Phenylalanine, Tyrosine, and DOPA Are bona fide Substrates for Bambusa oldhamii BoPAL4
Abstract
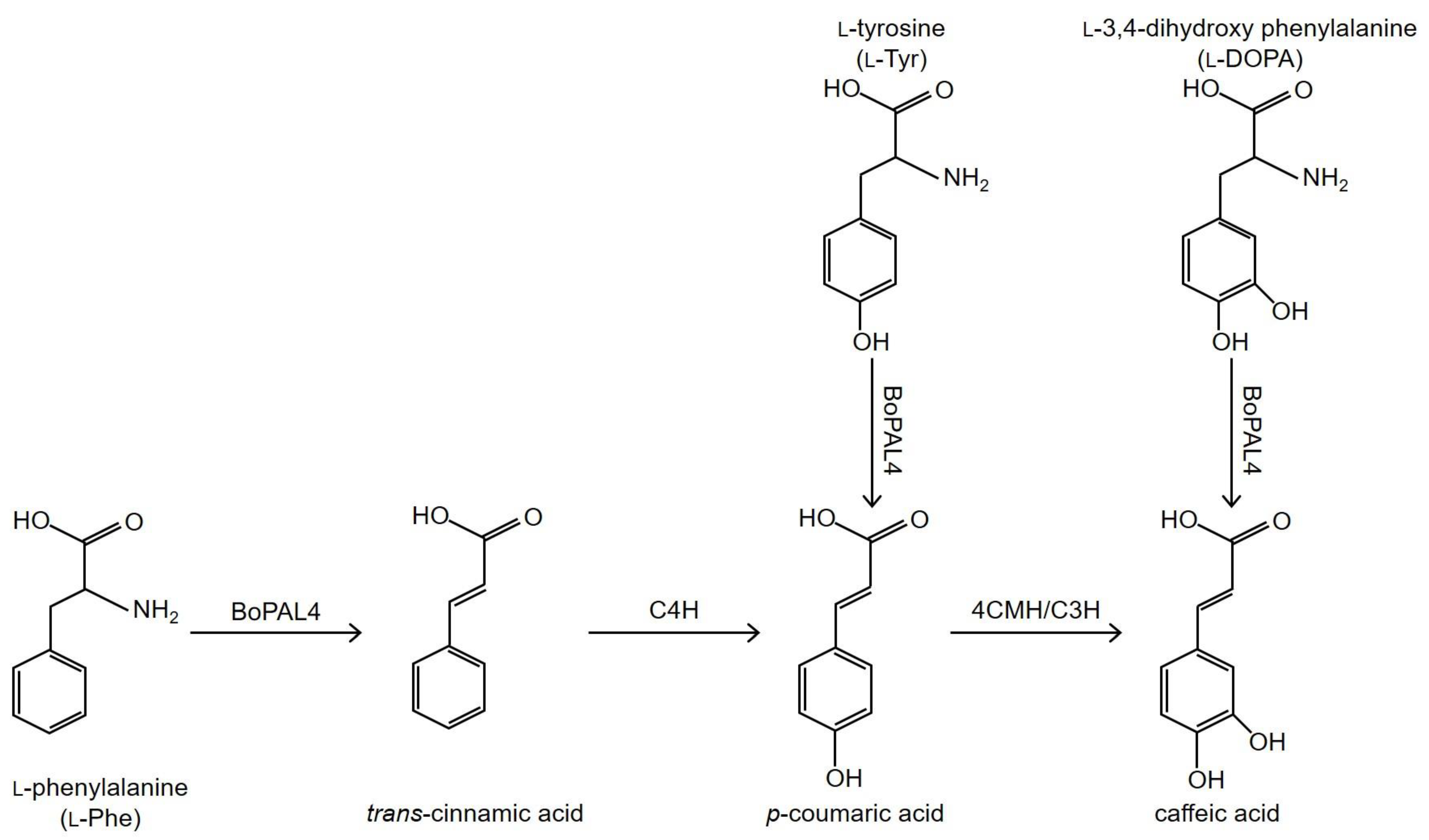
:1. Introduction
2. Results
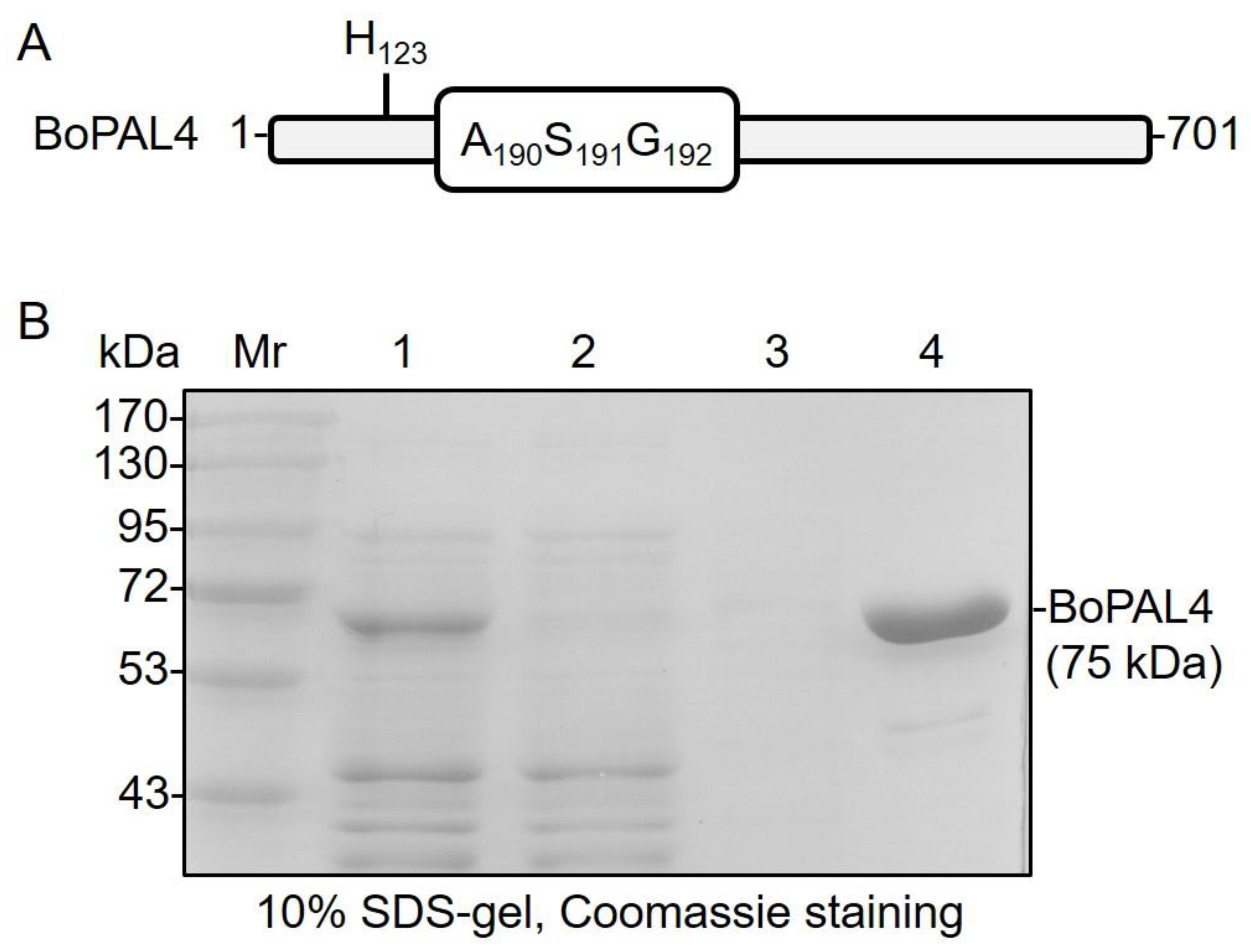
2.1. Expression and Purification of Recombinant BoPAL4 in Escherichia coli
2.2. Optimum pH and Temperature for PAL, TAL and DAL Activities of BoPAL4
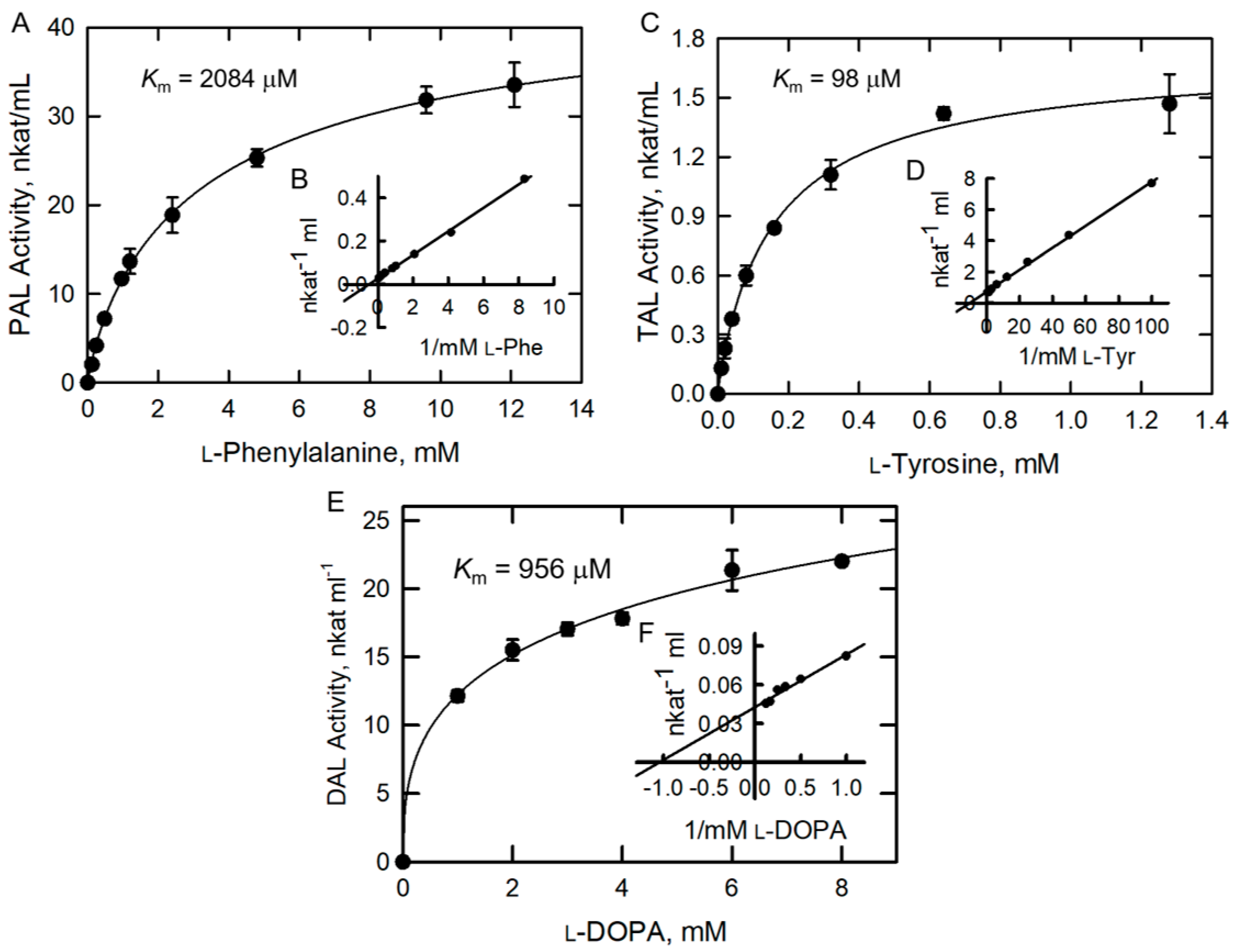
2.3. Kinetic Parameters for PAL, TAL, and DAL Activities of BoPAL4
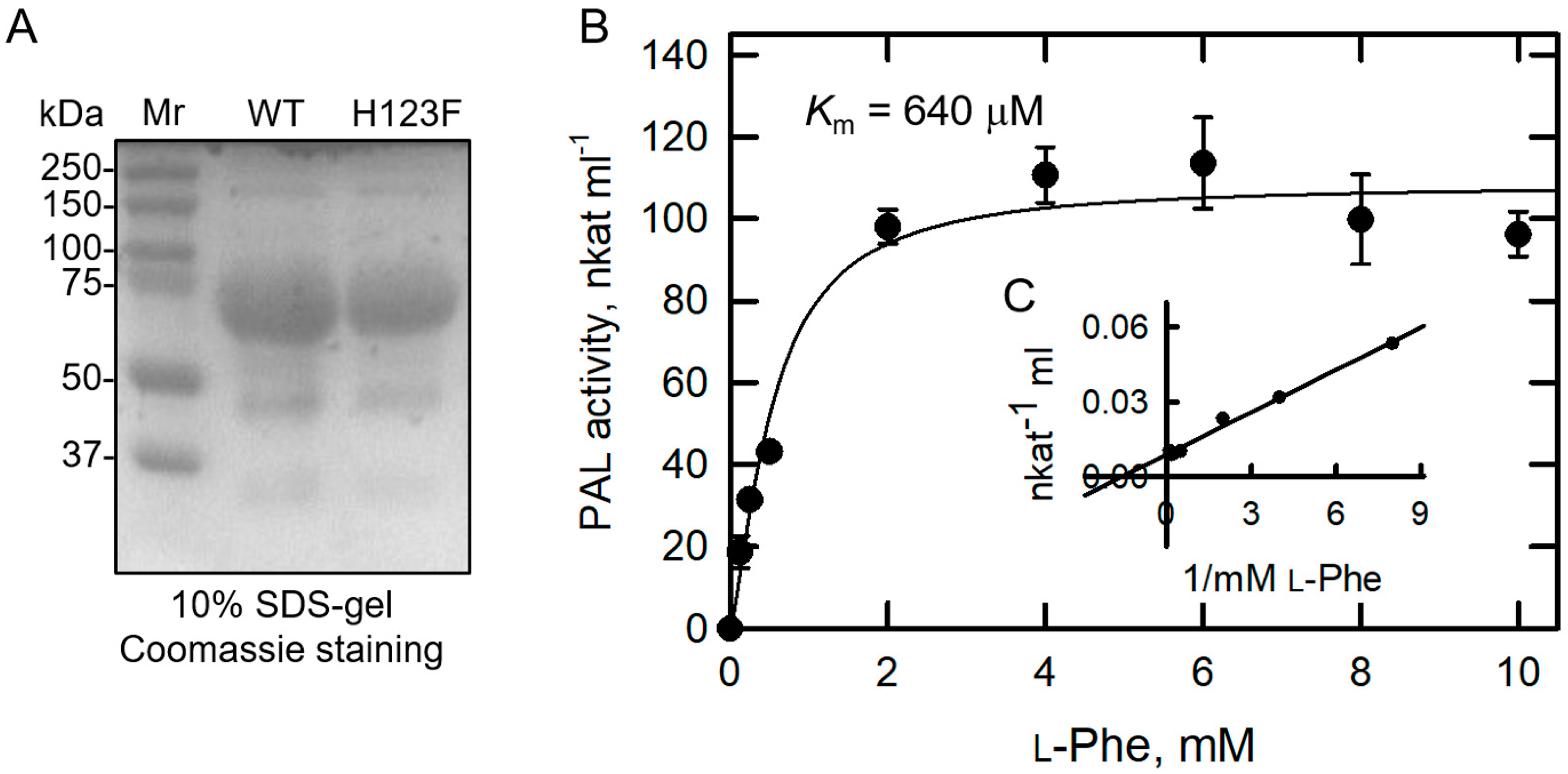
2.4. Kinetic Parameters for PAL Activity of BoPAL4-H123F
2.5. Comparison of Specific Activities in Wild-Type BoPAL4 and BoPAL4-H123F Mutants
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Bacterial Strains, Plasmids Construction, Site-Directed Mutagenesis, and Bacterial Growth Conditions
4.3. Preparations of BoPAL4 Enzymes
4.4. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
4.5. PAL Activity Assay
4.6. TAL Activity Assay
4.7. DAL Activity Assay
4.8. Biochemical Properties and Enzyme Kinetic
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Koukol, J.; Conn, E.E. Metabolism of aromatic compounds in higher plants. IV. Purification and properties of phenylalanine deaminase of Hordeum vulgare. J. Biol. Chem. 1961, 236, 2692–2698. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Shanfa, L. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Bell-Lelong, D.A.; Cusumano, J.C.; Meyer, K.; Chapple, C. Cinnamate 4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol. 1997, 113, 729–738. [Google Scholar] [CrossRef] [Green Version]
- Achnine, L.; Blancaflor, E.B.; Rasmussen, S.; Dixon, R.A. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoids biosynthesis. Plant Cell 2004, 16, 3098–3109. [Google Scholar] [CrossRef] [PubMed]
