HCV Pit Stop at the Lipid Droplet: Refuel Lipids and Put on a Lipoprotein Coat before Exit
Abstract
:1. Introduction
2. A Chip off the Old Block: HCV Particle Properties Reflect Interactions with the Lipoprotein Production Pathway
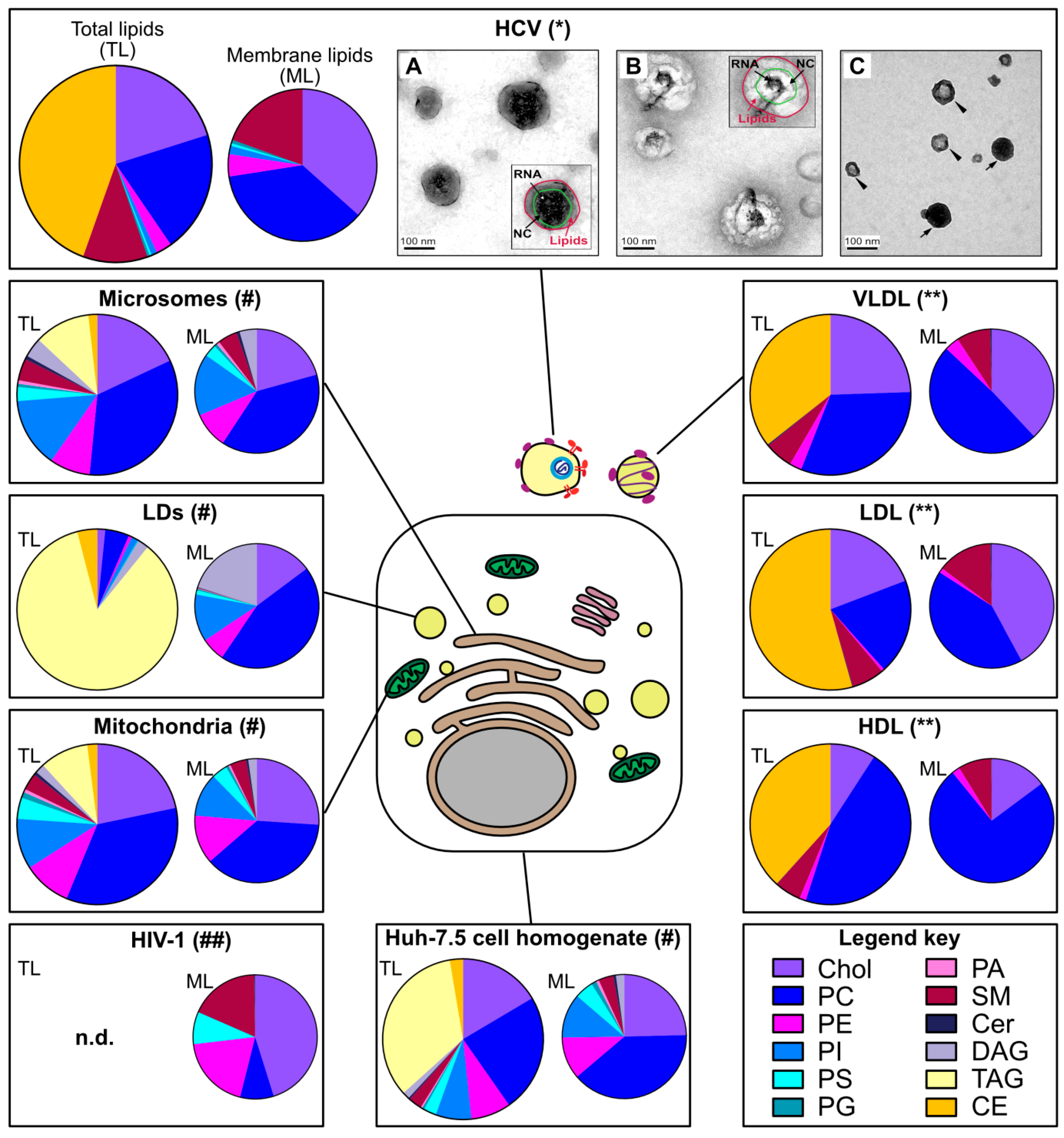
2.1. HCV Particles Resemble Host Serum Lipoproteins
2.2. HCV Lipidome Exhibits an Excess of Neutral Lipids
2.3. Apolipoproteins Make an Important Part of the Virion Proteome
2.4. Several HCV Proteins Colocalize with Lipid Droplets
3. From the ER, HCV Takes a Grip on the Lipid Droplet: Building an Interface between Replication and Assembly Complexes
3.1. Structural Basis for the Association of HCV Proteins with the Lipid Droplet Monolayer
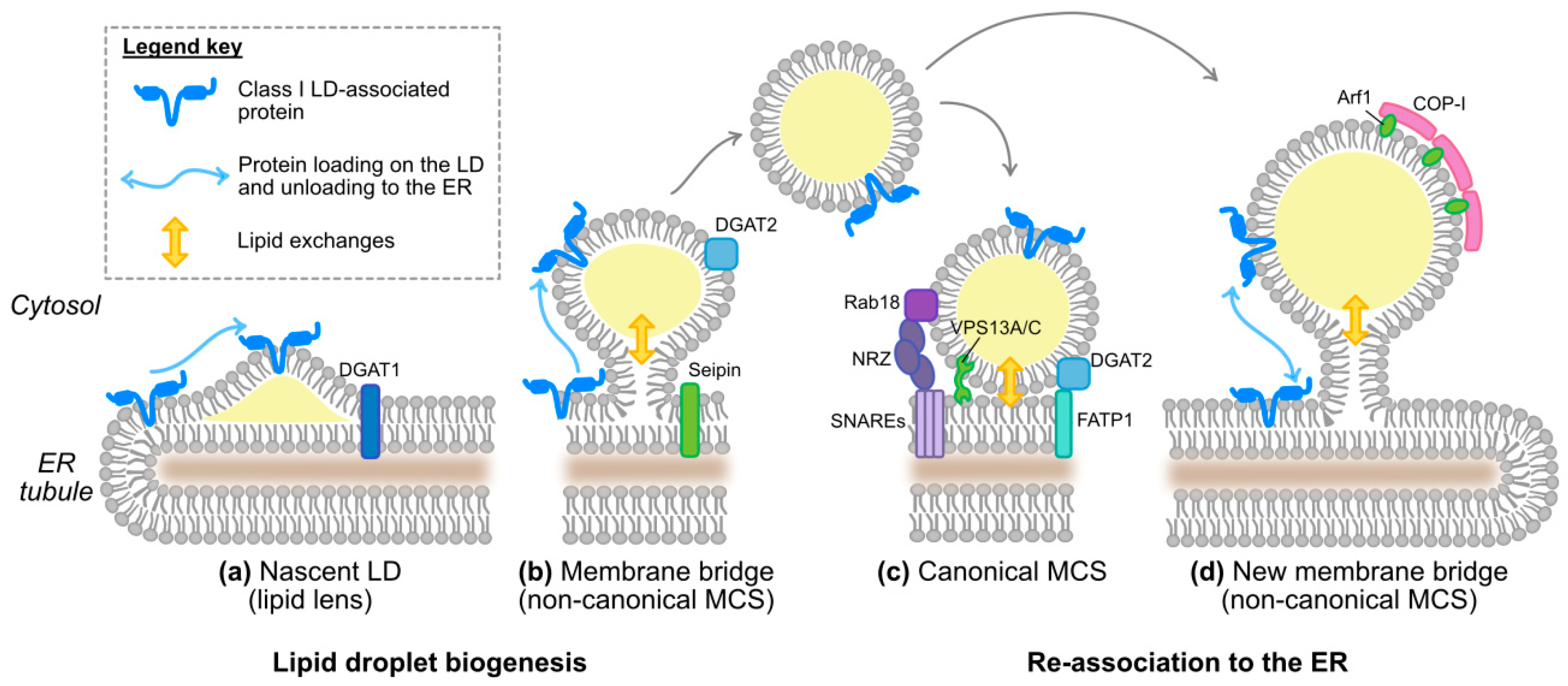
3.1.1. The Physiological Case: Several Ways to Bind a Lipid Droplet
3.1.2. HCV Dispatches Some of Its Proteins onto the LD Surface
3.2. Transfer of HCV Proteins from the ER to the LD Surface
3.2.1. The Physiological Case
3.2.2. HCV Case: Dynamics of HCV Hijacking of the Lipid Droplet Organelle
4. Lipid Droplets as an Entry Door for HCV to Reach the Site of Lipoprotein Biogenesis
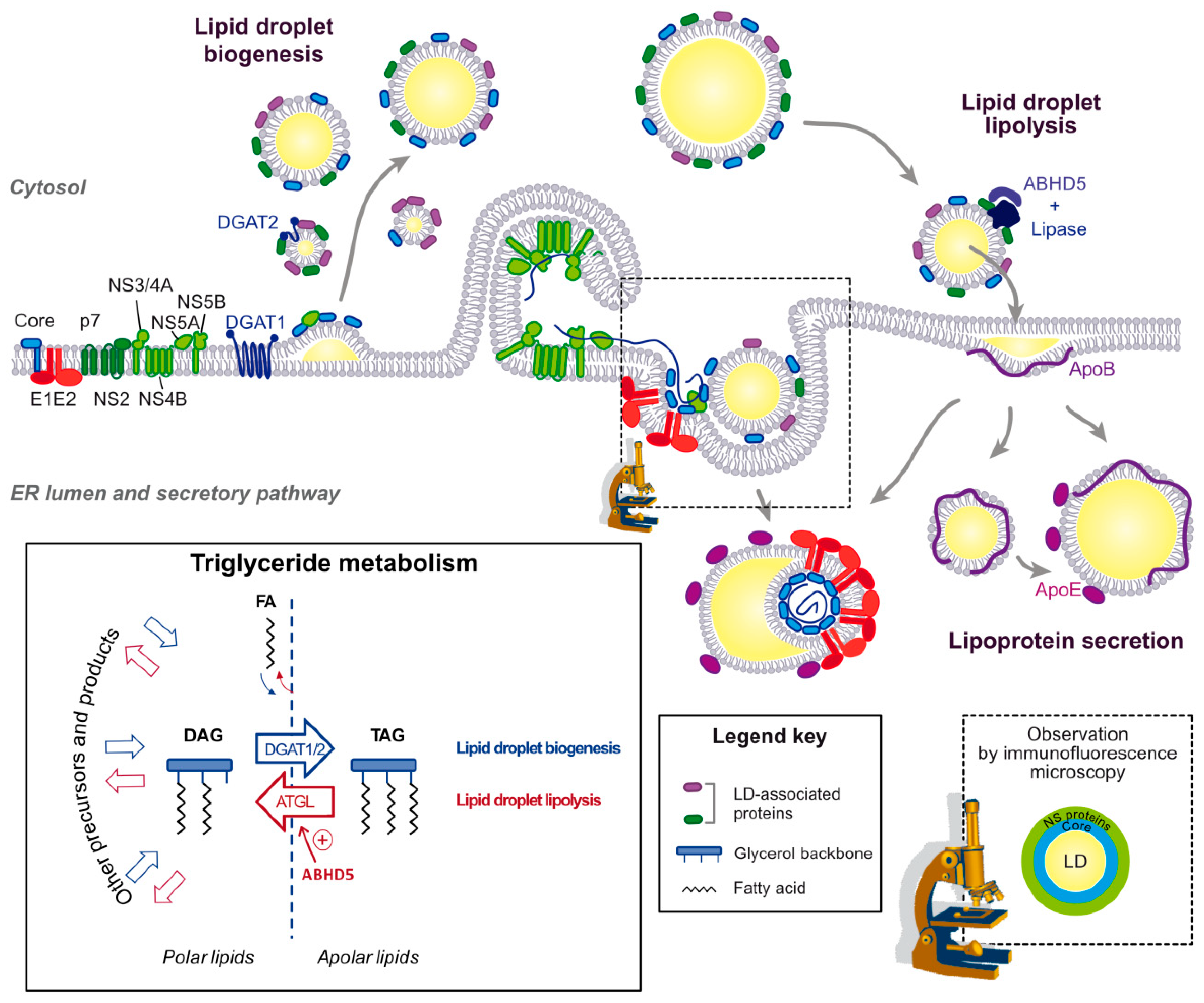
4.1. Lipid Droplets Are a Stopover during the Assembly Process
4.2. The Interface between LDs and ER as an Assembly Platform
4.3. Importance of Triglyceride Hydrolysis for HCV Morphogenesis
4.4. Key Role of Apolipoproteins in HCV Production in Liver Cells
5. Plasticity and Maturation of HCV Lipo-Viro-Particle
6. Apolipoproteins: The Only Hitchhike for HCV to Exit? A Range of Proteins Can Substitute for Apolipoproteins in HCV Assembly and Egress
7. Connections and Bifurcations between HCV and Lipoprotein Secretion Pathways
8. The Function of Apolipoproteins or Mimics in HCV Production—Hints and Guesses
9. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Walther, T.C.; Chung, J.; Farese, R.V., Jr. Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 2017, 33, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Frayn, K.N.; Arner, P.; Yki-Jarvinen, H. Fatty acid metabolism in adipose tissue, muscle and liver in health and disease. Essays Biochem. 2006, 42, 89–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welte, M.A. Expanding roles for lipid droplets. Curr. Biol. 2015, 25, R470–R481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krahmer, N.; Farese, R.V., Jr.; Walther, T.C. Balancing the fat: Lipid droplets and human disease. EMBO Mol. Med. 2013, 5, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Herker, E.; Ott, M. Emerging role of lipid droplets in host/pathogen interactions. J. Biol. Chem. 2012, 287, 2280–2287. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Madan, V.; Bartenschlager, R. Hepatitis c virus RNA replication and assembly: Living on the fat of the land. Cell Host Microbe 2014, 16, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Ploss, A.; Evans, M.J. Hepatitis c virus host cell entry. Curr. Opin. Virol. 2012, 2, 14–19. [Google Scholar] [CrossRef]
- Fukuhara, T.; Matsuura, Y. Role of mir-122 and lipid metabolism in hcv infection. J. Gastroenterol. 2013, 48, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. The ins and outs of hepatitis c virus entry and assembly. Nat. Rev. Microbiol. 2013, 11, 688–700. [Google Scholar] [CrossRef]
- Podevin, P.; Carpentier, A.; Pene, V.; Aoudjehane, L.; Carriere, M.; Zaidi, S.; Hernandez, C.; Calle, V.; Meritet, J.F.; Scatton, O.; et al. Production of infectious hepatitis c virus in primary cultures of human adult hepatocytes. Gastroenterology 2010, 139, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Felmlee, D.J.; Sheridan, D.A.; Bridge, S.H.; Nielsen, S.U.; Milne, R.W.; Packard, C.J.; Caslake, M.J.; McLauchlan, J.; Toms, G.L.; Neely, R.D.; et al. Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis c virus particles. Gastroenterology 2010, 139, 1774–1783. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Meuleman, P.; Ploss, A.; Vanwolleghem, T.; Syder, A.J.; McKeating, J.A.; Lanford, R.E.; Feinstone, S.M.; Major, M.E.; Leroux-Roels, G.; et al. Cell culture-grown hepatitis c virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA 2006, 103, 3805–3809. [Google Scholar] [CrossRef] [PubMed]
- Roingeard, P. Viral detection by electron microscopy: Past, present and future. Biol. Cell 2008, 100, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Gastaminza, P.; Dryden, K.A.; Boyd, B.; Wood, M.R.; Law, M.; Yeager, M.; Chisari, F.V. Ultrastructural and biophysical characterization of hepatitis c virus particles produced in cell culture. J. Virol. 2010, 84, 10999–11009. [Google Scholar] [CrossRef] [PubMed]
- Merz, A.; Long, G.; Hiet, M.S.; Brugger, B.; Chlanda, P.; Andre, P.; Wieland, F.; Krijnse-Locker, J.; Bartenschlager, R. Biochemical and morphological properties of hepatitis c virus particles and determination of their lipidome. J. Biol. Chem. 2011, 286, 3018–3032. [Google Scholar] [CrossRef] [PubMed]
- Catanese, M.T.; Uryu, K.; Kopp, M.; Edwards, T.J.; Andrus, L.; Rice, W.J.; Silvestry, M.; Kuhn, R.J.; Rice, C.M. Ultrastructural analysis of hepatitis c virus particles. Proc. Natl. Acad. Sci. USA 2013, 110, 9505–9510. [Google Scholar] [CrossRef] [PubMed]
- Piver, E.; Boyer, A.; Gaillard, J.; Bull, A.; Beaumont, E.; Roingeard, P.; Meunier, J.C. Ultrastructural organisation of hcv from the bloodstream of infected patients revealed by electron microscopy after specific immunocapture. Gut 2017, 66, 1487–1495. [Google Scholar] [CrossRef]
- Gastaminza, P.; Kapadia, S.B.; Chisari, F.V. Differential biophysical properties of infectious intracellular and secreted hepatitis c virus particles. J. Virol. 2006, 80, 11074–11081. [Google Scholar] [CrossRef]
- Kalvodova, L.; Sampaio, J.L.; Cordo, S.; Ejsing, C.S.; Shevchenko, A.; Simons, K. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 2009, 83, 7996–8003. [Google Scholar] [CrossRef]
