Increasing the Activity of Sugarcane Sucrose Phosphate Synthase Enhanced Growth and Grain Yields in Transgenic Indica Rice
Abstract
:1. Introduction
2. Materials and Methods
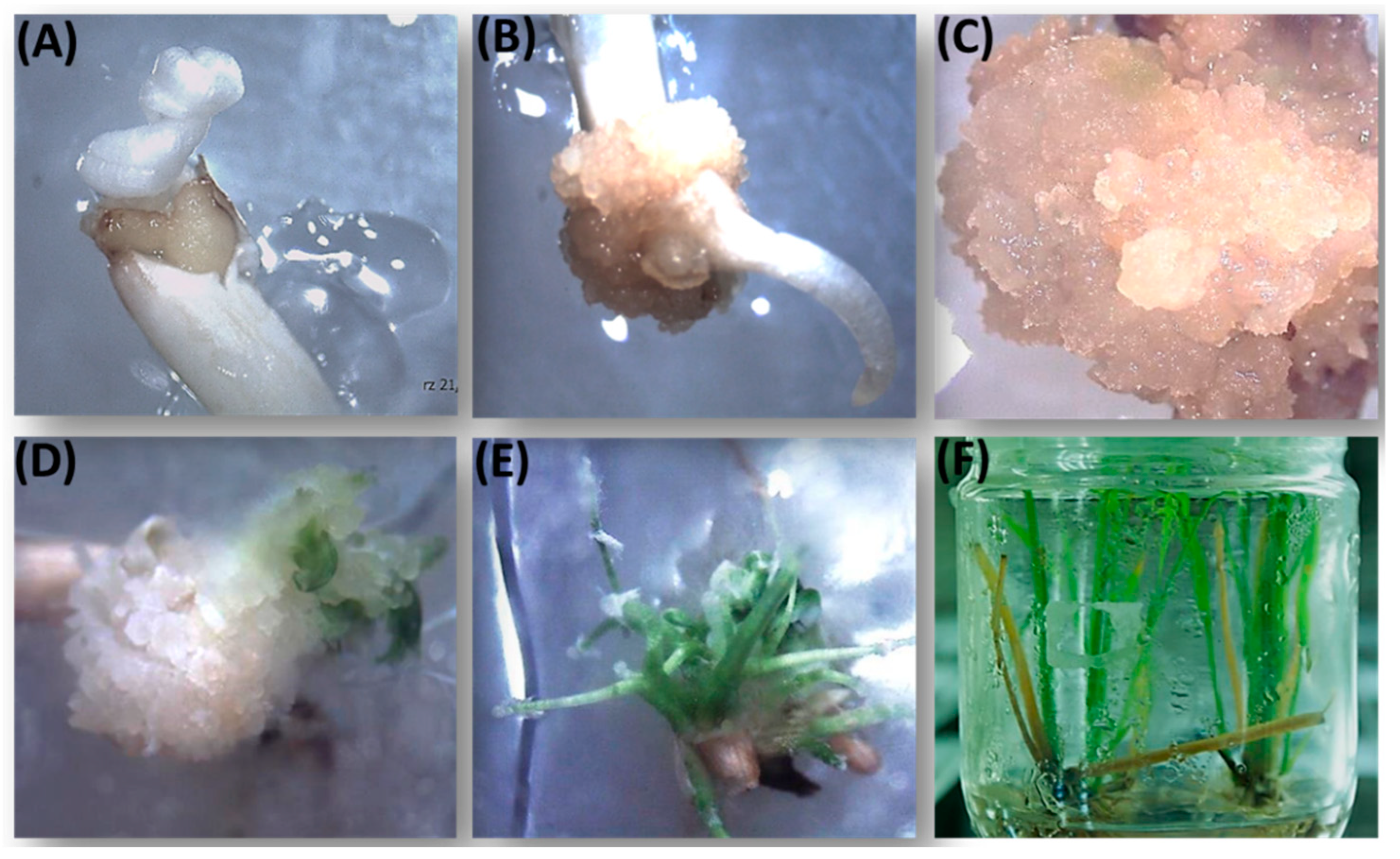
2.1. Plant Material and Agrobacterium-Mediated Transformation
2.2. Genomic PCR Analysis
2.3. Transcript Levels of Sucrose Phosphate Synthase Gene
2.4. Protein Extraction, Enzyme Assay, and Immunoblotting
2.5. Sucrose Analysis in Rice Leaves
2.6. Analysis of Amylose Content in Rice Grain
2.7. Plant Growth and Productivity
2.8. Statistical Analysis
3. Results
3.1. Rice Transformation and Molecular Analysis of the Transformants
3.2. The RNA Transcript and Protein Expression of Sucrose-Phosphate Synthase
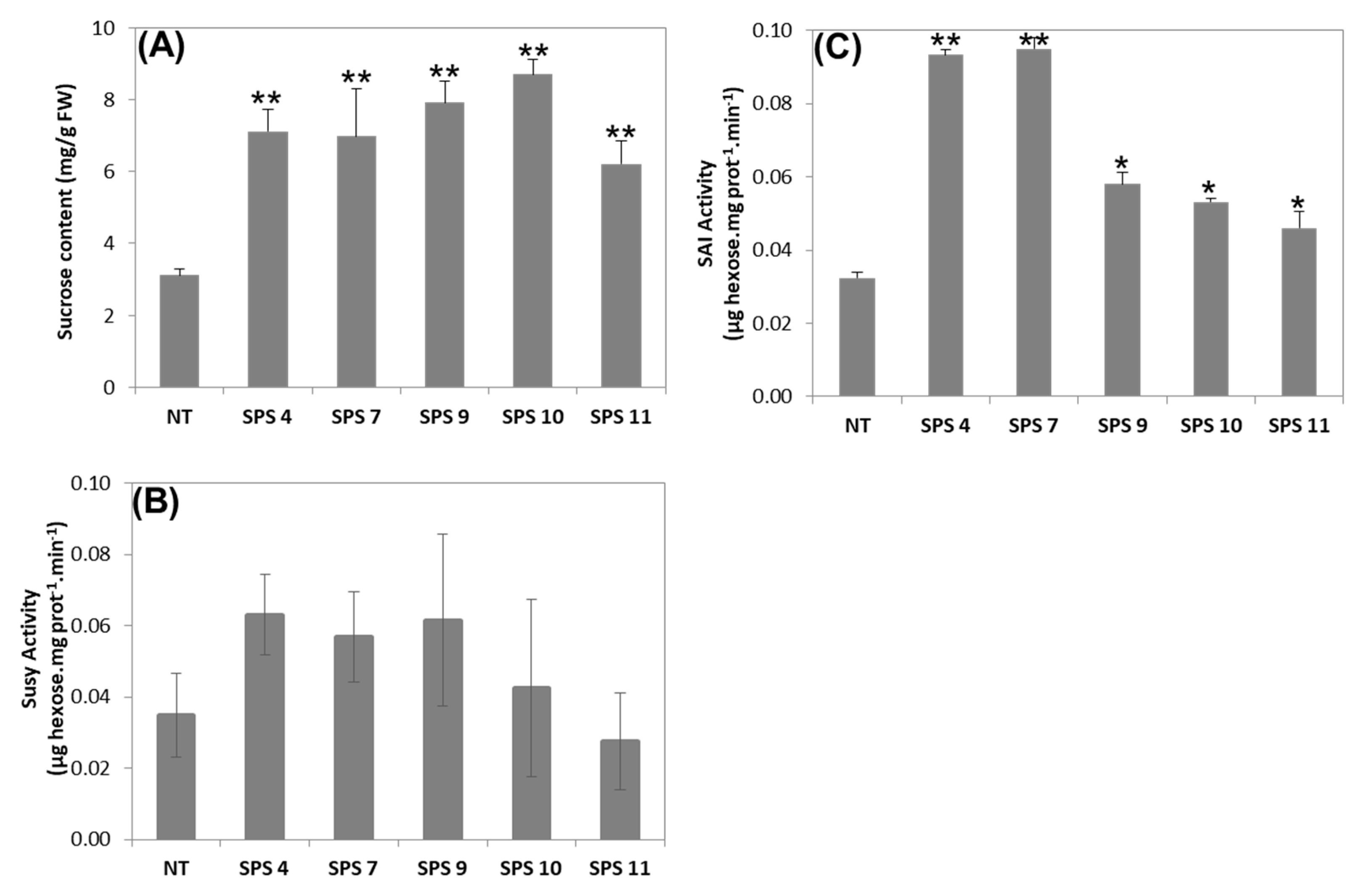
3.3. Sucrose Content and Activity of Sucrose Degrading Enzymes
3.4. Plant Growth and Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohanty, A.; Kathuria, H.; Ferjani, A.; Sakamoto, A.; Mohanty, P.; Murata, N.; Tyagi, A. Transgenics of an Elite Indica Rice Variety Pusa Basmati 1 Harbouring the CodA Gene Are Highly Tolerant to Salt Stress. Theor. Appl. Genet. 2002, 106, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Nagadhara, D.; Ramesh, S.; Pasalu, I.C.; Rao, Y.K.; Krishnaiah, N.V.; Sarma, N.P.; Bown, D.P.; Gatehouse, J.A.; Reddy, V.D.; Rao, K.V. Transgenic Indica Rice Resistant to Sap-Sucking Insects: Transgenic Rice Resistant to Sap-Sucking Insects. Plant Biotechnol. J. 2003, 1, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Ishimaru, K.; Aoki, N.; Takahashi, S.; Ozawa, K.; Ohkawa, Y.; Ohsugi, R. Characterization of a Maize Sucrose–Phosphate Synthase Protein and Its Effect on Carbon Partitioning in Transgenic Rice Plants. Plant Prod. Sci. 1999, 2, 172–177. [Google Scholar] [CrossRef]
- Okamura, M.; Hirose, T.; Hashida, Y.; Ohsugi, R.; Aoki, N. Suppression of Starch Accumulation in ‘Sugar Leaves’ of Rice Affects Plant Productivity under Field Conditions. Plant Prod. Sci. 2017, 20, 102–110. [Google Scholar] [CrossRef] [Green Version]