- Chen, O.; Deng, L.; Ruan, C.; Yi, L.; Zeng, K. Pichia galeiformis induced resistance in postharvest citrus by activating the phenylpropanoid biosynthesis pathway. J. Agri. Food Chem. 2021, 69, 2619–2631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lu, J.; Li, J.; Yu, Y.; Zhang, J.; He, Z.; Ismail, O.M.; Wu, J.; Xie, X.; Li, X.; et al. Efficiency of chitosan application against Phytophthora infestans and the activation of defence mechanisms in potato. Int. J. Biol. Macromol. 2021, 182, 1670–1680. [Google Scholar] [CrossRef]
- Maroga, G.M.; Soundy, P.; Sivakumar, D. Different postharvest responses of fresh-cut sweet peppers related to quality and antioxidant and phenylalanine ammonia lyase activities during exposure to light-emitting diode treatment. Foods 2019, 8, 359. [Google Scholar] [CrossRef] [Green Version]
- Mitani, T.; Ota, K.; Inaba, N.; Kishida, K.; Koyama, H.A. Antimicrobial activity of the phenolic compounds of Prunus mume against enterobacteria. Biol. Pharm. Bull. 2018, 41, 208–212. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Ge, S.; Li, S.; Lin, H.; Lin, S. Anti-obesity effect of trans-cinnamic acid on HepG2 cells and HFD-fed mice. Food Chem. Toxicol. 2020, 137, 111148. [Google Scholar] [CrossRef]
- Barros, J.; Escamilla-Trevino, L.; Song, L.; Rao, X.; Serrani-Yarce, J.C.; Palacios, M.D.; Engle, N.; Choudhury, F.K.; Tschalinski, T.J.; Venables, B.J.; et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019, 10, 1994. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, F.B.; Fetoui, H.; Fakhfakh, F.; Keskes, L. Caffeic acid and quercetin protect erythrocytes against the oxidative stress and the genotoxic effects of lambda-cyhalothrin in vitro. Hum. Exp. Toxicol. 2012, 31, 92–100. [Google Scholar] [CrossRef]
- Lima, V.N.; Oliveira-Tintino, C.D.M.; Santos, E.S.; Morais, L.P.; Tintino, S.R.; Freitas, T.S.; Geraldo, Y.S.; Pereira, R.L.S.; Cruz, R.P.; Menezes, I.R.A.; et al. Antimicrobial and enhancement of the antibiotic activity by phenolic compounds: Gallic acid, caffeic acid and pyrogallol. Microb. Pathog. 