- Liu, S.T.; Sharon-Friling, R.; Ivanova, P.; Milne, S.B.; Myers, D.S.; Rabinowitz, J.D.; Brown, H.A.; Shenk, T. Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for snare machinery in virion egress. Proc. Natl. Acad. Sci. USA 2011, 108, 12869–12874. [Google Scholar] [CrossRef]
- Hofmann, S.; Krajewski, M.; Scherer, C.; Scholz, V.; Mordhorst, V.; Truschow, P.; Schobel, A.; Reimer, R.; Schwudke, D.; Herker, E. Complex lipid metabolic remodeling is required for efficient hepatitis c virus replication. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Cantin, R.; Methot, S.; Tremblay, M.J. Plunder and stowaways: Incorporation of cellular proteins by enveloped viruses. J. Virol. 2005, 79, 6577–6587. [Google Scholar] [CrossRef] [PubMed]
- Wrensch, F.; Crouchet, E.; Ligat, G.; Zeisel, M.B.; Keck, Z.Y.; Foung, S.K.H.; Schuster, C.; Baumert, T.F. Hepatitis c virus (hcv)-apolipoprotein interactions and immune evasion and their impact on hcv vaccine design. Front. Immunol. 2018, 9, 1436. [Google Scholar] [CrossRef]
- Nielsen, S.U.; Bassendine, M.F.; Burt, A.D.; Martin, C.; Pumeechockchai, W.; Toms, G.L. Association between hepatitis c virus and very-low-density lipoprotein (vldl)/ldl analyzed in iodixanol density gradients. J. Virol. 2006, 80, 2418–2428. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.S.; Jiang, J.; Cai, Z.; Luo, G. Human apolipoprotein e is required for infectivity and production of hepatitis c virus in cell culture. J. Virol. 2007, 81, 13783–13793. [Google Scholar] [CrossRef]
- Jiang, J.; Luo, G. Apolipoprotein e but not b is required for the formation of infectious hepatitis c virus particles. J. Virol. 2009, 83, 12680–12691. [Google Scholar] [CrossRef]
- Lussignol, M.; Kopp, M.; Molloy, K.; Vizcay-Barrena, G.; Fleck, R.A.; Dorner, M.; Bell, K.L.; Chait, B.T.; Rice, C.M.; Catanese, M.T. Proteomics of hcv virions reveals an essential role for the nucleoporin nup98 in virus morphogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, 2484–2489. [Google Scholar] [CrossRef] [PubMed]
- Coller, K.E.; Heaton, N.S.; Berger, K.L.; Cooper, J.D.; Saunders, J.L.; Randall, G. Molecular determinants and dynamics of hepatitis c virus secretion. PLoS Pathog. 2012, 8, e1002466. [Google Scholar] [CrossRef]
- Moradpour, D.; Wakita, T.; Tokushige, K.; Carlson, R.I.; Krawczynski, K.; Wands, J.R. Characterization of three novel monoclonal antibodies against hepatitis c virus core protein. J. Med. Virol. 1996, 48, 234–241. [Google Scholar] [CrossRef]
- Rouille, Y.; Helle, F.; Delgrange, D.; Roingeard, P.; Voisset, C.; Blanchard, E.; Belouzard, S.; McKeating, J.; Patel, A.H.; Maertens, G.; et al. Subcellular localization of hepatitis c virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 2006, 80, 2832–2841. [Google Scholar] [CrossRef]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The lipid droplet is an important organelle for hepatitis c virus production. Nat. Cell Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Boulant, S.; Targett-Adams, P.; McLauchlan, J. Disrupting the association of hepatitis c virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 2007, 88, 2204–2213. [Google Scholar] [CrossRef] [PubMed]
- Andre, P.; Komurian-Pradel, F.; Deforges, S.; Perret, M.; Berland, J.L.; Sodoyer, M.; Pol, S.; Brechot, C.; Paranhos-Baccala, G.; Lotteau, V. Characterization of low- and very-low-density hepatitis c virus rna-containing particles. J. Virol. 2002, 76, 6919–6928. [Google Scholar] [CrossRef] [PubMed]
- Brugger, B.; Glass, B.; Haberkant, P.; Leibrecht, I.; Wieland, F.T.; Krausslich, H.G. The hiv lipidome: A raft with an unusual composition. Proc. Natl. Acad. Sci. USA 2006, 103, 2641–2646. [Google Scholar] [CrossRef]
- Wiesner, P.; Leidl, K.; Boettcher, A.; Schmitz, G.; Liebisch, G. Lipid profiling of fplc-separated lipoprotein fractions by electrospray ionization tandem mass spectrometry. J. Lipid Res. 2009, 50, 574–585. [Google Scholar] [CrossRef]
- Kory, N.; Farese, R.V., Jr.; Walther, T.C. Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol. 2016, 26, 535–546. [Google Scholar] [CrossRef]
- Abell, B.M.; Holbrook, L.A.; Abenes, M.; Murphy, D.J.; Hills, M.J.; Moloney, M.M. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 1997, 9, 1481–1493. [Google Scholar] [CrossRef]
- Huang, C.Y.; Huang, A.H.C. Unique motifs and length of hairpin in oleosin target the cytosolic side of endoplasmic reticulum and budding lipid droplet. Plant Physiol. 2017, 174, 2248–2260. [Google Scholar] [CrossRef]
- Wilfling, F.; Wang, H.; Haas, J.T.; Krahmer, N.; Gould, T.J.; Uchida, A.; Cheng, J.X.; Graham, M.; Christiano, R.; Frohlich, F.; et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the er to lipid droplets. Dev. Cell 2013, 24, 384–399. [Google Scholar] [CrossRef]
- Stevanovic, A.; Thiele, C. Monotopic topology is required for lipid droplet targeting of ancient ubiquitous protein 1. J. Lipid Res. 2013, 54, 503–513. [Google Scholar] [CrossRef]
- Huang, A.H. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996, 110, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- Moradpour, D.; Penin, F. Hepatitis c virus proteins: From structure to function. Curr. Top. Microbiol. Immunol. 2013, 369, 113–142. [Google Scholar]
- McLauchlan, J.; Lemberg, M.K.; Hope, G.; Martoglio, B. Intramembrane proteolysis promotes trafficking of hepatitis c virus core protein to lipid droplets. EMBO J. 2002, 21, 3980–3988. [Google Scholar] [CrossRef] [PubMed]
- Targett-Adams, P.; Hope, G.; Boulant, S.; McLauchlan, J. Maturation of hepatitis c virus core protein by signal peptide peptidase is required for virus production. J. Biol. Chem. 2008, 283, 16850–16859. [Google Scholar] [CrossRef] [PubMed]
- Boulant, S.; Montserret, R.; Hope, R.G.; Ratinier, M.; Targett-Adams, P.; Lavergne, J.P.; Penin, F.; McLauchlan, J. Structural determinants that target the hepatitis c virus core protein to lipid droplets. J. Biol. Chem. 2006, 281, 22236–22247. [Google Scholar] [CrossRef]