- Gesch, R.W.; Vu, J.C.V.; Boote, K.J.; Hartwell Allen, L.; Bowes, G. Sucrose-phosphate Synthase Activity in Mature Rice Leaves Following Changes in Growth CO 2 Is Unrelated to Sucrose Pool Size. New Phytol. 2002, 154, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Lu, Q.; Wen, X.; Lu, C. Enhanced Sucrose Loading Improves Rice Yield by Increasing Grain Size. Plant Physiol. 2015, 169, 01170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutschera, U.; Heiderich, A. Sucrose Metabolism and Cellulose Biosynthesis in Sunflower Hypocotyls. Physiol. Plant. 2002, 114, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Maloney, V.J.; Park, J.-Y.; Unda, F.; Mansfield, S.D. Sucrose Phosphate Synthase and Sucrose Phosphate Phosphatase Interact in Planta and Promote Plant Growth and Biomass Accumulation. J. Exp. Bot. 2015, 66, 4383–4394. [Google Scholar] [CrossRef] [Green Version]
- Sturm, A. Invertases. Primary Structures, Functions, and Roles in Plant Development and Sucrose Partitioning. Plant Physiol. 1999, 121, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Huber, S.C.; Huber, J.L. Role And Regulation of Sucrose-Phosphate Synthase in Higher Plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1996, 47, 431–444. [Google Scholar] [CrossRef]
- Salerno, G. Origin of Sucrose Metabolism in Higher Plants: When, How and Why? Trends Plant Sci. 2003, 8, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Koch, K. Sucrose Metabolism: Regulatory Mechanisms and Pivotal Roles in Sugar Sensing and Plant Development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Canam, T.; Kang, K.-Y.; Ellis, D.D.; Mansfield, S.D. Over-Expression of an Arabidopsis Family A Sucrose Phosphate Synthase (SPS) Gene Alters Plant Growth and Fibre Development. Transgenic Res. 2008, 17, 181–192. [Google Scholar] [CrossRef]
- Hashida, Y.; Hirose, T.; Okamura, M.; Hibara, K.; Ohsugi, R.; Aoki, N. A Reduction of Sucrose Phosphate Synthase (SPS) Activity Affects Sucrose/Starch Ratio in Leaves but Does Not Inhibit Normal Plant Growth in Rice. Plant Sci. 2016, 253, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Worrell, A.C.; Bruneau, J.M.; Summerfelt, K.; Boersig, M.; Voelker, T.A. Expression of a Maize Sucrose Phosphate Synthase in Tomato Alters Leaf Carbohydrate Partitioning. Plant Cell 1991, 3, 1121–1130. [Google Scholar] [CrossRef] [Green Version]