2016, 99, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Shull, T.E.; Kurepa, J.; Miller, R.D.; Martinez-Ochoa, N.; Smalle, J.A. Inhibition of Fusarium oxysporum f. sp. nicotianae growth by phenylpropanoid pathway intermediates. Plant Pathol. J. 2020, 36, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.F.; Rétey, J.; Schulz, G.E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 1999, 38, 5355–5361. [Google Scholar] [CrossRef] [PubMed]
- Rétey, J. Discovery and role of methylidene imidazolone, a highly electrophilic prosthetic group. Biochim. Biophys. Acta. 2003, 1647, 179–184. [Google Scholar] [CrossRef]
- Ritter, H.; Schulz, G.E. Structural basis for the entrance into the phenylpropanoid metabolism catalyzes by phenylalanine ammonia-lyase. Plant Cell 2004, 16, 3426–3436. [Google Scholar] [CrossRef] [PubMed]
- Sulis, D.; Wang, J.P. Regulation of lignin biosynthesis by post-translational protein modifications. Front Plant Sci. 2020, 11, 914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gou, M.; Liu, C.-J. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 2013, 25, 4994–5010. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Li, D.; Yang, D.; Xue, Z.; Li, J.; Xing, B.; Yan, K.; Han, R.; Liang, Z. SmKFB5 protein regulates phenolic acid biosynthesis by controlling the degradation the degradation of phenylalanine ammonia-lyase in Salvia miltiorrhiza. J. Exp. Bot. 2021, 72, 4915–4929. [Google Scholar] [CrossRef]
- Bolwell, G.P. A role for phosphorylation in the down-regulation of phenylalanine ammonia-lyase in suspension-cultured cells of French bean. Phytochemistry 1992, 31, 4081–4086. [Google Scholar] [CrossRef]
- Cheng, S.H.; Sheen, J.; Gerrish, C.; Bolwell, G.P. Molecular identification of phenylalanine ammonia-lyase as a substrate of a specific constitutively active Arabidopsis CDPK expressed in maize protoplasts. FEBS Lett. 2001, 503, 185–188. [Google Scholar] [CrossRef] [Green Version]
- Allwood, E.G.; Davies, D.R.; Gerrish, C.; Bolwell, G.P. Regulation of CDPKs, including identification of PAL kinase, in biotically stressed cells of French bean. Plant Mol. Biol. 2002, 49, 533–544. [Google Scholar] [CrossRef]
