Li, M.O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 2016, 354, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Bailis, W.; Shyer, J.A.; Zhao, J.; Canaveras, J.C.G.; Al Khazal, F.J.; Qu, R.; Steach, H.R.; Bielecki, P.; Khan, O.; Jackson, R.; et al. Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 2019, 571, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, S.E.; Singer, B.D.; Steinert, E.M.; Martinez, C.A.; Mehta, M.M.; Martinez-Reyes, I.; Gao, P.; Helmin, K.A.; Abdala-Valencia, H.; Sena, L.A.; et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 2019, 565, 495–499. [Google Scholar] [CrossRef] [PubMed]
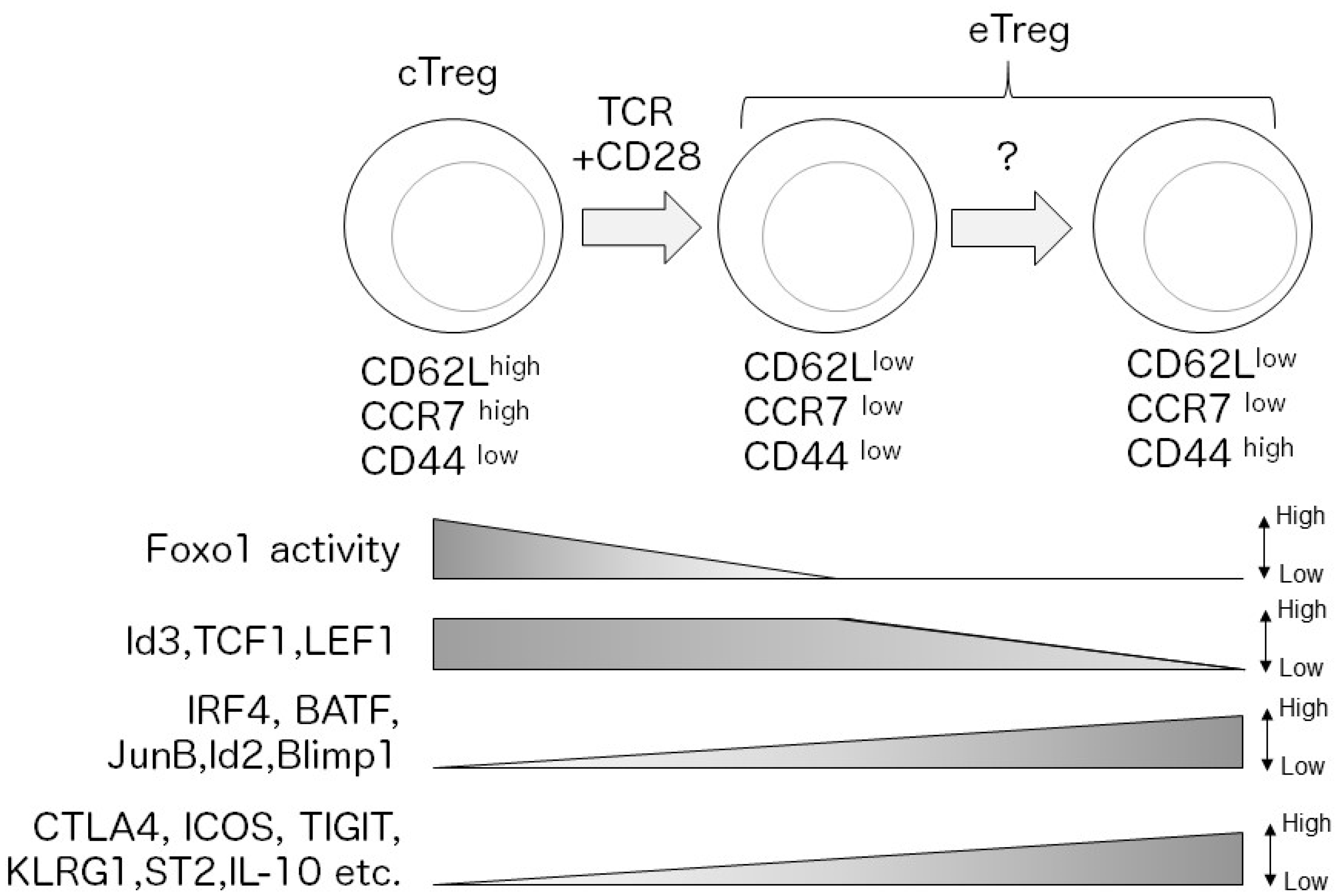
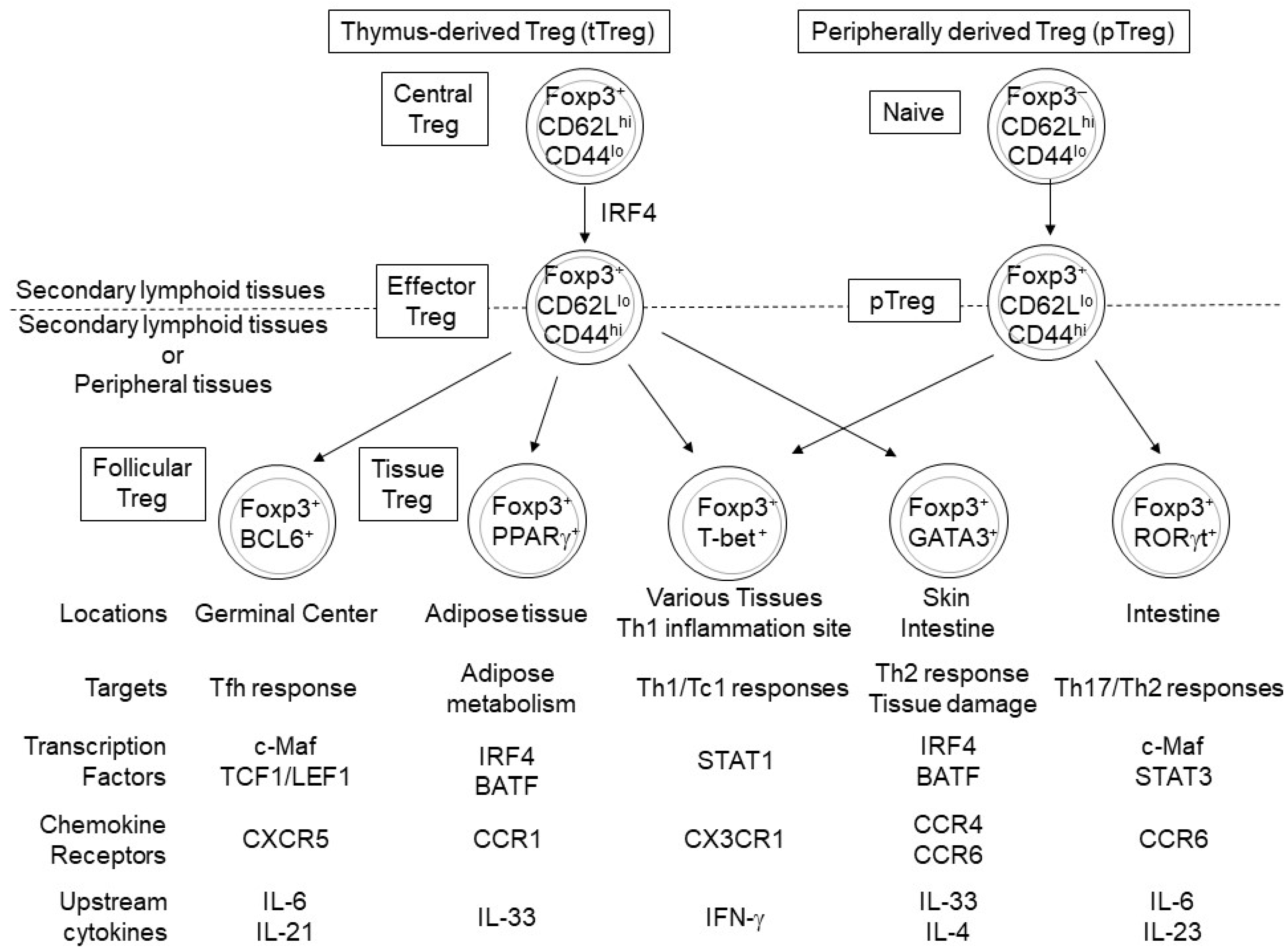
| Name | Expression | Function | Target | Upstream Regulator | Ref. |
|---|---|---|---|---|---|
| IRF4 | eTreg | Regulates eTreg differentiation | ICOS, Blimp1, FGL2, ST2, GATA3, CCR8, CTLA4, KLRG1, IL-10, CCR6 | TCR+CD28+IL-2 | [16,53,54] |
| BATF | eTreg | Promotes expression of eTreg-related genes | ICOS, GATA3, ST2, KLRG1, CCR4, CCR6 | TCR+CD28+IL-2 | [17,54,55] |
| JunB | eTreg | Promotes expression of eTreg-related genes | ICOS, FGL2, CTLA4, TIGIT, KLRG1 | TCR+CD28+IL-2 | [23,56,57] |
| Blimp-1 | eTreg | Promotes expression of eTreg-related genes | IL-10, KLRG1, Ebi3, CCR6, BCL2 | TCR+CD28+IL-2 | [58] |
