Recombinant Production of Arginyl Dipeptides by l-Amino Acid Ligase RizA Coupled with ATP Regeneration
Abstract
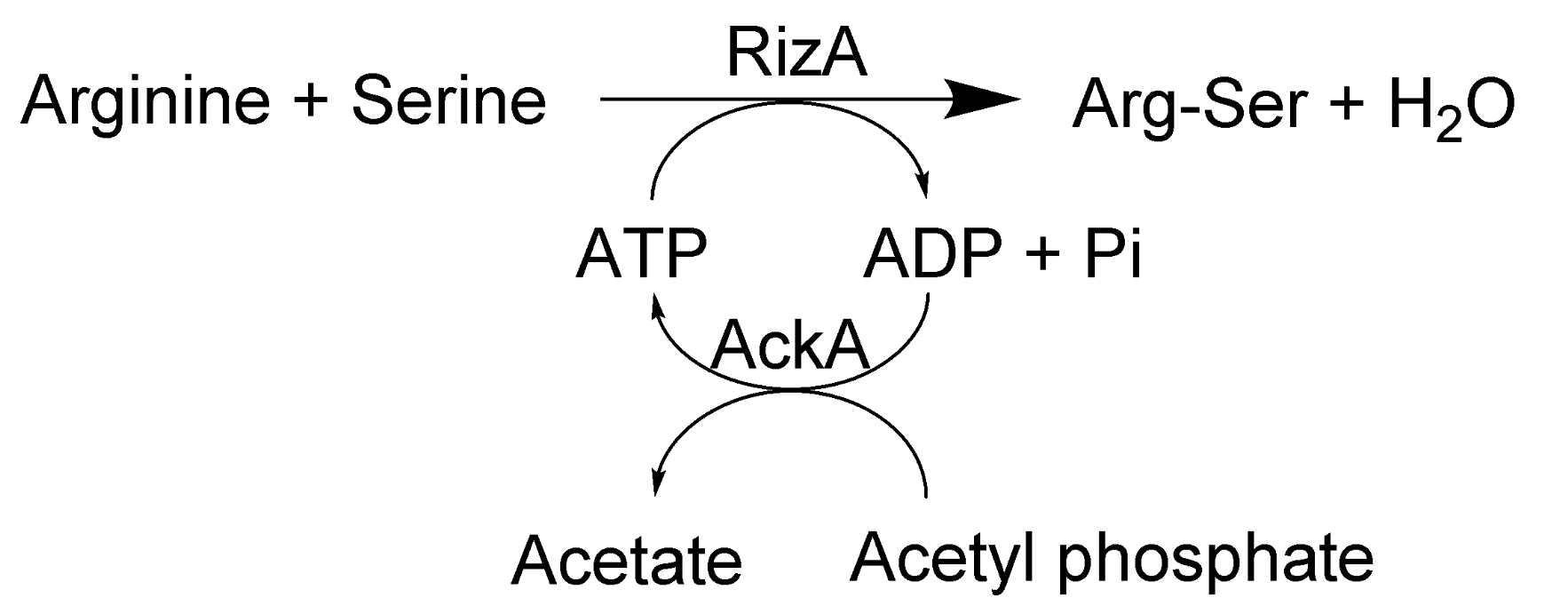
:1. Introduction
2. Results and Discussion
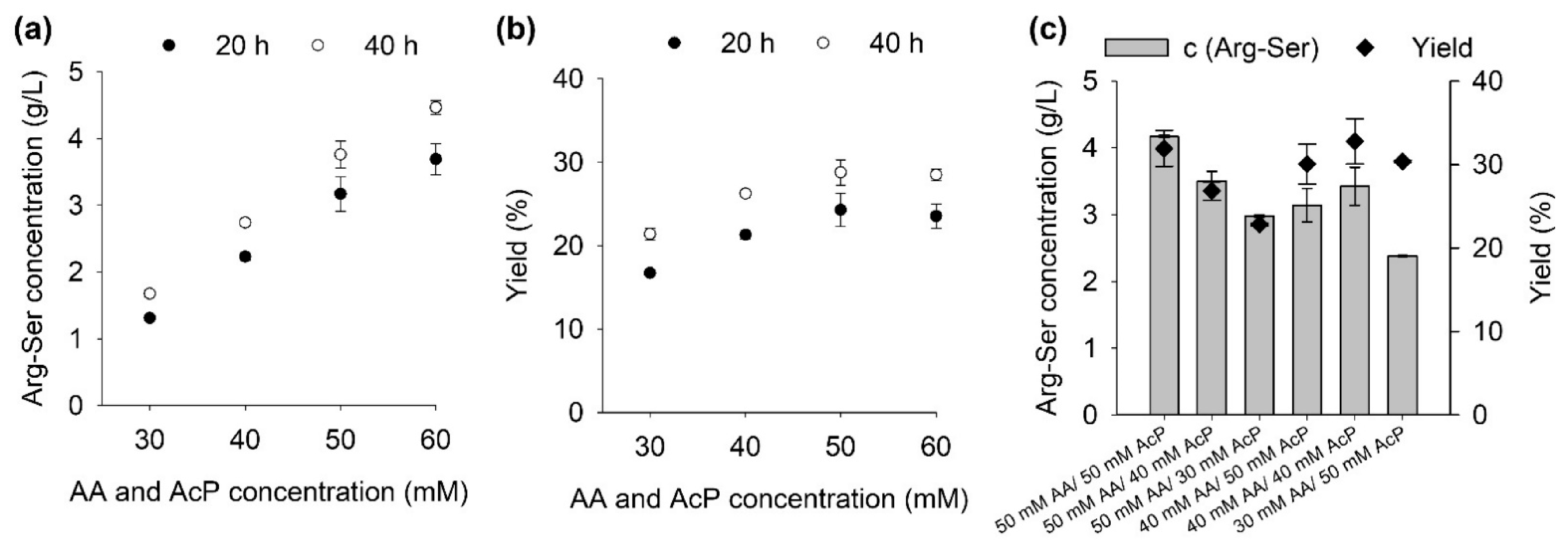
2.1. Influence of Substrate Concentrations
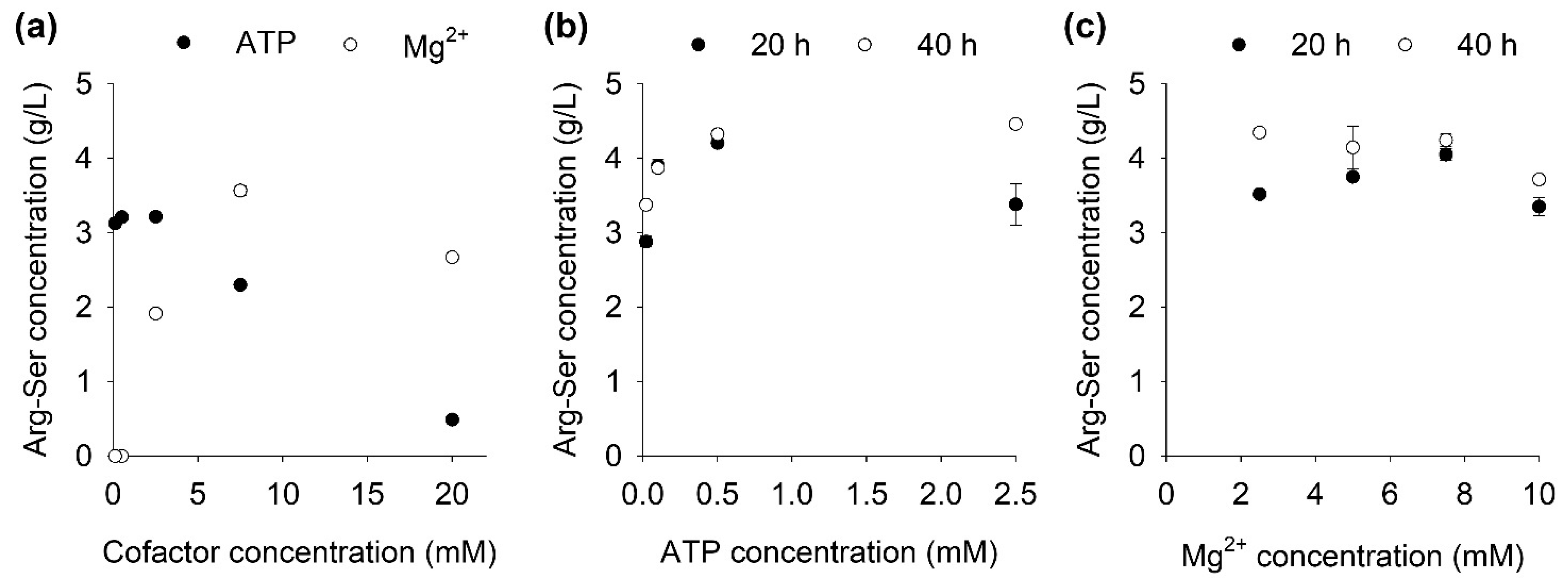
2.2. Influence of Cofactor Concentrations