- Hope, R.G.; Murphy, D.J.; McLauchlan, J. The domains required to direct core proteins of hepatitis c virus and gb virus-b to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 2002, 277, 4261–4270. [Google Scholar] [CrossRef] [PubMed]
- Brass, V.; Bieck, E.; Montserret, R.; Wolk, B.; Hellings, J.A.; Blum, H.E.; Penin, F.; Moradpour, D. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis c virus nonstructural protein 5a. J. Biol. Chem. 2002, 277, 8130–8139. [Google Scholar] [CrossRef]
- Brass, V.; Pal, Z.; Sapay, N.; Deleage, G.; Blum, H.E.; Penin, F.; Moradpour, D. Conserved determinants for membrane association of nonstructural protein 5a from hepatitis c virus and related viruses. J. Virol. 2007, 81, 2745–2757. [Google Scholar] [CrossRef] [PubMed]
- Hinson, E.R.; Cresswell, P. The antiviral protein, viperin, localizes to lipid droplets via its n-terminal amphipathic alpha-helix. Proc. Natl. Acad. Sci. USA 2009, 106, 20452–20457. [Google Scholar] [CrossRef]
- Vogt, D.A.; Camus, G.; Herker, E.; Webster, B.R.; Tsou, C.L.; Greene, W.C.; Yen, T.S.; Ott, M. Lipid droplet-binding protein tip47 regulates hepatitis c virus rna replication through interaction with the viral ns5a protein. PLoS Pathog. 2013, 9, e1003302. [Google Scholar] [CrossRef]
- Salloum, S.; Wang, H.; Ferguson, C.; Parton, R.G.; Tai, A.W. Rab18 binds to hepatitis c virus ns5a and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 2013, 9, e1003513. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Suzuki, R.; Murakami, K.; Aizaki, H.; Ishii, K.; Murayama, A.; Date, T.; Matsuura, Y.; Miyamura, T.; Wakita, T.; et al. Interaction of hepatitis c virus nonstructural protein 5a with core protein is critical for the production of infectious virus particles. J. Virol. 2008, 82, 7964–7976. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.; Baugh, J.; Chatterji, U.; Lim, P.J.; Bobardt, M.D.; Gallay, P.A. Hcv core residues critical for infectivity are also involved in core-ns5a complex formation. PLoS ONE 2014, 9, e88866. [Google Scholar] [CrossRef] [PubMed]
- Camus, G.; Herker, E.; Modi, A.A.; Haas, J.T.; Ramage, H.R.; Farese, R.V., Jr.; Ott, M. Diacylglycerol acyltransferase-1 localizes hepatitis c virus ns5a protein to lipid droplets and enhances ns5a interaction with the viral capsid core. J. Biol. Chem. 2013, 288, 9915–9923. [Google Scholar] [CrossRef] [PubMed]
- Gouttenoire, J.; Penin, F.; Moradpour, D. Hepatitis c virus nonstructural protein 4b: A journey into unexplored territory. Rev. Med. Virol. 2010, 20, 117–129. [Google Scholar] [CrossRef]
- Tanaka, T.; Kuroda, K.; Ikeda, M.; Wakita, T.; Kato, N.; Makishima, M. Hepatitis c virus ns4b targets lipid droplets through hydrophobic residues in the amphipathic helices. J. Lipid Res. 2013, 54, 881–892. [Google Scholar] [CrossRef]
- Cornell, R.B.; Northwood, I.C. Regulation of ctp:Phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci 2000, 25, 441–447. [Google Scholar] [CrossRef]
- Mól, A.R.; Castro, M.S.; Fontes, W. Netwheels: A web application to create high quality peptide helical wheel and net projections. bioRxiv 2018. [Google Scholar] [CrossRef]
- Kassan, A.; Herms, A.; Fernandez-Vidal, A.; Bosch, M.; Schieber, N.L.; Reddy, B.J.; Fajardo, A.; Gelabert-Baldrich, M.; Tebar, F.; Enrich, C.; et al. Acyl-coa synthetase 3 promotes lipid droplet biogenesis in er microdomains. J. Cell Biol. 2013, 203, 985–1001. [Google Scholar] [CrossRef]
- Wu, H.; Carvalho, P.; Voeltz, G.K. Here, there, and everywhere: The importance of er membrane contact sites. Science 2018, 361, eaan5835. [Google Scholar] [CrossRef]
- Xu, N.; Zhang, S.O.; Cole, R.A.; McKinney, S.A.; Guo, F.; Haas, J.T.; Bobba, S.; Farese, R.V., Jr.; Mak, H.Y. The fatp1-dgat2 complex facilitates lipid droplet expansion at the er-lipid droplet interface. J. Cell Biol. 2012, 198, 895–911. [Google Scholar] [CrossRef]
- Ozeki, S.; Cheng, J.; Tauchi-Sato, K.; Hatano, N.; Taniguchi, H.; Fujimoto, T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci. 2005, 118, 2601–2611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Li, Y.; Wu, L.; Zhao, D.; Yu, J.; Huang, T.; Ferguson, C.; Parton, R.G.; Yang, H.; Li, P. Rab18 promotes lipid droplet (ld) growth by tethering the er to lds through snare and nrz interactions. J. Cell Biol. 2018, 217, 975–995. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. Vps13a and vps13c are lipid transport proteins differentially localized at er contact sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [PubMed]
- Beller, M.; Sztalryd, C.; Southall, N.; Bell, M.; Jackle, H.; Auld, D.S.; Oliver, B. Copi complex is a regulator of lipid homeostasis. PLoS Biol. 2008, 6, e292. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.G.; Mardones, G.A.; Sougrat, R.; Smirnova, E.; Jackson, C.L.; Bonifacino, J.S. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 2009, 122, 1834–1841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilfling, F.; Thiam, A.R.; Olarte, M.J.; Wang, J.; Beck, R.; Gould, T.J.; Allgeyer, E.S.; Pincet, F.; Bewersdorf, J.; Farese, R.V., Jr.; et al. Arf1/copi machinery acts directly on lipid droplets and enables their connection to the er for protein targeting. eLife 2014, 3, e01607. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, K.M.; Binns, D.; Bartz, R.; Grishin, N.V.; Li, W.P.; Agarwal, A.K.; Garg, A.; Anderson, R.G.; Goodman, J.M. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA 2007, 104, 20890–20895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salo, V.T.; Belevich, I.; Li, S.; Karhinen, L.; Vihinen, H.; Vigouroux, C.; Magre, J.; Thiele, C.; Holtta-Vuori, M.; Jokitalo, E.; et al. Seipin regulates er-lipid droplet contacts and cargo delivery. EMBO J. 2016, 35, 2699–2716. [Google Scholar] [CrossRef]