- Galtier, N.; Foyer, C.H.; Huber, J.; Voelker, T.A.; Huber, S.C. Effects of Elevated Sucrose-Phosphate Synthase Activity on Photosynthesis, Assimilate Partitioning, and Growth in Tomato (Lycopersicon Esculentum Var UC82B). Plant Physiol. 1993, 101, 535–543. [Google Scholar] [CrossRef] [Green Version]
- Signora, L.; Galtier, N.; Skot, L.; Lucas, H.; Foyer, C.H. Over-Expression of Sucrose Phosphate Synthase in Arabidopsis thaliana Results in Increased Foliar Sucrose/Starch Ratios and Favours Decreased Foliar Carbohydrate Accumulation in Plants after Prolonged Growth with CO2 Enrichment. J. Exp. Bot. 1998, 49, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Haigler, C.H.; Singh, B.; Zhang, D.; Hwang, S.; Wu, C.; Cai, W.X.; Hozain, M.; Kang, W.; Kiedaisch, B.; Strauss, R.E.; et al. Transgenic Cotton Over-Producing Spinach Sucrose Phosphate Synthase Showed Enhanced Leaf Sucrose Synthesis and Improved Fiber Quality under Controlled Environmental Conditions. Plant Mol. Biol. 2007, 63, 815–832. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirotsu, N.; Kashiwagi, T.; Madoka, Y.; Nagasuga, K.; Ono, K.; Ohsugi, R. Overexpression of a Maize SPS Gene Improves Yield Characters of Potato under Field Conditions. Plant Prod. Sci. 2008, 11, 104–107. [Google Scholar] [CrossRef]
- Sugiharto, B.; Sakakibara, H.; Sumadi; Sugiyama, T. Differential Expression of Two Genes for Sucrose-Phosphate Synthase in Sugarcane: Molecular Cloning of the CDNAs and Comparative Analysis of Gene Expression. Plant Cell Physiol. 1997, 38, 961–965. [Google Scholar] [CrossRef]
- Sawitri, W.D.; Narita, H.; Ishizaka-Ikeda, E.; Sugiharto, B.; Hase, T.; Nakagawa, A. Purification and Characterization of Recombinant Sugarcane Sucrose Phosphate Synthase Expressed in E. coli and Insect Sf9 Cells: An Importance of the N-Terminal Domain for an Allosteric Regulatory Property. J. Biochem. 2016, 159, 599–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anur, R.M.; Mufithah, N.; Sawitri, W.D.; Sakakibara, H.; Sugiharto, B. Overexpression of Sucrose Phosphate Synthase Enhanced Sucrose Content and Biomass Production in Transgenic Sugarcane. Plants 2020, 9, 200. [Google Scholar] [CrossRef] [Green Version]
