An Efficient Decoder for the Recognition of Event-Related Potentials in High-Density MEG Recordings †
Abstract
:1. Introduction
2. Materials and Methods
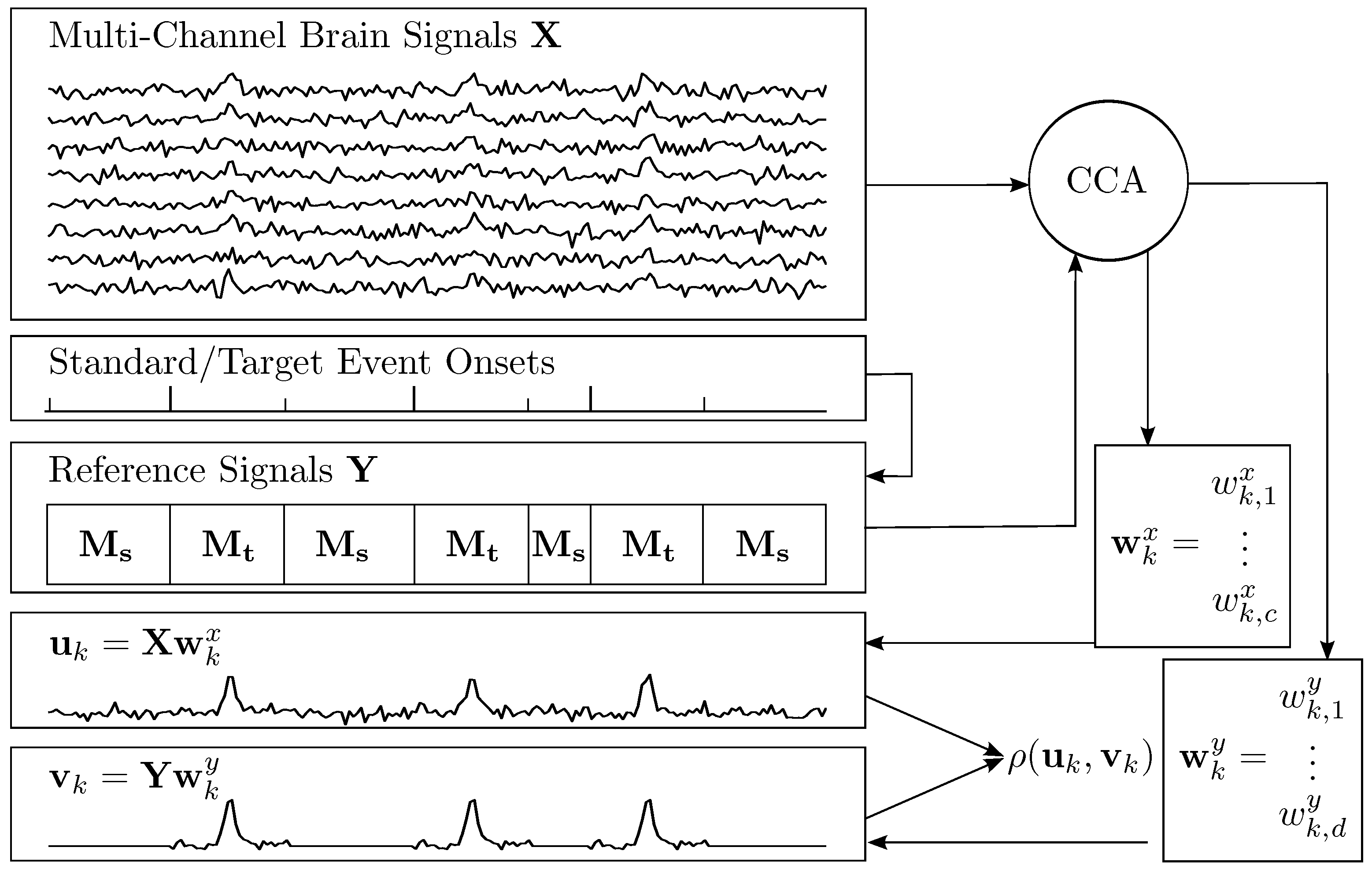
2.1. Canonical Correlation Analysis
2.2. Constructing Spatial Filters for ERP Extraction
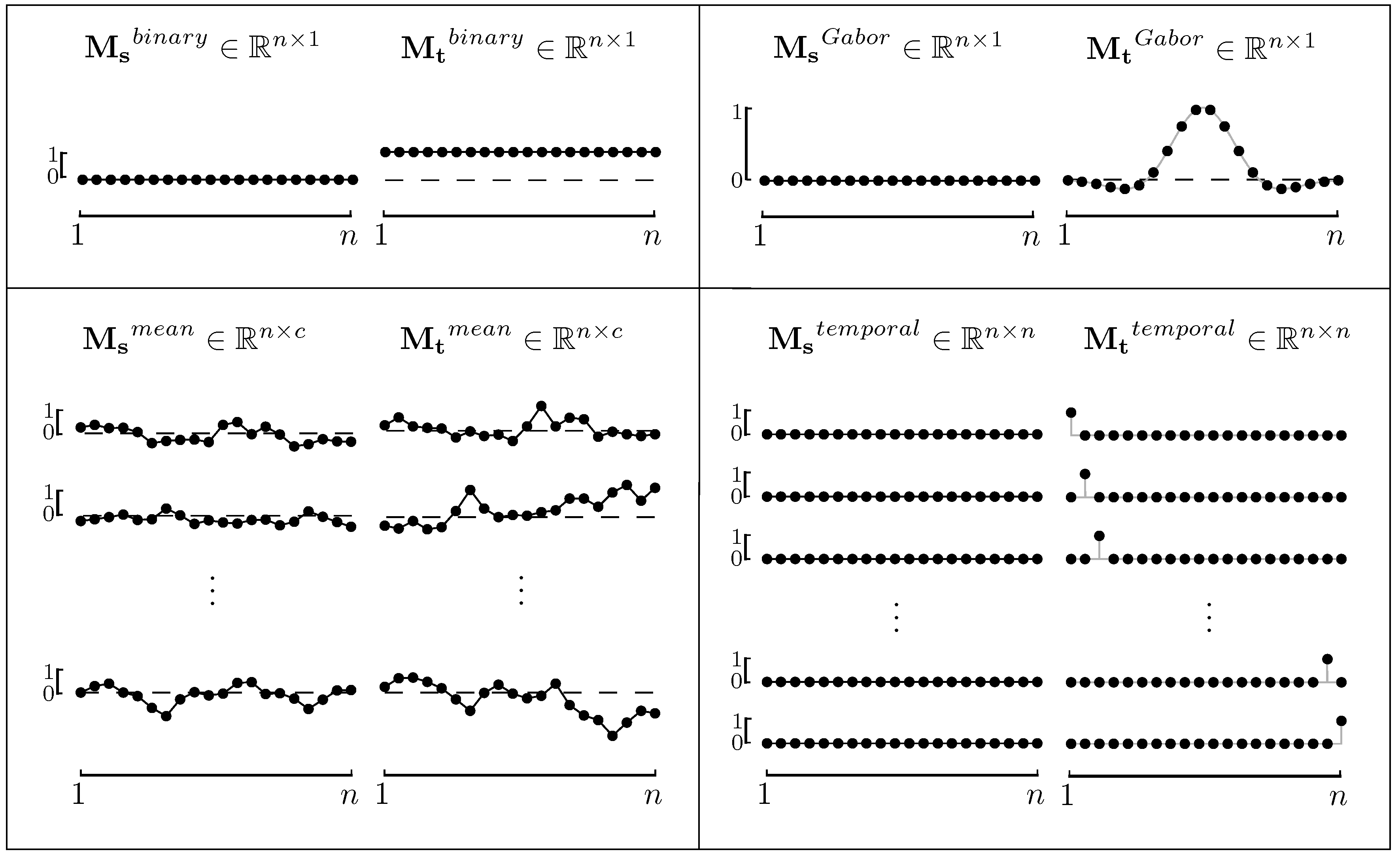
2.3. Model Functions for ERPs
2.4. A Spatio-Temporal Filter
2.5. Recognition of ERP Sequences
2.6. Experimental Data
2.7. Processing of Experimental Data
3. Results
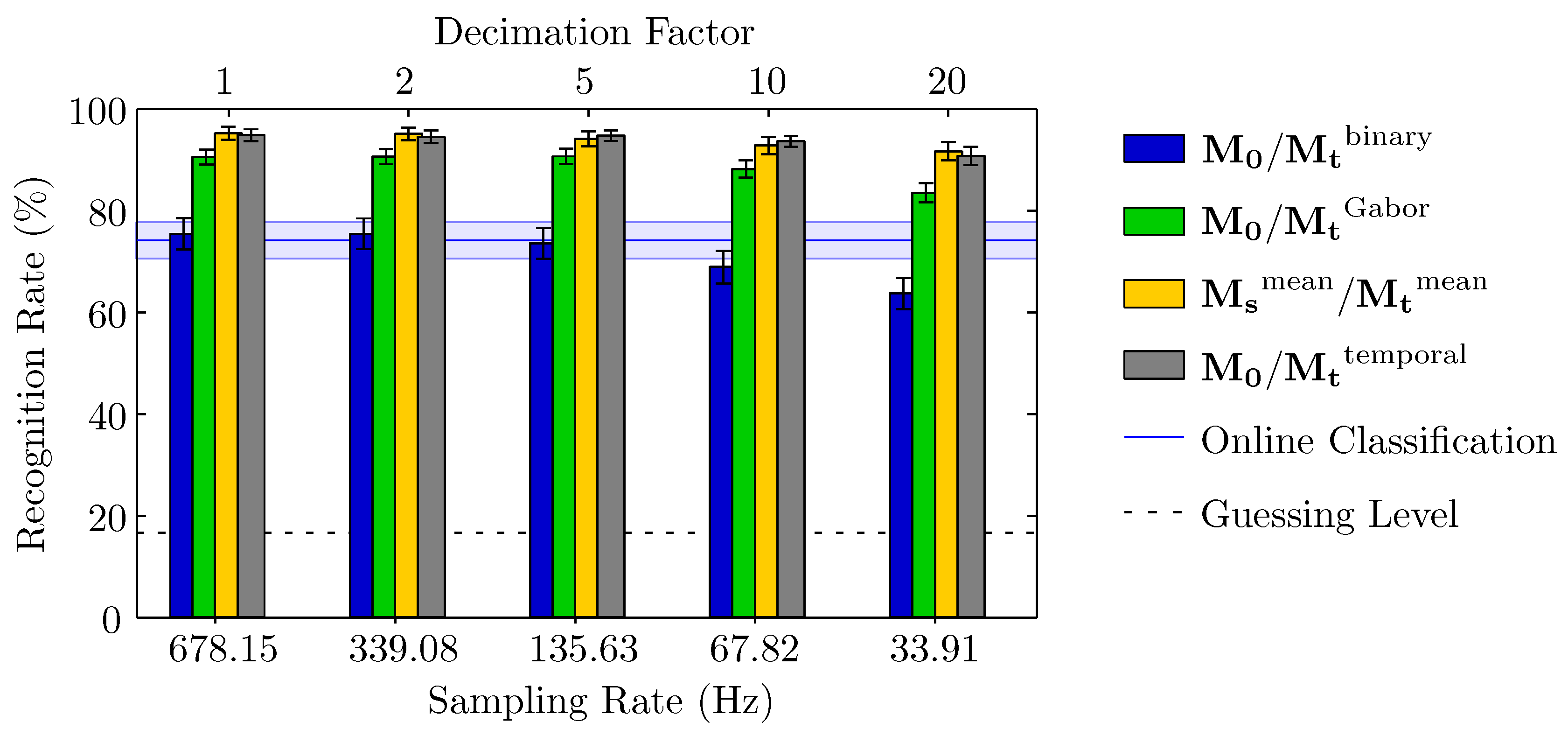
3.1. Accuracy of Recognition
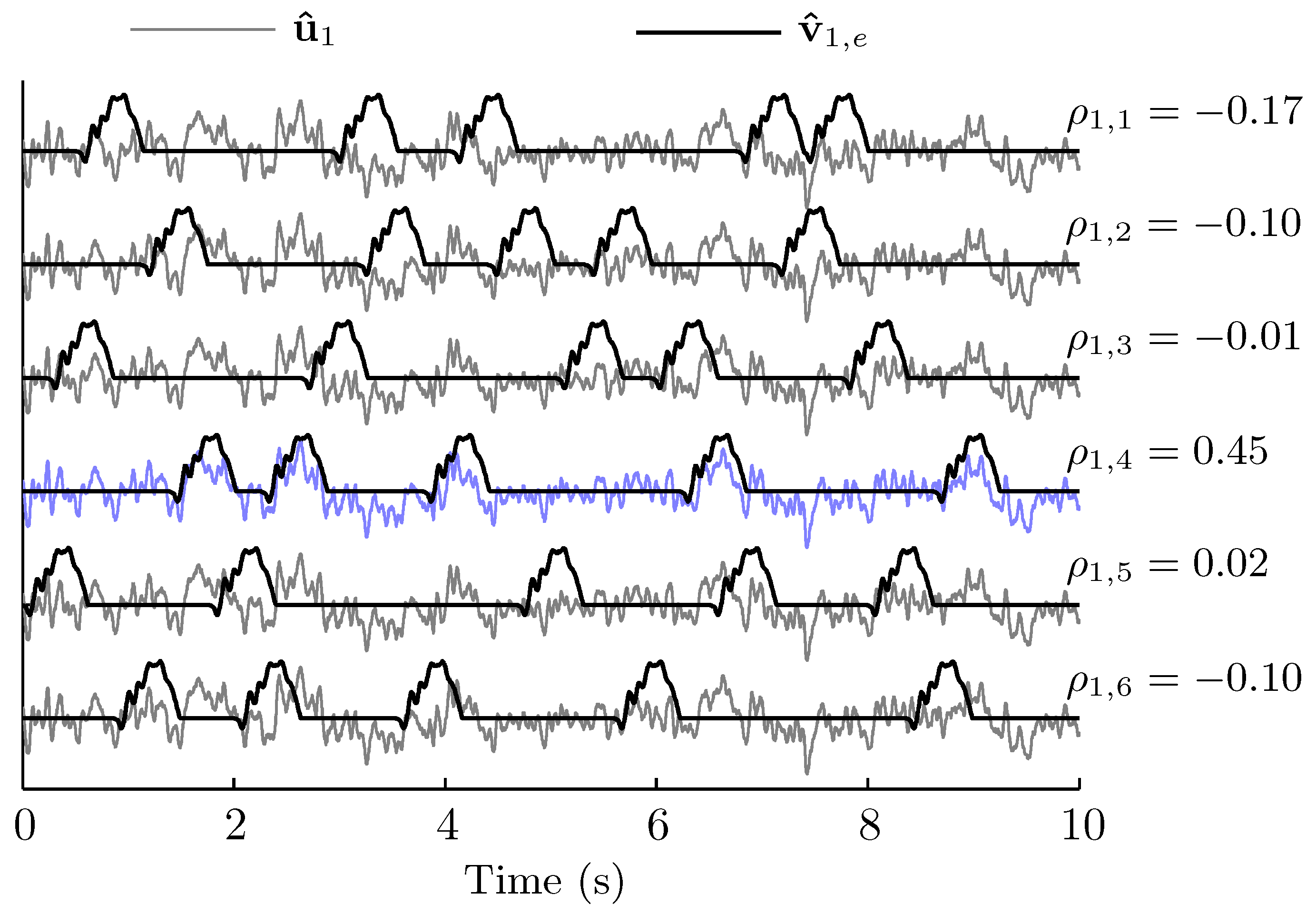
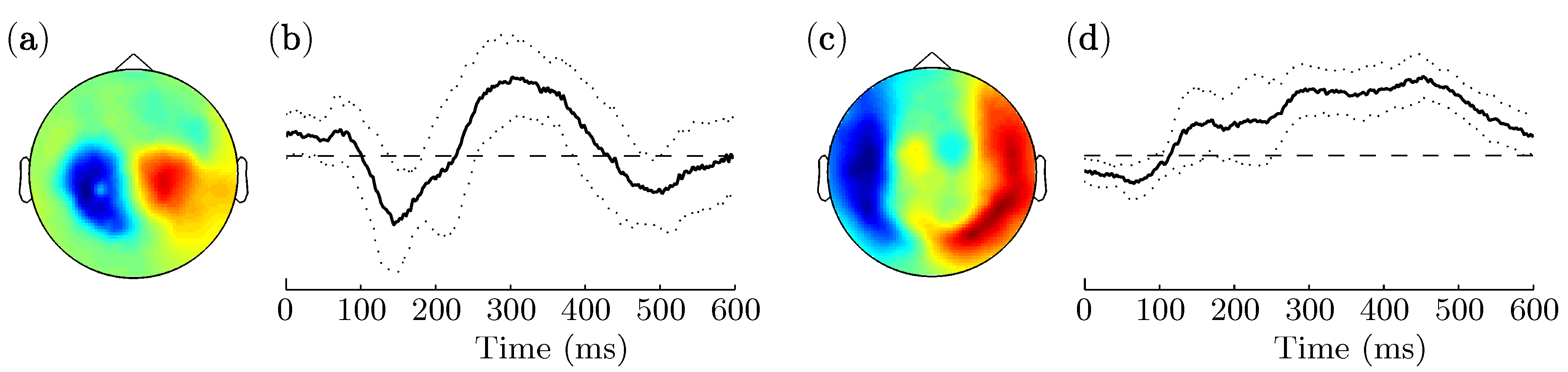