- Herker, E.; Harris, C.; Hernandez, C.; Carpentier, A.; Kaehlcke, K.; Rosenberg, A.R.; Farese, R.V., Jr.; Ott, M. Efficient hepatitis c virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 2010, 16, 1295–1298. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.L.; Stone, S.J.; Koliwad, S.; Harris, C.; Farese, R.V., Jr. Thematic review series: Glycerolipids. Dgat enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008, 49, 2283–2301. [Google Scholar] [CrossRef] [PubMed]
- Menzel, N.; Fischl, W.; Hueging, K.; Bankwitz, D.; Frentzen, A.; Haid, S.; Gentzsch, J.; Kaderali, L.; Bartenschlager, R.; Pietschmann, T. Map-kinase regulated cytosolic phospholipase a2 activity is essential for production of infectious hepatitis c virus particles. PLoS Pathog. 2012, 8, e1002829. [Google Scholar] [CrossRef] [PubMed]
- Pol, A.; Gross, S.P.; Parton, R.G. Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 2014, 204, 635–646. [Google Scholar] [CrossRef]
- Vieyres, G.; Thomas, X.; Descamps, V.; Duverlie, G.; Patel, A.H.; Dubuisson, J. Characterization of the envelope glycoproteins associated with infectious hepatitis c virus. J. Virol. 2010, 84, 10159–10168. [Google Scholar] [CrossRef] [PubMed]
- Gastaminza, P.; Cheng, G.; Wieland, S.; Zhong, J.; Liao, W.; Chisari, F.V. Cellular determinants of hepatitis c virus assembly, maturation, degradation, and secretion. J. Virol. 2008, 82, 2120–2129. [Google Scholar] [CrossRef]
- Zehmer, J.K.; Bartz, R.; Bisel, B.; Liu, P.; Seemann, J.; Anderson, R.G. Targeting sequences of ubxd8 and aam-b reveal that the er has a direct role in the emergence and regression of lipid droplets. J. Cell Sci. 2009, 122, 3694–3702. [Google Scholar] [CrossRef] [PubMed]
- Vieyres, G.; Welsch, K.; Gerold, G.; Gentzsch, J.; Kahl, S.; Vondran, F.W.; Kaderali, L.; Pietschmann, T. Abhd5/cgi-58, the chanarin-dorfman syndrome protein, mobilises lipid stores for hepatitis c virus production. PLoS Pathog. 2016, 12, e1005568. [Google Scholar] [CrossRef]
- Shavinskaya, A.; Boulant, S.; Penin, F.; McLauchlan, J.; Bartenschlager, R. The lipid droplet binding domain of hepatitis c virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 2007, 282, 37158–37169. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Granio, O.; Bartenschlager, R.; Cosset, F.L. A concerted action of hepatitis c virus p7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog. 2011, 7, e1002144. [Google Scholar] [CrossRef] [PubMed]
- Counihan, N.A.; Rawlinson, S.M.; Lindenbach, B.D. Trafficking of hepatitis c virus core protein during virus particle assembly. PLoS Pathog. 2011, 7, e1002302. [Google Scholar] [CrossRef]
- Gentzsch, J.; Brohm, C.; Steinmann, E.; Friesland, M.; Menzel, N.; Vieyres, G.; Perin, P.M.; Frentzen, A.; Kaderali, L.; Pietschmann, T. Hepatitis c virus p7 is critical for capsid assembly and envelopment. PLoS Pathog. 2013, 9, e1003355. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, T.; Wada, M.; Nakamura, S.; Ono, C.; Shiokawa, M.; Yamamoto, S.; Motomura, T.; Okamoto, T.; Okuzaki, D.; Yamamoto, M.; et al. Amphipathic alpha-helices in apolipoproteins are crucial to the formation of infectious hepatitis c virus particles. PLoS Pathog. 2014, 10, e1004534. [Google Scholar] [CrossRef] [PubMed]
- Vieyres, G.; Dubuisson, J.; Pietschmann, T. Incorporation of hepatitis c virus e1 and e2 glycoproteins: The keystones on a peculiar virion. Viruses 2014, 6, 1149–1187. [Google Scholar] [CrossRef]
- Yan, Y.; He, Y.; Boson, B.; Wang, X.; Cosset, F.L.; Zhong, J. A point mutation in the n-terminal amphipathic helix alpha0 in ns3 promotes hepatitis c virus assembly by altering core localization to the endoplasmic reticulum and facilitating virus budding. J. Virol. 2017, 91, e02399-16. [Google Scholar] [CrossRef] [PubMed]
- Boulant, S.; Douglas, M.W.; Moody, L.; Budkowska, A.; Targett-Adams, P.; McLauchlan, J. Hepatitis c virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 2008, 9, 1268–1282. [Google Scholar] [CrossRef]
- Akil, A.; Peng, J.; Omrane, M.; Gondeau, C.; Desterke, C.; Marin, M.; Tronchere, H.; Taveneau, C.; Sar, S.; Briolotti, P.; et al. Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by hcv. Nat. Commun. 2016, 7, 12203. [Google Scholar] [CrossRef] [PubMed]
- Horner, S.M.; Liu, H.M.; Park, H.S.; Briley, J.; Gale, M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (mam) form innate immune synapses and are targeted by hepatitis c virus. Proc. Natl. Acad. Sci. USA 2011, 108, 14590–14595. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, I.K.; Lee, J.Y.; Tabata, K.; Romero-Brey, I.; Paul, D.; Schult, P.; Lohmann, V.; Kaderali, L.; Bartenschlager, R. Hepatitis c virus replication depends on endosomal cholesterol homeostasis. J. Virol. 2017, 92, e01196-17. [Google Scholar] [CrossRef]
- Amako, Y.; Syed, G.H.; Siddiqui, A. Protein kinase d negatively regulates hepatitis c virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein. J. Biol. Chem. 2011, 286, 11265–11274. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Perry, J.W.; Lauring, A.S.; Neddermann, P.; De Francesco, R.; Tai, A.W. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for hcv replication membrane integrity and cholesterol trafficking. Gastroenterology 2014, 146, 1373–1385. [Google Scholar] [CrossRef] [PubMed]
- Altan-Bonnet, N.; Balla, T. Phosphatidylinositol 4-kinases: Hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 2012, 37, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Dansako, H.; Hiramoto, H.; Ikeda, M.; Wakita, T.; Kato, N. Rab18 is required for viral assembly of hepatitis c virus through trafficking of the core protein to lipid droplets. Virology 2014, 462–463, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Ploen, D.; Hafirassou, M.L.; Himmelsbach, K.; Sauter, D.; Biniossek, M.L.; Weiss, T.S.; Baumert, T.F.; Schuster, C.; Hildt, E. Tip47 plays a crucial role in the life cycle of hepatitis c virus. J. Hepatol. 2013, 58, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tai, A.W. Continuous de novo generation of spatially segregated hepatitis c virus replication organelles revealed by pulse-chase imaging. J. Hepatol. 2017, 66, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, G.F.; Islam, K.; Pease, R.J. Mobilisation of triacylglycerol stores. Biochim. Biophys. Acta 2000, 1483, 37–57. [Google Scholar] [CrossRef]