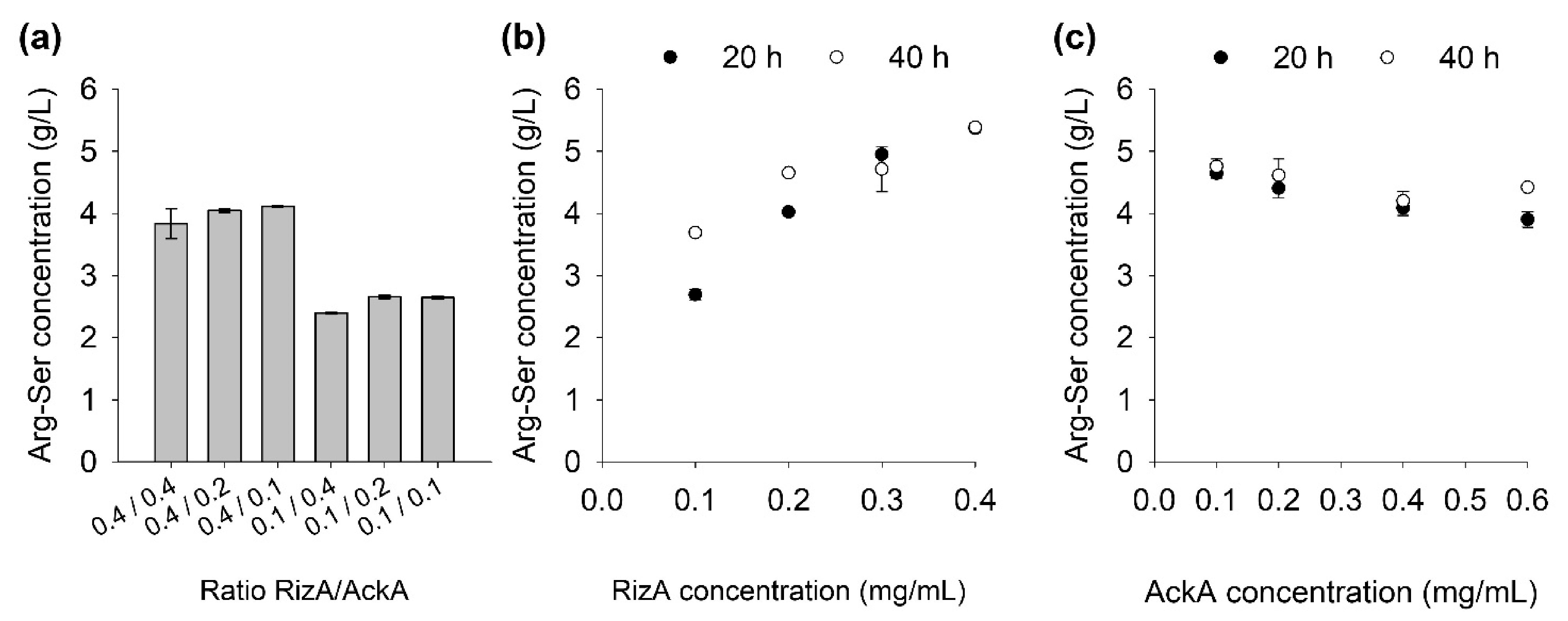
2.3. Influence of Enzyme Concentrations
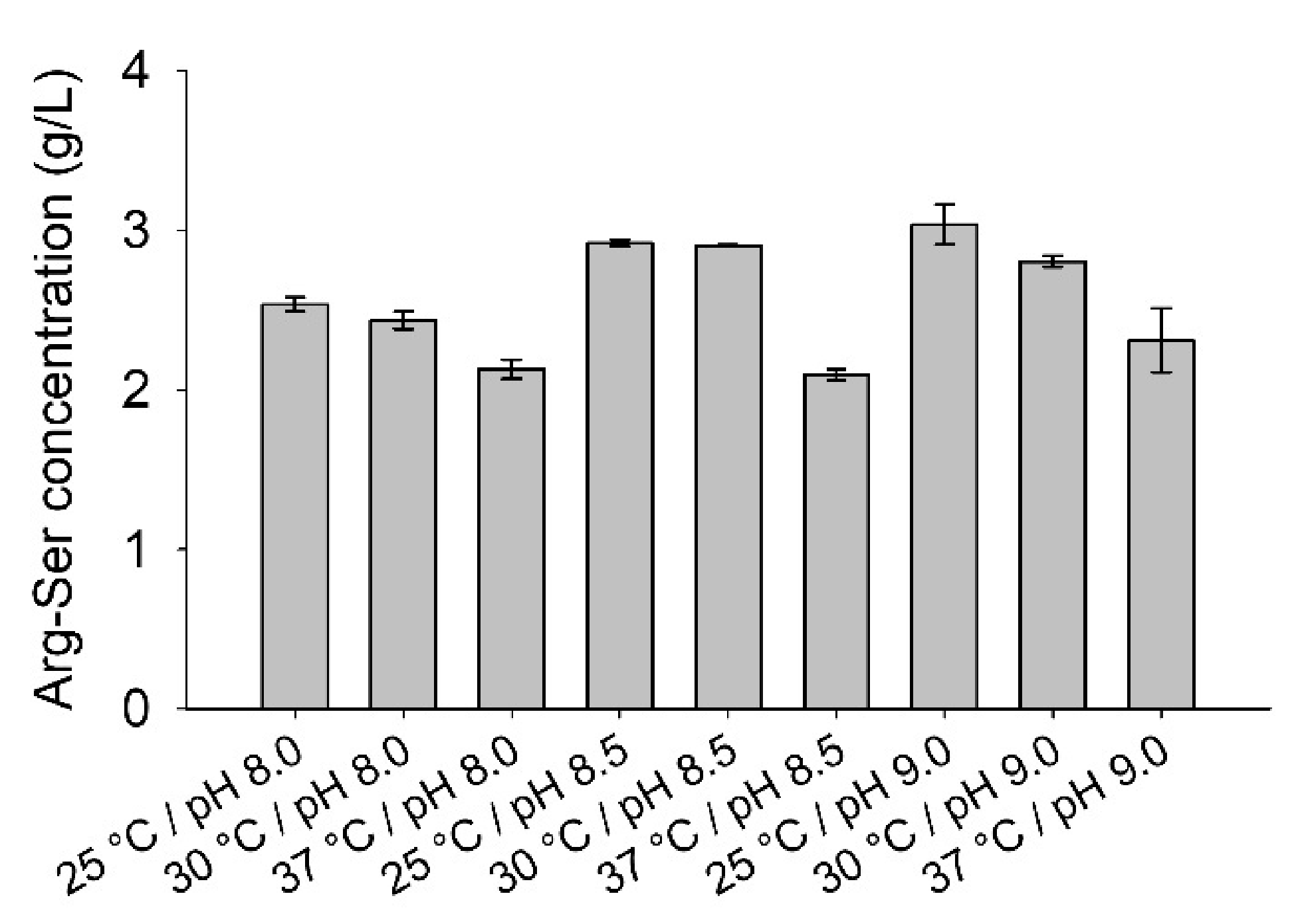
2.4. Influence of Temperature and pH
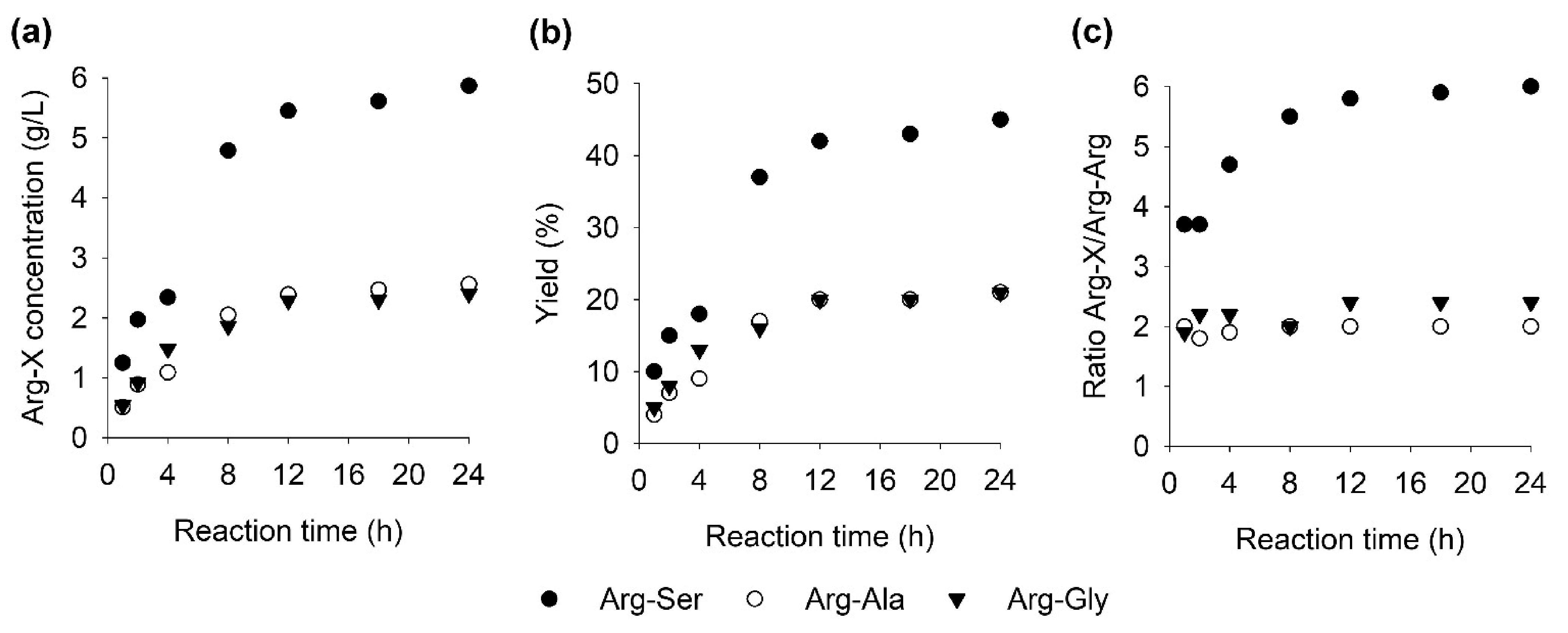
2.5. Time Course of Formation of Arg-Ser, Arg-Ala, Arg-Gly, and Arg-Arg
3. Materials and Methods
3.1. Chemicals, Reagents and Strains
3.2. Construction of pET28a_his6-rizA and pET28a_his6-ackA Constructs
3.3. Cultivation and Expression
3.4. Purification
3.5. Biocatalysis
3.6. Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- He, F.J.; MacGregor, G.A. A comprehensive review on salt and health and current experience of worldwide salt reduction programmes. J. Hum. Hypertens. 2009, 23, 363–384. [Google Scholar] [CrossRef] [PubMed]
- Trieu, K.; Neal, B.; Hawkes, C.; Dunford, E.; Campbell, N.; Rodriguez-Fernandez, R.; Legetic, B.; McLaren, L.; Barberio, A.; Webster, J. Salt Reduction Initiatives around the World–A Systematic Review of Progress towards the Global Target. PLoS ONE 2015, 10, e0130247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strazzullo, P.; D’Elia, L.; Kandala, N.-B.; Cappuccio, F.P. Salt intake, stroke, and cardiovascular disease: Meta-analysis of prospective studies. BMJ 2009, 339, b4567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO Guideline: Sodium Intake for Adults and Children. Available online: https://www.who.int/publications/i/item/9789241504836 (accessed on 29 September 2021).