3.2. Spatio-Temporal Patterns
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ADHD | Attention deficit hyperactivity disorder |
| BCI | Brain–computer interface |
| CCA | Canonical correlation analysis |
| CSP | Common spatial pattern |
| EEG | Electroencephalogram |
| ERP | Event-related potential |
| ICA | Independent component analysis |
| ITR | Information transfer rate |
| MEG | Magnetoencephalogram |
| P300 | Positive deflection, peaking at 300 ms |
| PCA | Principal component analysis |
| SD | Standard deviation |
| ANR | Signal-to-noise ratio |
| SVM | Support vector machine |
Nomenclature
| c | number of variables (channels) in |
| d | number of variables (reference functions) in |
| e | index for event sequence |
| f | averaged Fisher z-transformed correlation coefficients |
| i | general iteration index |
| k | index for columns in (virtual channel/component) |
| m | element in |
| n | number of observations (sampling points) |
| p | p-value |
| q | number of selected components |
| s | refers to standard event |
| t | refers to target event |
| w | element in |
| x | refers to / element in |
| y | refers to / element in |
| column vector in | |
| column vector in | |
| column vector in | |
| identity matrix of size | |
| matrix of model functions | |
| matrix of filtered | |
| matrix of filtered | |
| filter matrix (weights for linear combination) | |
| matrix of observations (brain signals) | |
| matrix of observations (reference signals) | |
| ρ | (canonical) correlation coefficient |
| μ | Gaussian mean |
| σ | standard deviation |
| ω | scaling parameter |
| denotes test data |
References
- Birbaumer, N.; Cohen, L.G. Brain-computer interfaces: Communication and restoration of movement in paralysis. J. Physiol. 2007, 579, 621–636. [Google Scholar] [CrossRef] [PubMed]
- Farwell, L.A.; Donchin, E. Talking off the top of your head: Toward a mental prosthesis utilizing event-related brain potentials. Electroencephalogr. Clin. Neurophysiol. 1988, 70, 510–523. [Google Scholar] [CrossRef]
- Serby, H.; Yom-Tov, E.; Inbar, G.F. An improved P300-based brain-computer interface. IEEE Trans. Neural Syst. Rehabil. Eng. 2005, 13, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Krusienski, D.; Sellers, E.; Vaughan, T. Common spatio-temporal patterns for the P300 speller. In Proceedings of the CNE ’07. 3rd International IEEE/EMBS Conference on Neural Engineering, Kohala Coast, HI, USA, 2–5 May 2007; pp. 421–424.