- Lass, A.; Zimmermann, R.; Haemmerle, G.; Riederer, M.; Schoiswohl, G.; Schweiger, M.; Kienesberger, P.; Strauss, J.G.; Gorkiewicz, G.; Zechner, R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by cgi-58 and defective in chanarin-dorfman syndrome. Cell Metab. 2006, 3, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lu, X.; Lombes, M.; Rha, G.B.; Chi, Y.I.; Guerin, T.M.; Smart, E.J.; Liu, J. The g(0)/g(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010, 11, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, M.; Lass, A.; Zimmermann, R.; Eichmann, T.O.; Zechner, R. Neutral lipid storage disease: Genetic disorders caused by mutations in adipose triglyceride lipase/pnpla2 or cgi-58/abhd5. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E289–E296. [Google Scholar] [CrossRef]
- Guo, M.; Pei, R.; Yang, Q.; Cao, H.; Wang, Y.; Wu, C.; Chen, J.; Zhou, Y.; Hu, X.; Lu, M.; et al. Phosphatidylserine-specific phospholipase a1 involved in hepatitis c virus assembly through ns2 complex formation. J. Virol. 2015, 89, 2367–2377. [Google Scholar] [CrossRef]
- Li, X.; Jiang, H.; Qu, L.; Yao, W.; Cai, H.; Chen, L.; Peng, T. Hepatocyte nuclear factor 4alpha and downstream secreted phospholipase a2 gxiib regulate production of infectious hepatitis c virus. J. Virol. 2014, 88, 612–627. [Google Scholar] [CrossRef]
- Xu, S.; Pei, R.; Guo, M.; Han, Q.; Lai, J.; Wang, Y.; Wu, C.; Zhou, Y.; Lu, M.; Chen, X. Cytosolic phospholipase a2 gamma is involved in hepatitis c virus replication and assembly. J. Virol. 2012, 86, 13025–13037. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, M.; Douglas, D.N.; Pu, C.H.; Lewis, J.T.; Kawahara, T.; Lisboa, L.F.; Wei, E.; Asthana, S.; Quiroga, A.D.; Law, L.M.; et al. Arylacetamide deacetylase: A novel host factor with important roles in the lipolysis of cellular triacylglycerol stores, vldl assembly and hcv production. J. Hepatol. 2013, 59, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.; Cornaciu, I.; Lass, A.; Schweiger, M.; Poeschl, M.; Eder, C.; Kumari, M.; Schoiswohl, G.; Wolinski, H.; Kohlwein, S.D.; et al. The n-terminal region of comparative gene identification-58 (cgi-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J. Biol. Chem. 2010, 285, 12289–12298. [Google Scholar] [CrossRef] [PubMed]
- Oberer, M.; Boeszoermenyi, A.; Nagy, H.M.; Zechner, R. Recent insights into the structure and function of comparative gene identification-58. Curr. Opin. Lipidol. 2011, 22, 149–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, C.C.; Brown, J.M. Distinct roles for alpha-beta hydrolase domain 5 (abhd5/cgi-58) and adipose triglyceride lipase (atgl/pnpla2) in lipid metabolism and signaling. Adipocyte 2012, 1, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.C.; Ferguson, D.; Thomas, G.; Brown, A.L.; Schugar, R.C.; Burrows, A.; Gromovsky, A.D.; Betters, J.; Neumann, C.; Sacks, J.; et al. Regulation of hepatic triacylglycerol metabolism by cgi-58 does not require atgl co-activation. Cell Rep. 2016, 16, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Ong, K.T.; Mashek, M.T.; Bu, S.Y.; Greenberg, A.S.; Mashek, D.G. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology 2011, 53, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Wang, S.P.; Alvarez, F.; Casavant, S.; Gauthier, N.; Abed, L.; Soni, K.G.; Yang, G.; Mitchell, G.A. Deficiency of liver adipose triglyceride lipase in mice causes progressive hepatic steatosis. Hepatology 2011, 54, 122–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zechner, R.; Zimmermann, R.; Eichmann, T.O.; Kohlwein, S.D.; Haemmerle, G.; Lass, A.; Madeo, F. Fat signals--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012, 15, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Wurie, H.R.; Buckett, L.; Zammit, V.A. Evidence that diacylglycerol acyltransferase 1 (dgat1) has dual membrane topology in the endoplasmic reticulum of hepg2 cells. J. Biol. Chem. 2011, 286, 36238–36247. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.J.; Levin, M.C.; Farese, R.V., Jr. Membrane topology and identification of key functional amino acid residues of murine acyl-coa:Diacylglycerol acyltransferase-2. J. Biol. Chem. 2006, 281, 40273–40282. [Google Scholar] [CrossRef] [PubMed]
- Boyer, A.; Park, S.B.; de Boer, Y.S.; Li, Q.; Liang, T.J. Tm6sf2 promotes lipidation and secretion of hepatitis c virus in infected hepatocytes. Gastroenterology 2018, 155, 1923–1935. [Google Scholar] [CrossRef] [PubMed]
- Smagris, E.; Gilyard, S.; BasuRay, S.; Cohen, J.C.; Hobbs, H.H. Inactivation of tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J. Biol. Chem. 2016, 291, 10659–10676. [Google Scholar] [CrossRef] [PubMed]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjaerg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a tm6sf2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef]
- Bartenschlager, R.; Penin, F.; Lohmann, V.; Andre, P. Assembly of infectious hepatitis c virus particles. Trends Microbiol. 2011, 19, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yao, W.; Li, L.; Li, X.; Hu, L.; Mai, R.; Peng, T. Cell-death-inducing dffa-like effector b contributes to the assembly of hepatitis c virus (hcv) particles and interacts with hcv ns5a. Sci. Rep. 2016, 6, 27778. [Google Scholar] [CrossRef] [PubMed]
- Hueging, K.; Weller, R.; Doepke, M.; Vieyres, G.; Todt, D.; Wolk, B.; Vondran, F.W.; Geffers, R.; Lauber, C.; Kaderali, L.; et al. Several human liver cell expressed apolipoproteins complement hcv virus production with varying efficacy conferring differential specific infectivity to released viruses. PLoS ONE 2015, 10, e0134529. [Google Scholar] [CrossRef]