- Allwood, E.G.; Davies, D.R.; Gerrish, C.; Ellis, B.E.; Bolwell, G.P. Phosphorylation of phenylalanine ammonia-lyase: Evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 1999, 457, 47–52. [Google Scholar] [CrossRef] [Green Version]
- Reichert, A.I.; He, X.Z.; Dixon, R.A. Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): Characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem. J. 2009, 424, 233–242. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, L.-S.; Yeh, C.-S.; Cheng, C.-Y.; Yang, C.-C.; Lee, P.-D. Cloning and expression of a phenylalanine ammonia-lyase gene (BoPAL2) from Bambusa oldhamii in Escherichia coli and Pichia pastoris. Protein Expr. Purif. 2010, 71, 224–230. [Google Scholar] [CrossRef]
- Hsieh, L.-S.; Ma, G.-J.; Yang, C.-C.; Lee, P.-D. Cloning, expression, site-directed mutagenesis and immunolocalization of phenylalanine ammonia-lyase in Bambusa oldhamii. Phytochemistry 2010, 71, 1999–2009. [Google Scholar] [CrossRef]
- Appert, C.; Logemann, E.; Hahlbrock, K.; Schmid, J.; Amrhein, N. Structural and catalytic properties of the four phenylalanine ammonia-lyases from parsley (Petroselinum crispum Nym). Eur. J. Biochem. 1994, 225, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Dixon, R.A. Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci. 2020, 25, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Moisă, M.E.; Amariei, D.A.; Nagy, E.Z.A.; Szarvas, N.; Toșa, M.I.; Paizs, C.; Bencze, L.C. Fluorescent enzyme-coupled activity assay for phenylalanine ammonia-lyases. Sci. Rep. 2020, 10, 18418. [Google Scholar] [CrossRef] [PubMed]
- Rösler, J.; Krekel, F.; Amrhein, N.; Schmid, J. Maize phenylalanine ammonia lyase has tyrosine ammonia-lyase activity. Plant Physiol. 1997, 113, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, C.-Y.; Huang, Y.-H.; Lin, Z.-Y.; Hsieh, L.-S. Insights into the substrate selectivity of Bambusa oldhamii phenylalanine ammonia-lyase 1 and 2 through mutational analysis. Phytochem. Lett. 2020, 38, 140–143. [Google Scholar] [CrossRef]