- Farquhar, J.; Hill, N.J. Interactions between pre-processing and classification methods for event-related-potential classification: Best-practice guidelines for brain-computer interfacing. Neuroinformatics 2013, 11, 175–192. [Google Scholar] [CrossRef] [PubMed]
- Spüler, M.; Walter, A.; Rosenstiel, W.; Bogdan, M. Spatial filtering based on canonical correlation analysis for classification of evoked or event-related potentials in EEG data. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Rivet, B.; Souloumiac, A.; Attina, V.; Gibert, G. xDAWN algorithm to enhance evoked potentials: Application to brain-computer interface. IEEE Trans. Biomed. Eng. 2009, 56, 2035–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, G.; Nunes, U.; Castelo-Branco, M. Statistical spatial filtering for a P300-based BCI: Tests in able-bodied, and patients with cerebral palsy and amyotrophic lateral sclerosis. J. Neurosci. Methods 2011, 195, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Reichert, C.; Kennel, M.; Kruse, R.; Heinze, H.J.; Schmucker, U.; Hinrichs, H.; Rieger, J.W. Brain-controlled selection of objects combined with autonomous robotic grasping. In Neurotechnology, Electronics, and Informatics; Springer: Cham, Switzerland, 2015; Volume 13, pp. 65–78. [Google Scholar]
- Quandt, F.; Reichert, C.; Hinrichs, H.; Heinze, H.J.; Knight, R.T.; Rieger, J.W. Single trial discrimination of individual finger movements on one hand: A combined MEG and EEG study. Neuroimage 2012, 59, 3316–3324. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, L.; Sami, S.; Hillebrand, A.; Fawcett, I.P.; Quitadamo, L.R.; Seri, S. Which physiological components are more suitable for visual ERP based brain-computer interface? A preliminary MEG/EEG study. Brain Topogr. 2010, 23, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Hotelling, H. The most predictable criterion. J. Educ. Psychol. 1935, 26, 139–142. [Google Scholar] [CrossRef]
- Blankertz, B.; Tomioka, R.; Lemm, S.; Kawanabe, M.; Müller, K.R. Optimizing spatial filters for robust EEG single-trial analysis. IEEE Signal Process. Mag. 2008, 25, 41–56. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, C.; Wu, W.; Gao, X. Frequency recognition based on canonical correlation analysis for SSVEP-based BCIs. IEEE Trans. Biomed. Eng. 2006, 53, 2610–2614. [Google Scholar] [CrossRef] [PubMed]
- Reichert, C.; Kennel, M.; Kruse, R.; Hinrichs, H.; Rieger, J.W. Efficiency of SSVEF recognition from the magnetoencephalogram—A Comparison of spectral feature classification and CCA-based prediction. In Proceedings of the International Congress on Neurotechnology, Electronics and Informatics, Algarve, Portugal, 18–20 September 2013; pp. 233–237.
- Sielużycki, C.; König, R.; Matysiak, A.; Kuś, R.; Ircha, D.; Durka, P. Single-trial evoked brain responses modeled by multivariate matching pursuit. IEEE Trans. Biomed. Eng. 2009, 56, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Matlab implementation of CCA. Available online: www.mathworks.com/help/stats/canoncorr.html (accessed on 12 April 2016).