- Meex, S.J.; Andreo, U.; Sparks, J.D.; Fisher, E.A. Huh-7 or hepg2 cells: Which is the better model for studying human apolipoprotein-b100 assembly and secretion? J. Lipid Res. 2011, 52, 152–158. [Google Scholar] [CrossRef]
- Schobel, A.; Rosch, K.; Herker, E. Functional innate immunity restricts hepatitis c virus infection in induced pluripotent stem cell-derived hepatocytes. Sci. Rep. 2018, 8, 3893. [Google Scholar] [CrossRef]
- Gural, N.; Mancio-Silva, L.; He, J.; Bhatia, S.N. Engineered livers for infectious diseases. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 131–144. [Google Scholar] [CrossRef]
- Da Costa, D.; Turek, M.; Felmlee, D.J.; Girardi, E.; Pfeffer, S.; Long, G.; Bartenschlager, R.; Zeisel, M.B.; Baumert, T.F. Reconstitution of the entire hepatitis c virus life cycle in nonhepatic cells. J. Virol. 2012, 86, 11919–11925. [Google Scholar] [CrossRef] [PubMed]
- Hueging, K.; Doepke, M.; Vieyres, G.; Bankwitz, D.; Frentzen, A.; Doerrbecker, J.; Gumz, F.; Haid, S.; Wolk, B.; Kaderali, L.; et al. Apolipoprotein e codetermines tissue tropism of hepatitis c virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. J. Virol. 2014, 88, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lee, E.M.; Hammack, C.; Robotham, J.M.; Basu, M.; Lang, J.; Brinton, M.A.; Tang, H. Cell death-inducing dffa-like effector b is required for hepatitis c virus entry into hepatocytes. J. Virol. 2014, 88, 8433–8444. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.E.; Fisher, E.A. Lipoprotein metabolism, dyslipidemia, and nonalcoholic fatty liver disease. Semin. Liver Dis. 2013, 33, 380–388. [Google Scholar]
- Feingold, K.R.; Grunfeld, C. Introduction to Lipids and Lipoproteins. In Endotext; De Groot, L.J., Chrousos, G., Dungan, K., Feingold, K.R., Grossman, A., Hershman, J.M., Koch, C., Korbonits, M., McLachlan, R., New, M., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Hatters, D.M.; Peters-Libeu, C.A.; Weisgraber, K.H. Apolipoprotein e structure: Insights into function. Trends Biochem. Sci. 2006, 31, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Q.; Wang, J. Topology of human apolipoprotein e3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA 2011, 108, 14813–14818. [Google Scholar] [CrossRef] [PubMed]
- Andreo, U.; de Jong, Y.P.; Scull, M.A.; Xiao, J.W.; Vercauteren, K.; Quirk, C.; Mommersteeg, M.C.; Bergaya, S.; Menon, A.; Fisher, E.A.; et al. Analysis of hepatitis c virus particle heterogeneity in immunodeficient human liver chimeric fah-/- mice. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 405–417. [Google Scholar] [CrossRef]
- Bankwitz, D.; Doepke, M.; Hueging, K.; Weller, R.; Bruening, J.; Behrendt, P.; Lee, J.Y.; Vondran, F.W.R.; Manns, M.P.; Bartenschlager, R.; et al. Maturation of secreted hcv particles by incorporation of secreted apoe protects from antibodies by enhancing infectivity. J. Hepatol. 2017, 67, 480–489. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Chi, X.; Zhao, F.; Guo, J.; Ma, P.; Zhong, J.; Niu, J.; Pan, X.; Long, G. Neglected but important role of apolipoprotein e exchange in hepatitis c virus infection. J. Virol. 2016, 90, 9632–9643. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Bi, Y.; Zhang, H.; Yao, Y.; Li, Q.; Cun, W.; Dong, S. Extracellular interactions between hepatitis c virus and secreted apolipoprotein e. J. Virol. 2017, 91, e02227-16. [Google Scholar] [CrossRef]
- Meunier, J.C.; Russell, R.S.; Engle, R.E.; Faulk, K.N.; Purcell, R.H.; Emerson, S.U. Apolipoprotein c1 association with hepatitis c virus. J. Virol. 2008, 82, 9647–9656. [Google Scholar] [CrossRef] [PubMed]
- Meunier, J.C.; Engle, R.E.; Faulk, K.; Zhao, M.; Bartosch, B.; Alter, H.; Emerson, S.U.; Cosset, F.L.; Purcell, R.H.; Bukh, J. Evidence for cross-genotype neutralization of hepatitis c virus pseudo-particles and enhancement of infectivity by apolipoprotein c1. Proc. Natl. Acad. Sci. USA 2005, 102, 4560–4565. [Google Scholar] [CrossRef]
- Dreux, M.; Boson, B.; Ricard-Blum, S.; Molle, J.; Lavillette, D.; Bartosch, B.; Pecheur, E.I.; Cosset, F.L. The exchangeable apolipoprotein apoc-i promotes membrane fusion of hepatitis c virus. J. Biol. Chem. 2007, 282, 32357–32369. [Google Scholar] [CrossRef] [PubMed]
- Puig-Basagoiti, F.; Fukuhara, T.; Tamura, T.; Ono, C.; Uemura, K.; Kawachi, Y.; Yamamoto, S.; Mori, H.; Kurihara, T.; Okamoto, T.; et al. Human cathelicidin compensates for the role of apolipoproteins in hepatitis c virus infectious particle formation. J. Virol. 2016, 90, 8464–8477. [Google Scholar] [CrossRef] [PubMed]
- Malle, E.; Steinmetz, A.; Raynes, J.G. Serum amyloid a (saa): An acute phase protein and apolipoprotein. Atherosclerosis 1993, 102, 131–146. [Google Scholar] [CrossRef]
- Wilson, P.G.; Thompson, J.C.; Shridas, P.; McNamara, P.J.; de Beer, M.C.; de Beer, F.C.; Webb, N.R.; Tannock, L.R. Serum amyloid a is an exchangeable apolipoprotein. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 1890–1900. [Google Scholar] [CrossRef]
- Lavie, M.; Voisset, C.; Vu-Dac, N.; Zurawski, V.; Duverlie, G.; Wychowski, C.; Dubuisson, J. Serum amyloid a has antiviral activity against hepatitis c virus by inhibiting virus entry in a cell culture system. Hepatology 2006, 44, 1626–1634. [Google Scholar] [CrossRef]
- Wang, G. Structures of human host defense cathelicidin ll-37 and its smallest antimicrobial peptide kr-12 in lipid micelles. J. Biol. Chem. 2008, 283, 32637–32643. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, O.; Bratt, T.; Johnsen, A.H.; Madsen, M.T.; Borregaard, N. The human antibacterial cathelicidin, hcap-18, is bound to lipoproteins in plasma. J. Biol. Chem. 1999, 274, 22445–22451. [Google Scholar] [CrossRef]