- Suherman; Wijayanto, S.I.; Anur, R.M.; Neliana, I.R.; Dewanti, P.; Sugiharto, B. Field Evaluation on Growth and Productivity of the Transgenic Sugarcane Lines Overexpressing Sucrose-Phosphate Synthase. Sugar Tech 2022, 24, 1689–1698. [Google Scholar] [CrossRef]
- Azizah, K.A.; Restanto, D.P.; Sugiharto, B. Enhancement of Regeneration Efficiency through Callus Induction Media Using 2,4-Dicholorophenoxyacetic Acid in Indica Rice (Oryza sativa L. Var. Ciherang). J. Ilmu Dasar 2017, 18, 91–98. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Jiang, J.; Oard, J.H. Structure, Expression and Promoter Activity of Two Polyubiquitin Genes from Rice (Oryza sativa L.). Plant Sci. 2000, 156, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.-F.; Li, H.; Qin, R.-Y.; Li, J.; Qiu, C.-H.; Yang, Y.-C.; Ma, H.; Li, L.; Wei, P.-C.; Yang, J.-B. Generation of Inheritable and “Transgene Clean” Targeted Genome-Modified Rice in Later Generations Using the CRISPR/Cas9 System. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apriasti, R.; Widyaningrum, S.; Hidayati, W.N.; Sawitri, W.D.; Darsono, N.; Hase, T.; Sugiharto, B. Full Sequence of the Coat Protein Gene Is Required for the Induction of Pathogen-Derived Resistance against Sugarcane Mosaic Virus in Transgenic Sugarcane. Mol. Biol. Rep. 2018, 45, 2749–2758. [Google Scholar] [CrossRef]
- Buttery, R.G.; Ling, L.C.; Juliano, B.O.; Turnbaugh, J.G. Cooked Rice Aroma and 2-Acetyl-1-Pyrroline. J. Agric. Food Chem. 1983, 31, 823–826. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. An Overview of Sucrose Synthases in Plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Park, I.M.; Ibáñez, A.M.; Shoemaker, C.F. Rice Starch Molecular Size and Its Relationship with Amylose Content. Starch—Stärke 2007, 59, 69–77. [Google Scholar] [CrossRef]