- Robinson, S.E. Environmental noise cancellation for biomagnetic measurements. In Advances in Biomagnetism; Springer: New York, NY, USA, 1989; pp. 721–724. [Google Scholar]
- Congedo, M.; Gouy-Pailler, C.; Jutten, C. On the blind source separation of human electroencephalogram by approximate joint diagonalization of second order statistics. Clin. Neurophysiol. 2008, 119, 2677–2686. [Google Scholar] [CrossRef] [PubMed]
- Mecklinger, A.; Maess, B.; Opitz, B.; Pfeifer, E.; Cheyne, D.; Weinberg, H. A MEG analysis of the P300 in visual discrimination tasks. Electroencephalogr. Clin. Neurophysiol. 1998, 108, 45–56. [Google Scholar] [CrossRef]
- Spencer, K.M.; Dien, J.; Donchin, E. Spatiotemporal analysis of the late ERP responses to deviant stimuli. Psychophysiology 2001, 38, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Krusienski, D.J.; Sellers, E.W.; Cabestaing, F.; Bayoudh, S.; McFarland, D.J.; Vaughan, T.M.; Wolpaw, J.R. A comparison of classification techniques for the P300 speller. J. Neural Eng. 2006, 3, 299–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, U.; Vesin, J.M.; Ebrahimi, T.; Diserens, K. An efficient P300-based brain-computer interface for disabled subjects. J. Neurosci. Methods 2008, 167, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Donchin, E.; Spencer, K.M.; Wijesinghe, R. The mental prosthesis: Assessing the speed of a P300-based brain-computer interface. IEEE Trans. Rehabil. Eng. 2000, 8, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Lenhardt, A.; Kaper, M.; Ritter, H.J. An adaptive P300-based online brain-computer interface. IEEE Trans. Neural Syst. Rehabil. Eng. 2008, 16, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.N.; Bonnet, S.; Charbonnier, S.; Jallon, P.; Campagne, A. A comparison of ERP spatial filtering methods for optimal mental workload estimation. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 7254–7257.
- Dähne, S.; Meinecke, F.C.; Haufe, S.; Höhne, J.; Tangermann, M.; Müller, K.R.; Nikulin, V.V. SPoC: A novel framework for relating the amplitude of neuronal oscillations to behaviorally relevant parameters. Neuroimage 2014, 86, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Dähne, S.; Nikulin, V.V.; Ramírez, D.; Schreier, P.J.; Müller, K.R.; Haufe, S. Finding brain oscillations with power dependencies in neuroimaging data. Neuroimage 2014, 96, 334–348. [Google Scholar] [CrossRef] [PubMed]

| Decimation Factor | / | / | / | / |
|---|---|---|---|---|
| 1 | 16.09 (1.23) | 16.10 (1.29) | 33.95 (2.70) | 48.80 (3.87) |
| 2 | 8.33 (1.01) | 8.23 (0.97) | 17.97 (3.14) | 15.52 (1.39) |
| 5 | 3.05 (0.29) | 3.19 (0.49) | 6.46 (0.58) | 3.93 (0.33) |
| 10 | 1.41 (0.13) | 1.41 (0.16) | 2.93 (0.27) | 1.53 (0.16) |
| 20 | 0.61 (0.14) | 0.63 (0.11) | 1.27 (0.15) | 0.64 (0.11) |
| Decimation Factor | / | / | / | / |
|---|---|---|---|---|
| 1 | 1.0 (0.0) | 1.0 (0.0) | 13.6 (1.2) | 5.0 (0.8) |
| 2 | 1.0 (0.0) | 1.0 (0.0) | 14.8 (1.2) | 5.1 (0.8) |
| 5 | 1.0 (0.0) | 1.0 (0.0) | 7.7 (1.2) | 4.9 (0.8) |
| 10 | 1.0 (0.0) | 1.0 (0.0) | 7.6 (1.3) | 4.8 (1.0) |
| 20 | 1.0 (0.0) | 1.0 (0.0) | 7.3 (1.4) | 4.6 (1.1) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reichert, C.; Dürschmid, S.; Kruse, R.; Hinrichs, H. An Efficient Decoder for the Recognition of Event-Related Potentials in High-Density MEG Recordings. Computers 2016, 5, 5. https://doi.org/10.3390/computers5020005
Reichert C, Dürschmid S, Kruse R, Hinrichs H. An Efficient Decoder for the Recognition of Event-Related Potentials in High-Density MEG Recordings. Computers. 2016; 5(2):5. https://doi.org/10.3390/computers5020005
Chicago/Turabian StyleReichert, Christoph, Stefan Dürschmid, Rudolf Kruse, and Hermann Hinrichs. 2016. "An Efficient Decoder for the Recognition of Event-Related Potentials in High-Density MEG Recordings" Computers 5, no. 2: 5. https://doi.org/10.3390/computers5020005