- Andersson, E.; Rydengard, V.; Sonesson, A.; Morgelin, M.; Bjorck, L.; Schmidtchen, A. Antimicrobial activities of heparin-binding peptides. Eur. J. Biochem. 2004, 271, 1219–1226. [Google Scholar] [CrossRef] [Green Version]
- Henzler Wildman, K.A.; Lee, D.K.; Ramamoorthy, A. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, ll-37. Biochemistry 2003, 42, 6545–6558. [Google Scholar] [CrossRef] [PubMed]
- Fukuhara, T.; Tamura, T.; Ono, C.; Shiokawa, M.; Mori, H.; Uemura, K.; Yamamoto, S.; Kurihara, T.; Okamoto, T.; Suzuki, R.; et al. Host-derived apolipoproteins play comparable roles with viral secretory proteins erns and ns1 in the infectious particle formation of flaviviridae. PLoS Pathog. 2017, 13, e1006475. [Google Scholar] [CrossRef]
- Rumenapf, T.; Unger, G.; Strauss, J.H.; Thiel, H.J. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 1993, 67, 3288–3294. [Google Scholar] [PubMed]
- Tews, B.A.; Meyers, G. The pestivirus glycoprotein erns is anchored in plane in the membrane via an amphipathic helix. J. Biol. Chem. 2007, 282, 32730–32741. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.; Muhle-Goll, C.; Burck, J.; Wolf, M.; Reisser, S.; Luy, B.; Wenzel, W.; Ulrich, A.S.; Meyers, G. Structure of the membrane anchor of pestivirus glycoprotein e(rns), a long tilted amphipathic helix. PLoS Pathog. 2014, 10, e1003973. [Google Scholar] [CrossRef] [PubMed]
- Tautz, N.; Tews, B.A.; Meyers, G. The molecular biology of pestiviruses. Adv. Virus Res. 2015, 93, 47–160. [Google Scholar]
- Iqbal, M.; Flick-Smith, H.; McCauley, J.W. Interactions of bovine viral diarrhoea virus glycoprotein e(rns) with cell surface glycosaminoglycans. J. Gen. Virol. 2000, 81, 451–459. [Google Scholar] [CrossRef]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus ns1 structures reveal surfaces for associations with membranes and the immune system. Science 2014, 343, 881–885. [Google Scholar] [CrossRef]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus ns1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef]
- Gutsche, I.; Coulibaly, F.; Voss, J.E.; Salmon, J.; d’Alayer, J.; Ermonval, M.; Larquet, E.; Charneau, P.; Krey, T.; Megret, F.; et al. Secreted dengue virus nonstructural protein ns1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. USA 2011, 108, 8003–8008. [Google Scholar] [CrossRef]
- Saito, H.; Lund-Katz, S.; Phillips, M.C. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog. Lipid Res. 2004, 43, 350–380. [Google Scholar] [CrossRef]
- Avirutnan, P.; Zhang, L.; Punyadee, N.; Manuyakorn, A.; Puttikhunt, C.; Kasinrerk, W.; Malasit, P.; Atkinson, J.P.; Diamond, M.S. Secreted ns1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate e. PLoS Pathog. 2007, 3, e183. [Google Scholar] [CrossRef]
- Copic, A.; Antoine-Bally, S.; Gimenez-Andres, M.; La Torre Garay, C.; Antonny, B.; Manni, M.M.; Pagnotta, S.; Guihot, J.; Jackson, C.L. A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nat. Commun. 2018, 9, 1332. [Google Scholar] [CrossRef]
- Luo, C.C.; Li, W.H.; Moore, M.N.; Chan, L. Structure and evolution of the apolipoprotein multigene family. J. Mol. Biol. 1986, 187, 325–340. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Rose, A.S.; Bradley, A.R.; Valasatava, Y.; Duarte, J.M.; Prlic, A.; Rose, P.W. Ngl viewer: Web-based molecular graphics for large complexes. Bioinformatics 2018, 34, 3755–3758. [Google Scholar] [CrossRef]
- Sayle, R.A.; Milner-White, E.J. Rasmol: Biomolecular graphics for all. Trends Biochem. Sci. 1995, 20, 374. [Google Scholar] [CrossRef]
- Tiwari, S.; Siddiqi, S.A. Intracellular trafficking and secretion of vldl. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1079–1086. [Google Scholar] [CrossRef]
- Syed, G.H.; Khan, M.; Yang, S.; Siddiqui, A. Hepatitis c virus lipoviroparticles assemble in the endoplasmic reticulum (er) and bud off from the er to the golgi compartment in copii vesicles. J. Virol. 2017, 91, e00499-17. [Google Scholar] [CrossRef]
- Bishe, B.; Syed, G.H.; Field, S.J.; Siddiqui, A. Role of phosphatidylinositol 4-phosphate (pi4p) and its binding protein golph3 in hepatitis c virus secretion. J. Biol. Chem. 2012, 287, 27637–27647. [Google Scholar] [CrossRef]
- Rustaeus, S.; Lindberg, K.; Boren, J.; Olofsson, S.O. Brefeldin a reversibly inhibits the assembly of apob containing lipoproteins in mca-rh7777 cells. J. Biol. Chem. 1995, 270, 28879–28886. [Google Scholar] [CrossRef]
- Sarhan, M.A.; Abdel-Hakeem, M.S.; Mason, A.L.; Tyrrell, D.L.; Houghton, M. Glycogen synthase kinase 3beta inhibitors prevent hepatitis c virus release/assembly through perturbation of lipid metabolism. Sci. Rep. 2017, 7, 2495. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Li, J.; Cai, H.; Yao, W.; Xiao, J.; Li, Y.P.; Qiu, X.; Xia, H.; Peng, T. Avasimibe: A novel hepatitis c virus inhibitor that targets the assembly of infectious viral particles. Antivir. Res. 2017, 148, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Benedicto, I.; Gondar, V.; Molina-Jimenez, F.; Garcia-Buey, L.; Lopez-Cabrera, M.; Gastaminza, P.; Majano, P.L. Clathrin mediates infectious hepatitis c virus particle egress. J. Virol. 2015, 89, 4180–4190. [Google Scholar] [CrossRef] [PubMed]
- Corless, L.; Crump, C.M.; Griffin, S.D.; Harris, M. Vps4 and the escrt-iii complex are required for the release of infectious hepatitis c virus particles. J. Gen. Virol. 2010, 91, 362–372. [Google Scholar] [CrossRef]
- Elgner, F.; Ren, H.; Medvedev, R.; Ploen, D.; Himmelsbach, K.; Boller, K.; Hildt, E. The intracellular cholesterol transport inhibitor u18666a inhibits the exosome-dependent release of mature hepatitis c virus. J. Virol. 2016, 90, 11181–11196. [Google Scholar] [CrossRef] [PubMed]
- Barouch-Bentov, R.; Neveu, G.; Xiao, F.; Beer, M.; Bekerman, E.; Schor, S.; Campbell, J.; Boonyaratanakornkit, J.; Lindenbach, B.; Lu, A.; et al. Hepatitis c virus proteins interact with the endosomal sorting complex required for transport (escrt) machinery via ubiquitination to facilitate viral envelopment. mBio 2016, 7, e01456-16. [Google Scholar] [CrossRef]