- He, F.J.; MacGregor, G.A. Effect of modest salt reduction on blood pressure: A meta-analysis of randomized trials. Implications for public health. J. Hum. Hypertens. 2002, 16, 761–770. [Google Scholar] [CrossRef] [Green Version]
- Desmond, E. Reducing salt: A challenge for the meat industry. Meat Sci. 2006, 74, 188–196. [Google Scholar] [CrossRef]
- Breslin, P.A.S.; Beauchamp, G.K. Salt enhances flavour by suppressing bitterness. Nature 1997, 387, 563. [Google Scholar] [CrossRef]
- Yagasaki, M.; Hashimoto, S.-I. Synthesis and application of dipeptides; current status and perspectives. Appl. Microbiol. Biotechnol. 2008, 81, 13–22. [Google Scholar] [CrossRef]
- Harth, L.; Krah, U.; Linke, D.; Dunkel, A.; Hofmann, T.; Berger, R.G. Salt Taste Enhancing L-Arginyl Dipeptides from Casein and Lysozyme Released by Peptidases of Basidiomycota. J. Agric. Food Chem. 2018, 66, 2344–2353. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Y.-F.; Ning, L.-X.; Wang, Y.-F.; Liu, X.-H.; Li, R.; Chen, X.-E. L-amino acid ligase: A promising alternative for the biosynthesis of L-dipeptides. Enzyme Microb. Technol. 2020, 136, 109537. [Google Scholar] [CrossRef]
- Kino, K.; Kotanaka, Y.; Arai, T.; Yagasaki, M. A Novel L-Amino Acid Ligase from Bacillus subtilis NBRC3134, a Microorganism Producing Peptide-Antibiotic Rhizocticin. Biosci. Biotechnol. Biochem. 2009, 73, 901–907. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Yin, Z. Highly Efficient Synthesis of Glutathione via a Genetic Engineering Enzymatic Method Coupled with Yeast ATP Generation. Catalysts 2020, 10, 33. [Google Scholar] [CrossRef] [Green Version]
- Alissandratos, A.; Caron, K.; Loan, T.D.; Hennessy, J.E.; Easton, C.J. ATP Recycling with Cell Lysate for Enzyme-Catalyzed Chemical Synthesis, Protein Expression and PCR. ACS Chem. Biol. 2016, 11, 3289–3293. [Google Scholar] [CrossRef]
- Andexer, J.N.; Richter, M. Emerging Enzymes for ATP Regeneration in Biocatalytic Processes. ChemBioChem 2015, 16, 380–386. [Google Scholar] [CrossRef]
- Li, Z.; Ning, X.; Zhao, Y.; Zhang, X.; Xiao, C.; Li, Z. Efficient One-Pot Synthesis of Cytidine 5′-Monophosphate Using an Extremophilic Enzyme Cascade System. J. Agric. Food Chem. 2020, 68, 9188–9194. [Google Scholar] [CrossRef]
- Yan, B.; Ding, Q.; Ou, L.; Zou, Z. Production of glucose-6-phosphate by glucokinase coupled with an ATP regeneration system. World J. Microbiol. Biotechnol. 2014, 30, 1123–1128. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Y.-H.P.J. Enzymatic regeneration and conservation of ATP: Challenges and opportunities. Crit. Rev. Biotechnol. 2021, 41, 16–33. [Google Scholar] [CrossRef]
- Crans, D.C.; Whitesides, G.M. A convenient synthesis of disodium acetyl phosphate for use in in situ ATP cofactor regeneration. J. Org. Chem. 1983, 48, 3130–3132. [Google Scholar] [CrossRef]
- Kim, D.-M.; Swartz, J.R. Prolonging cell-free protein synthesis with a novel ATP regeneration system. Biotechnol. Bioeng. 1999, 66, 180–188. [Google Scholar] [CrossRef]
- Huffman, M.A.; Fryszkowska, A.; Alvizo, O.; Borra-Garske, M.; Campos, K.R.; Canada, K.A.; Devine, P.N.; Duan, D.; Forstater, J.H.; Grosser, S.T.; et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 2019, 366, 1255–1259. [Google Scholar] [CrossRef]