| Myb | eTreg | Promotes expression of eTreg-related genesin tTreg | ICOS, TIGIT, KLRG1, BCL2, MYC | TCR+CD28+IL-2 | [22] |
| RelA | All Tregs | Promotes expression of eTreg-related genes | TIGIT, ST2, KLRG1, CD103, CD30 | TNF-α, GITR | [59,60] |
| c-Rel | All Tregs | Promotes expression of eTreg-related genes | TIGIT, ST2, ICOS, EBI3,Blimp-1,IL-10 | Unknown | [59,61] |
| Id2 | eTreg | Suppresses Tfr differentiation | CXCR5, IL-10, HIF1-α, IKZF3, Myb, IL-10 rα, E2F2 | TCR+CD28+IL-2 | [62,63,64] |
| Id3 | cTreg, CD44lo eTreg | TCR+CD28+IL-2- Erk,PI3K/mTOR | [62,63] | ||
| E2A/ HEB | eTreg | Suppresses eTreg differentiation | ICOS, IRF4, CD103, KLRG1, RORγt | TCR+CD28 | [65] |
| TCF1/ LEF1 | cTreg, CD44lo eTreg | Regulates Tfr differentiationSuppresses expression of eTreg-related genes? | CD44, ICOS, TIGIT, IRF4, GATA3, Blimp-1, T-bet, Bcl6, CXCR5 | Unknown | [66,67] |
| Foxo1 | All Treg | Maintains expression of cTreg-related genes | CD62L, CCR7, Bim, GzmB | TCR+CD28-Akt | [18,68] |
| T-bet | eTreg, Tissue Tregs | Regulates migration to Th1-inflammatory sites | CXCR3 | IFNγ-STAT1 | [45,69] |
| GATA3 | Skin and Intestinal tTreg | Maintains Treg homeostasis Suppresses skin inflammation | ST2, Foxp3 | TCR, IL-4, IL-33 | [37,70,71] |