- Barros, J.; Serrani-Yarce, J.C.; Chen, F.; Baxter, D.; Venables, B.J.; Dixon, R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2016, 2, 16050. [Google Scholar] [CrossRef]
- Jun, S.Y.; Sattler, S.A.; Cortez, G.S.; Vermerris, W.; Sattler, S.E.; Kang, C. Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase. Plant Physiol. 2018, 176, 1452–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyndt, J.A.; Meyer, T.E.; Cusanovich, M.A.; Van Beeumen, J.J. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 2002, 512, 240–244. [Google Scholar] [CrossRef] [Green Version]
- Louie, G.V.; Bowman, M.E.; Moffitt, M.C.; Baiga, T.J.; Moore, B.S.; Noel, N.P. Structural determinants and modulation of substrate specificity in phenylalanine–tyrosine ammonia-lyase. Chem. Biol. 2006, 13, 1327–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moffitt, M.C.; Louie, G.V.; Bowman, M.E.; Pence, J.; Noel, J.P.; Moore, B.S. Discovery of two cyanobacterial phenylalanine ammonia-lyases: Kinetic and structural characterization. Biochemistry 2008, 46, 1004–1012. [Google Scholar] [CrossRef] [Green Version]
- Jendresen, C.B.; Stahlhut, S.G.; Li, M.; Gasper, P.; Siedler, S.; Förster, J.; Maury, J.; Borodina, I.; Nielsen, A.T. Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2015, 81, 4458–4476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacDonald, M.C.; Arivalagan, P.; Barre, D.E.; Maclnnis, J.A.; D’Cunha, G.B. Rhodotorula glutinis phenylalanine/tyrosine ammonia lyase enzyme catalyzed synthesis of the methyl ester of para-hydroxycinnamic acid and its potential antibacterial activity. Front Microbiol. 2016, 7, 281. [Google Scholar] [CrossRef] [Green Version]
- Feduraev, P.; Skrypnik, L.; Riabova, A.; Pungin, A.; Tokupova, E.; Maslennikov, P.; Chupakhina, G. Phenylalanine and tyrosine as exogenous precursors of wheat (Triticum aestivum L.) secondary metabolism through PAL-associate pathways. Plants 2020, 9, 476. [Google Scholar] [CrossRef] [Green Version]