- Kino, H.; Kino, K. Alteration of the substrate specificity of L-amino acid ligase and selective synthesis of Met-Gly as a salt taste enhancer. Biosci. Biotechnol. Biochem. 2015, 79, 1827–1832. [Google Scholar] [CrossRef]
- Tsuda, T.; Asami, M.; Koguchi, Y.; Kojima, S. Single Mutation Alters the Substrate Specificity of L-Amino Acid Ligase. Biochemistry 2014, 53, 2650–2660. [Google Scholar] [CrossRef]
- Kino, K.; Nakazawa, Y.; Yagasaki, M. Dipeptide synthesis by L-amino acid ligase from Ralstonia solanacearum. Biochem. Biophys. Res. Commun. 2008, 371, 536–540. [Google Scholar] [CrossRef]
- Kino, K.; Noguchi, A.; Nakazawa, Y.; Yagasaki, M. A novel L-amino acid ligase from Bacillus Licheniformis. J. Biosci. Bioeng. 2008, 106, 313–315. [Google Scholar] [CrossRef]
- Nakajima, H.; Suzuki, K.; Imahori, K. Purification and Properties of Acetate Kinase from Bacillus stearothermophilus. J. Biochem. 1978, 84, 193–203. [Google Scholar] [CrossRef]
- Dean, R.L. Kinetic studies with alkaline phosphatase in the presence and absence of inhibitors and divalent cations. Biochem. Mol. Biol. Educ. 2002, 30, 401–407. [Google Scholar] [CrossRef]
- Kino, H.; Nakajima, S.; Arai, T.; Kino, K. Effective production of Pro–Gly by mutagenesis of l-amino acid ligase. J. Biosci. Bioeng. 2016, 122, 155–159. [Google Scholar] [CrossRef]
- Burgener, S.; Luo, S.; McLean, R.; Miller, T.E.; Erb, T.J. A roadmap towards integrated catalytic systems of the future. Nat. Cat. 2020, 3, 186–192. [Google Scholar] [CrossRef]
- Kagebayashi, T.; Kontani, N.; Yamada, Y.; Mizushige, T.; Arai, T.; Kino, K.; Ohinata, K. Novel CCK-dependent vasorelaxing dipeptide, Arg-Phe, decreases blood pressure and food intake in rodents. Mol. Nutr. Food Res. 2012, 56, 1456–1463. [Google Scholar] [CrossRef]
- Yushkova, E.D.; Nazarova, E.A.; Matyuhina, A.V.; Noskova, A.O.; Shavronskaya, D.O.; Vinogradov, V.V.; Skvortsova, N.N.; Krivoshapkina, E.F. Application of Immobilized Enzymes in Food Industry. J. Agric. Food Chem. 2019, 67, 11553–11567. [Google Scholar] [CrossRef]
- El-Ashram, S.; Al Nasr, I.; Suo, X. Nucleic acid protocols: Extraction and optimization. Biotechnol. Rep. 2016, 12, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Rottmann, E.; Hauke, K.F.; Krings, U.; Berger, R.G. Enzymatic acrylamide mitigation in French fries–An industrial-scale case study. Food Control 2021, 123, 107739. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordewick, S.; Mast, T.A.; Berger, R.G.; Ersoy, F. Recombinant Production of Arginyl Dipeptides by l-Amino Acid Ligase RizA Coupled with ATP Regeneration. Catalysts 2021, 11, 1290. https://doi.org/10.3390/catal11111290
Bordewick S, Mast TA, Berger RG, Ersoy F. Recombinant Production of Arginyl Dipeptides by l-Amino Acid Ligase RizA Coupled with ATP Regeneration. Catalysts. 2021; 11(11):1290. https://doi.org/10.3390/catal11111290
Chicago/Turabian StyleBordewick, Sven, Tim A. Mast, Ralf G. Berger, and Franziska Ersoy. 2021. "Recombinant Production of Arginyl Dipeptides by l-Amino Acid Ligase RizA Coupled with ATP Regeneration" Catalysts 11, no. 11: 1290. https://doi.org/10.3390/catal11111290