- Falter, C.; Voigt, C.A. Improving Biomass Production and Saccharification in Brachypodium Distachyon through Overexpression of a Sucrose-Phosphate Synthase from Sugarcane. J. Plant Biochem. Biotechnol. 2016, 25, 311–318. [Google Scholar] [CrossRef] [Green Version]
- Barratt, D.H.P.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal Growth of Arabidopsis Requires Cytosolic Invertase but Not Sucrose Synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar Sensing and Signaling In Plants: Conserved and Novel Mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and Signaling Molecule. Phytochemistry 2010, 71, 1610–1614. [Google Scholar] [CrossRef]
- Eveland, A.L.; Jackson, D.P. Sugars, Signalling, and Plant Development. J. Exp. Bot. 2012, 63, 3367–3377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smeekens, S.; Hellmann, H.A. Sugar Sensing and Signaling in Plants. Front. Plant Sci. 2014, 5, S185–S205. [Google Scholar] [CrossRef] [Green Version]
- Gibson, S.I. Control of Plant Development and Gene Expression by Sugar Signaling. Curr. Opin. Plant Biol. 2005, 8, 93–102. [Google Scholar] [CrossRef]
- Geng, M.-T.; Yao, Y.; Wang, Y.-L.; Wu, X.-H.; Sun, C.; Li, R.-M.; Fu, S.-P.; Duan, R.-J.; Liu, J.; Hu, X.-W.; et al. Structure, Expression, and Functional Analysis of the Hexokinase Gene Family in Cassava. Int. J. Mol. Sci. 2017, 18, 1041. [Google Scholar] [CrossRef] [Green Version]
- Lastdrager, J.; Hanson, J.; Smeekens, S. Sugar Signals and the Control of Plant Growth and Development. J. Exp. Bot. 2014, 65, 799–807. [Google Scholar] [CrossRef]
- Aguilera-Alvarado, G.P.; Sánchez-Nieto, S. Plant Hexokinases Are Multifaceted Proteins. Plant Cell Physiol. 2017, 58, 1151–1160. [Google Scholar] [CrossRef]
- Cho, Y.-H.; Yoo, S.-D.; Sheen, J. Regulatory Functions of Nuclear Hexokinase1 Complex in Glucose Signaling. Cell 2006, 127, 579–589. [Google Scholar] [CrossRef] [PubMed]


| Lines | Tiller Number | Plant Height (cm) | ||||
|---|---|---|---|---|---|---|
| 1 MAP | 2 MAP | 3 MAP | 1 MAP | 2 MAP | 3 MAP | |
| NT | 8.67 ± 0.57 | 14.67 ± 1.52 | 25.67 ± 2.08 | 14.43 ± 1.01 | 35.61 ± 1.65 | 67.00 ± 2.00 |