- Maeda, E.A. Lignin biosynthesis: Tyrosine shortcut in grasses. Nat. Plants 2016, 2, 16080. [Google Scholar] [CrossRef] [PubMed]
- Watts, K.T.; Mijts, B.N.; Lee, P.C.; Manning, A.J.; Schmidt-Dannert, C. Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 2006, 13, 1317–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, L.-S.; Hsieh, Y.-L.; Yeh, C.-S.; Cheng, C.-Y.; Yang, C.-C.; Lee, P.-D. Molecular characterization of a phenylalanine ammonia-lyase gene (BoPAL1) from Bambusa oldhamii. Mol. Biol. Rep. 2011, 38, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhao, Y.; Feng, Y.; Lin, T.; Zhong, C.; Tan, Z.; Jia, S. Encapsulation of spherical cross-linked phenylalanine ammonia lyase aggregates in mesoporous biosilica. J. Agri. Food Chem. 2017, 65, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhao, Y.; Tan, Z.; Zhong, C.; Han, P.; Jia, S. Mesoporous phenylalanine ammonia lyase microspheres with improved stability through calcium carbonate templating. Int. J. Biol. Macromol. 2017, 98, 887–896. [Google Scholar] [CrossRef]
- Boros, K.; Moisă, M.E.; Nagy, C.L.; Paizs, C.; Toşa, M.I.; Bencze, L.C. Robust, site-specificity immobilized phenylalanine ammonia-lyases for the enantioselective ammonia addition of cinnamic acids. Catal. Sci. Technol. 2021, 11, 5553. [Google Scholar] [CrossRef]
- Hong, P.-Y.; Huang, Y.-H.; Lim, G.C.W.; Chen, Y.-P.; Hsiao, C.-J.; Chen, L.-H.; Ciou, J.-Y.; Hsieh, L.-S. Production of trans-cinnamic acid by immobilization of the Bambusa oldhamii BoPAL1 and BoPAL2 phenylalanine ammonia-lyases on electrospun nanofibers. Int. J. Mol. Sci. 2021, 22, 11184. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hsiao, C.-J.; Hsieh, C.-Y.; Hsieh, L.-S. Cloning and characterization of the Bambusa oldhamii BoMDH-encoded malate dehydrogenase. Protein Expr. Purif. 2020, 174, 105665. [Google Scholar] [CrossRef]