- Takacs, C.N.; Andreo, U.; Dao Thi, V.L.; Wu, X.; Gleason, C.E.; Itano, M.S.; Spitz-Becker, G.S.; Belote, R.L.; Hedin, B.R.; Scull, M.A.; et al. Differential regulation of lipoprotein and hepatitis c virus secretion by rab1b. Cell Rep. 2017, 21, 431–441. [Google Scholar] [CrossRef]
- Mankouri, J.; Walter, C.; Stewart, H.; Bentham, M.; Park, W.S.; Heo, W.D.; Fukuda, M.; Griffin, S.; Harris, M. Release of infectious hepatitis c virus from huh7 cells occurs via a trans-golgi network-to-endosome pathway independent of very-low-density lipoprotein secretion. J. Virol. 2016, 90, 7159–7170. [Google Scholar] [CrossRef]
- Lee, J.Y.; Acosta, E.G.; Stoeck, I.K.; Long, G.; Hiet, M.S.; Mueller, B.; Fackler, O.T.; Kallis, S.; Bartenschlager, R. Apolipoprotein e likely contributes to a maturation step of infectious hepatitis c virus particles and interacts with viral envelope glycoproteins. J. Virol. 2014, 88, 12422–12437. [Google Scholar] [CrossRef]
- Hishiki, T.; Shimizu, Y.; Tobita, R.; Sugiyama, K.; Ogawa, K.; Funami, K.; Ohsaki, Y.; Fujimoto, T.; Takaku, H.; Wakita, T.; et al. Infectivity of hepatitis c virus is influenced by association with apolipoprotein e isoforms. J. Virol. 2010, 84, 12048–12057. [Google Scholar] [CrossRef] [PubMed]
- Boyer, A.; Dumans, A.; Beaumont, E.; Etienne, L.; Roingeard, P.; Meunier, J.C. The association of hepatitis c virus glycoproteins with apolipoproteins e and b early in assembly is conserved in lipoviral particles. J. Biol. Chem. 2014, 289, 18904–18913. [Google Scholar] [CrossRef] [PubMed]
- Flamand, M.; Megret, F.; Mathieu, M.; Lepault, J.; Rey, F.A.; Deubel, V. Dengue virus type 1 nonstructural glycoprotein ns1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J. Virol. 1999, 73, 6104–6110. [Google Scholar] [PubMed]
- Branza-Nichita, N.; Lazar, C.; Dwek, R.A.; Zitzmann, N. Role of n-glycan trimming in the folding and secretion of the pestivirus protein e(rns). Biochem. Biophys. Res. Commun. 2004, 319, 655–662. [Google Scholar] [CrossRef]
- Icard, V.; Diaz, O.; Scholtes, C.; Perrin-Cocon, L.; Ramiere, C.; Bartenschlager, R.; Penin, F.; Lotteau, V.; Andre, P. Secretion of hepatitis c virus envelope glycoproteins depends on assembly of apolipoprotein b positive lipoproteins. PLoS ONE 2009, 4, e4233. [Google Scholar] [CrossRef] [PubMed]
- Welsch, S.; Muller, B.; Krausslich, H.G. More than one door—Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007, 581, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Drin, G.; Antonny, B. Amphipathic helices and membrane curvature. FEBS Lett. 2010, 584, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Wriggers, W.; Li, Z.; Jonas, A.; Schulten, K. Predicting the structure of apolipoprotein a-i in reconstituted high-density lipoprotein disks. Biophys. J. 1997, 73, 2337–2346. [Google Scholar] [CrossRef]
- Gimenez-Andres, M.; Copic, A.; Antonny, B. The many faces of amphipathic helices. Biomolecules 2018, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.; Zitzmann, N.; Dwek, R.A.; Branza-Nichita, N. The pestivirus e(rns) glycoprotein interacts with e2 in both infected cells and mature virions. Virology 2003, 314, 696–705. [Google Scholar] [CrossRef]
- Shelness, G.S.; Thornburg, J.T. Role of intramolecular disulfide bond formation in the assembly and secretion of apolipoprotein b-100-containing lipoproteins. J. Lipid Res. 1996, 37, 408–419. [Google Scholar]
- Elliott, D.A.; Halliday, G.M.; Garner, B. Apolipoprotein-e forms dimers in human frontal cortex and hippocampus. BMC Neurosci. 2010, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Rall, S.C., Jr.; Weisgraber, K.H.; Innerarity, T.L.; Mahley, R.W. Structural basis for receptor binding heterogeneity of apolipoprotein e from type iii hyperlipoproteinemic subjects. Proc. Natl. Acad. Sci. USA 1982, 79, 4696–4700. [Google Scholar] [CrossRef]
- Weller, R.; Hueging, K.; Brown, R.J.P.; Todt, D.; Joecks, S.; Vondran, F.W.R.; Pietschmann, T. Hepatitis c virus strain-dependent usage of apolipoprotein e modulates assembly efficiency and specific infectivity of secreted virions. J. Virol. 2017, 91, e00422-17. [Google Scholar] [CrossRef] [PubMed]
- Long, G.; Hiet, M.S.; Windisch, M.P.; Lee, J.Y.; Lohmann, V.; Bartenschlager, R. Mouse hepatic cells support assembly of infectious hepatitis c virus particles. Gastroenterology 2011, 141, 1057–1066. [Google Scholar] [CrossRef]
- Cun, W.; Jiang, J.; Luo, G. The c-terminal alpha-helix domain of apolipoprotein e is required for interaction with nonstructural protein 5a and assembly of hepatitis c virus. J. Virol. 2010, 84, 11532–11541. [Google Scholar] [CrossRef] [PubMed]
- Tews, B.A.; Schurmann, E.M.; Meyers, G. Mutation of cysteine 171 of pestivirus e rns rnase prevents homodimer formation and leads to attenuation of classical swine fever virus. J. Virol. 2009, 83, 4823–4834. [Google Scholar] [CrossRef]
- Akazawa, D.; Moriyama, M.; Yokokawa, H.; Omi, N.; Watanabe, N.; Date, T.; Morikawa, K.; Aizaki, H.; Ishii, K.; Kato, T.; et al. Neutralizing antibodies induced by cell culture-derived hepatitis c virus protect against infection in mice. Gastroenterology 2013, 145, 447–455. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vieyres, G.; Pietschmann, T. HCV Pit Stop at the Lipid Droplet: Refuel Lipids and Put on a Lipoprotein Coat before Exit. Cells 2019, 8, 233. https://doi.org/10.3390/cells8030233
Vieyres G, Pietschmann T. HCV Pit Stop at the Lipid Droplet: Refuel Lipids and Put on a Lipoprotein Coat before Exit. Cells. 2019; 8(3):233. https://doi.org/10.3390/cells8030233
Chicago/Turabian StyleVieyres, Gabrielle, and Thomas Pietschmann. 2019. "HCV Pit Stop at the Lipid Droplet: Refuel Lipids and Put on a Lipoprotein Coat before Exit" Cells 8, no. 3: 233. https://doi.org/10.3390/cells8030233