| SPS 4 | 11.33 ± 1.52 * | 23.33 ± 2.51 ** | 37.01 ± 2.64 ** | 19.97 ± 1.94 * | 45.33 ± 3.75 ** | 81.67 ± 3.79 ** |
| SPS 7 | 11.67 ± 1.15 * | 21.00 ± 2.00 ** | 39.33 ± 3.05 ** | 19.87 ± 2.10 * | 43.67 ± 1.15 ** | 86.67 ± 3.21 ** |
| SPS 9 | 12.00 ± 2.64 * | 24.67 ± 1.52 ** | 34.33 ± 3.05 ** | 17.23 ± 2.25 | 43.00 ± 0.87 ** | 79.33 ± 0.58 ** |
| SPS 10 | 10.67 ± 1.15 * | 21.67 ± 0.57 ** | 37.33 ± 3.05 ** | 17.27 ± 1.74 | 43.80 ± 1.45 ** | 81.00 ± 3.46 ** |
| SPS 11 | 9.67 ± 1.15 | 18.00 ± 2.00 | 35.33 ± 3.51 ** | 15.83 ± 1.26 | 38.33 ± 0.76* | 77.00 ± 1.00 ** |
| Lines | Amylose (%) | Grain Number Per Panicle | 1000-Grain Weight (g) | Dry Weight (g) |
|---|---|---|---|---|
| NT | 43.48 ± 2.78 | 94.67 ± 11.06 | 22.69 ± 0.51 | 56.41 ± 4.51 |
| SPS 4 | 57.08 ± 5.36 ** | 152.67 ± 15.53 ** | 24.89 ± 0.38 * | 76.78 ± 8.41 * |
| SPS 7 | 51.33 ± 2.67 ** | 169.67 ± 30.17 ** | 25.36 ± 0.91 * | 81.58 ± 11.35 * |
| SPS 9 | 49.37 ± 0.80 * | 152.33 ± 18.01 ** | 24.35 ± 0.15 * | 73.31 ± 15.56 |
| SPS 10 | 54.41 ± 1.61 ** | 133.67 ± 20.13 * | 24.39 ± 0.37 * | 70.45 ± 17.79 |
| SPS 11 | 49.84 ± 1.06 * | 138.33 ± 8.96 * | 24.95 ± 0.16 * | 66.69 ± 16.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulyatama, R.A.; Neliana, I.R.; Sawitri, W.D.; Sakakibara, H.; Kim, K.-M.; Sugiharto, B. Increasing the Activity of Sugarcane Sucrose Phosphate Synthase Enhanced Growth and Grain Yields in Transgenic Indica Rice. Agronomy 2022, 12, 2949. https://doi.org/10.3390/agronomy12122949
Mulyatama RA, Neliana IR, Sawitri WD, Sakakibara H, Kim K-M, Sugiharto B. Increasing the Activity of Sugarcane Sucrose Phosphate Synthase Enhanced Growth and Grain Yields in Transgenic Indica Rice. Agronomy. 2022; 12(12):2949. https://doi.org/10.3390/agronomy12122949
Chicago/Turabian StyleMulyatama, Reza Anugrah, Intan Ria Neliana, Widhi Dyah Sawitri, Hitoshi Sakakibara, Kyung-Min Kim, and Bambang Sugiharto. 2022. "Increasing the Activity of Sugarcane Sucrose Phosphate Synthase Enhanced Growth and Grain Yields in Transgenic Indica Rice" Agronomy 12, no. 12: 2949. https://doi.org/10.3390/agronomy12122949