| RORγt | Intestinal pTreg | Regulates migration to intestinal tissue | CCR6 | IL-6/IL-23-STAT3, Microbiota | [35,36,72] |
| BCL6 | Follicular Treg | Regulates Tfr differentiation | CXCR5, PD-1 | IL-21, IL-6 | [73,74] |
| c-Maf | eTreg | Regulates RORγτ+ Treg and Tfr differentiations | RORγt, CXCR5, IL-10 | TCR+CD28+IL-2, STAT3, Microbiota, Notch1/2 | [75,76,77] |
| STAT3 | All Tregs | Regulates RORγt+ Treg and Tfr differentiation | TCF1, RORγt | IL-6,IL-23,IL-21 | [35,74,75,78,79] |
| PPARγ | VAT-Treg | Regulates VAT-Treg differentiation | CCR1, PCYT1A, DGAT1, IL-10 | TCR,IL-33, Adipose tissue-derived factor? | [32,34,44,54] |
| RORα | Skin and Colon Tregs | Promotes expression of eTreg-related genes | CCR6, CD73, DR3 | Unknown | [52,80] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koizumi, S.-i.; Ishikawa, H. Transcriptional Regulation of Differentiation and Functions of Effector T Regulatory Cells. Cells 2019, 8, 939. https://doi.org/10.3390/cells8080939
Koizumi S-i, Ishikawa H. Transcriptional Regulation of Differentiation and Functions of Effector T Regulatory Cells. Cells. 2019; 8(8):939. https://doi.org/10.3390/cells8080939
Chicago/Turabian StyleKoizumi, Shin-ichi, and Hiroki Ishikawa. 2019. "Transcriptional Regulation of Differentiation and Functions of Effector T Regulatory Cells" Cells 8, no. 8: 939. https://doi.org/10.3390/cells8080939