- Hsu, W.-H.; Huang, Y.-H.; Chen, P.-R.; Hsieh, L.-S. NLIP and HAD-like domains of Pah1 and Lipin 1 phosphatidate phosphatases are essential for their catalytic activities. Molecules 2021, 26, 5470. [Google Scholar] [CrossRef]
- De Jong, F.; Hanley, S.J.; Beale, M.H.; Karp, A. Characterization of the willow phenylalanine ammonia-lyase (PAL) gene family reveals expression differences compared with poplar. Phytochemistry 2015, 117, 90–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaelis, L.; Menten, M.L. Die kinetik der invertinwirkung. Biochem. Z. 1913, 49, 333–369. [Google Scholar]
- Lineweavwer, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
 ) as substrate. Activities were measured under standard assay conditions in a range of temperatures from 25 to 80 °C. (B) Optimum pH of BoPAL4 using L-Phe (●), L-Tyr (●), and L-DOPA (
) as substrate. Activities were measured under standard assay conditions in a range of temperatures from 25 to 80 °C. (B) Optimum pH of BoPAL4 using L-Phe (●), L-Tyr (●), and L-DOPA (  ) as substrates. Activities were measured under standard assay conditions in a range of pH from 5 to 11. All experiments were performed in triplicate and expressed as average ± standard deviation (S.D., error bars).
) as substrates. Activities were measured under standard assay conditions in a range of pH from 5 to 11. All experiments were performed in triplicate and expressed as average ± standard deviation (S.D., error bars).
 ) as substrate. Activities were measured under standard assay conditions in a range of temperatures from 25 to 80 °C. (B) Optimum pH of BoPAL4 using L-Phe (●), L-Tyr (●), and L-DOPA (
) as substrate. Activities were measured under standard assay conditions in a range of temperatures from 25 to 80 °C. (B) Optimum pH of BoPAL4 using L-Phe (●), L-Tyr (●), and L-DOPA (  ) as substrates. Activities were measured under standard assay conditions in a range of pH from 5 to 11. All experiments were performed in triplicate and expressed as average ± standard deviation (S.D., error bars).
) as substrates. Activities were measured under standard assay conditions in a range of pH from 5 to 11. All experiments were performed in triplicate and expressed as average ± standard deviation (S.D., error bars).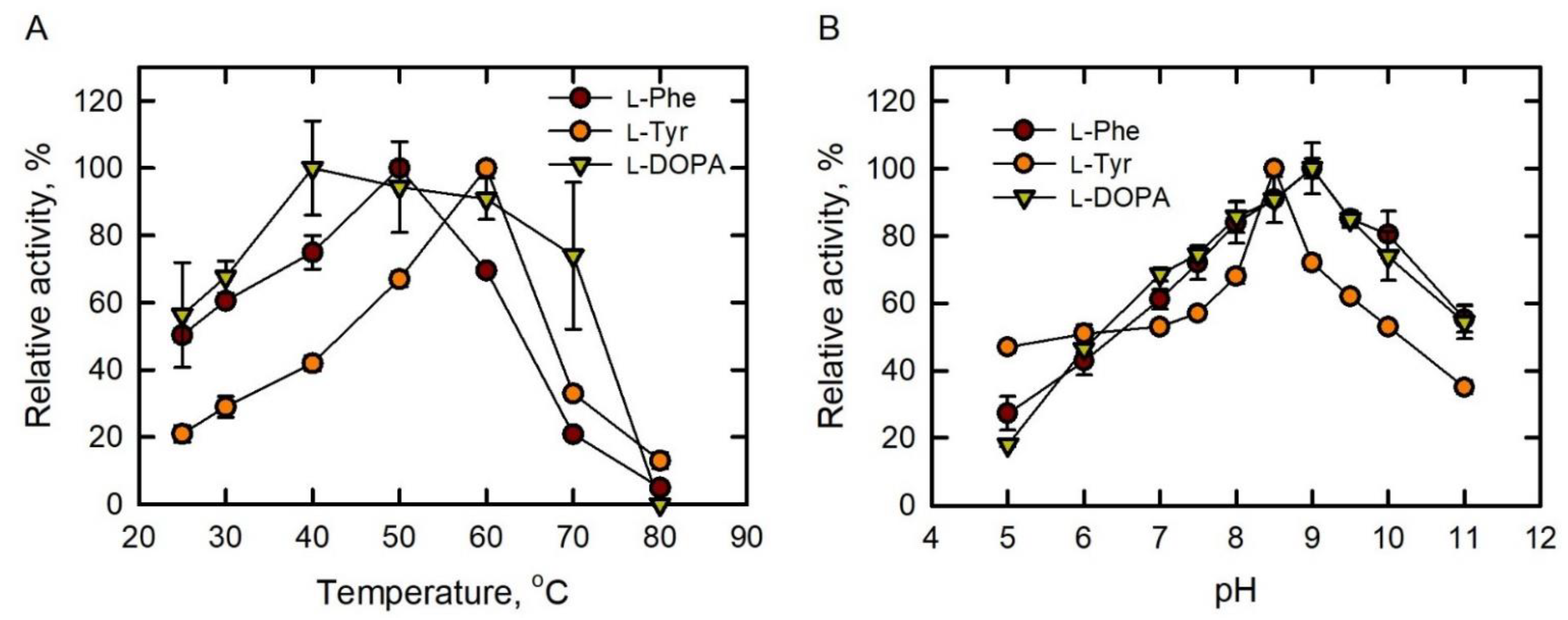

| Strain | Relevant Characteristics | Source or Ref. |
| E. coli Top10 | F– mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK λ– rpsL(StrR) endA1 nupG | Invitrogen |
| Plasmid | Relevant Characteristics | Source or Ref. |
| pTrcHisA | E. coli expression vector with N-terminal His6-tag fusion | Invitrogen |
| pTrcHisA-BoPAL4 | BoPAL4 coding sequence inserted into pTrcHisA | [27] |
| pTrcHisA-BoPAL4-H123F | Point mutation H123F derivative of pTrcHisA-BoPAL4 | This study |
| Protein | Substrate 1 | Optimum pH | Optimum Temp (°C) | kcat (s−1) | Km (μM) | kcat/Km (s−1 μM−1) |
|---|---|---|---|---|---|---|
| BoPAL4 | L-Phe | 9.0 | 50 | 1.44 | 2084 | 6.9 × 10−4 |
| L-Tyr | 8.5 | 60 | 0.18 | 98 | 18.4 × 10−4 | |
| L-DOPA | 9.0 | 40 | 0.06 | 956 | 0.6 × 10−4 | |
| BoPAL4-H123F | L-Phe | 9.0 | 50 | 1.87 | 640 | 29.2 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, C.-Y.; Huang, Y.-H.; Yeh, H.-H.; Hong, P.-Y.; Hsiao, C.-J.; Hsieh, L.-S. Phenylalanine, Tyrosine, and DOPA Are bona fide Substrates for Bambusa oldhamii BoPAL4. Catalysts 2021, 11, 1263. https://doi.org/10.3390/catal11111263
Hsieh C-Y, Huang Y-H, Yeh H-H, Hong P-Y, Hsiao C-J, Hsieh L-S. Phenylalanine, Tyrosine, and DOPA Are bona fide Substrates for Bambusa oldhamii BoPAL4. Catalysts. 2021; 11(11):1263. https://doi.org/10.3390/catal11111263
Chicago/Turabian StyleHsieh, Chun-Yen, Yi-Hao Huang, Hui-Hsuan Yeh, Pei-Yu Hong, Che-Jen Hsiao, and Lu-Sheng Hsieh. 2021. "Phenylalanine, Tyrosine, and DOPA Are bona fide Substrates for Bambusa oldhamii BoPAL4" Catalysts 11, no. 11: 1263. https://doi.org/10.3390/catal11111263






