Innovative In Vivo Imaging and Single Cell Expression from Tumor Bulk and Corpus Callosum Reveal Glioma Stem Cells with Unique Regulatory Programs
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Animal Studies
2.3. Tissue Clearing and Optical Imaging
2.4. Brain Dissection and GSC Isolation
2.5. Histology
2.6. Single-Cell Gene Expression
2.7. Survival Analysis
2.8. Data Availability
2.9. Statistical Analysis
3. Results
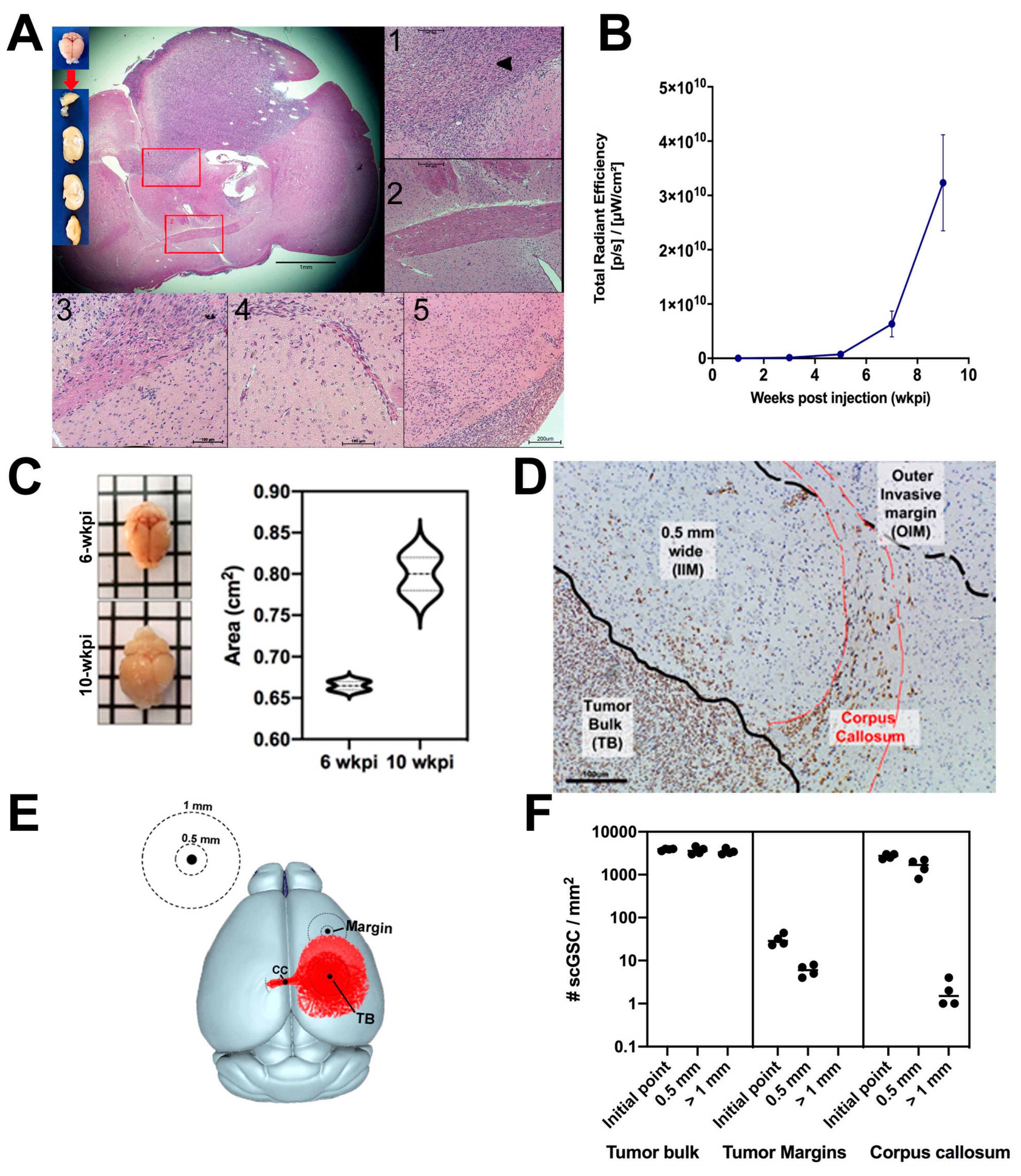
3.1. Establishment and Characterization of an Invasive GBM Mouse Model
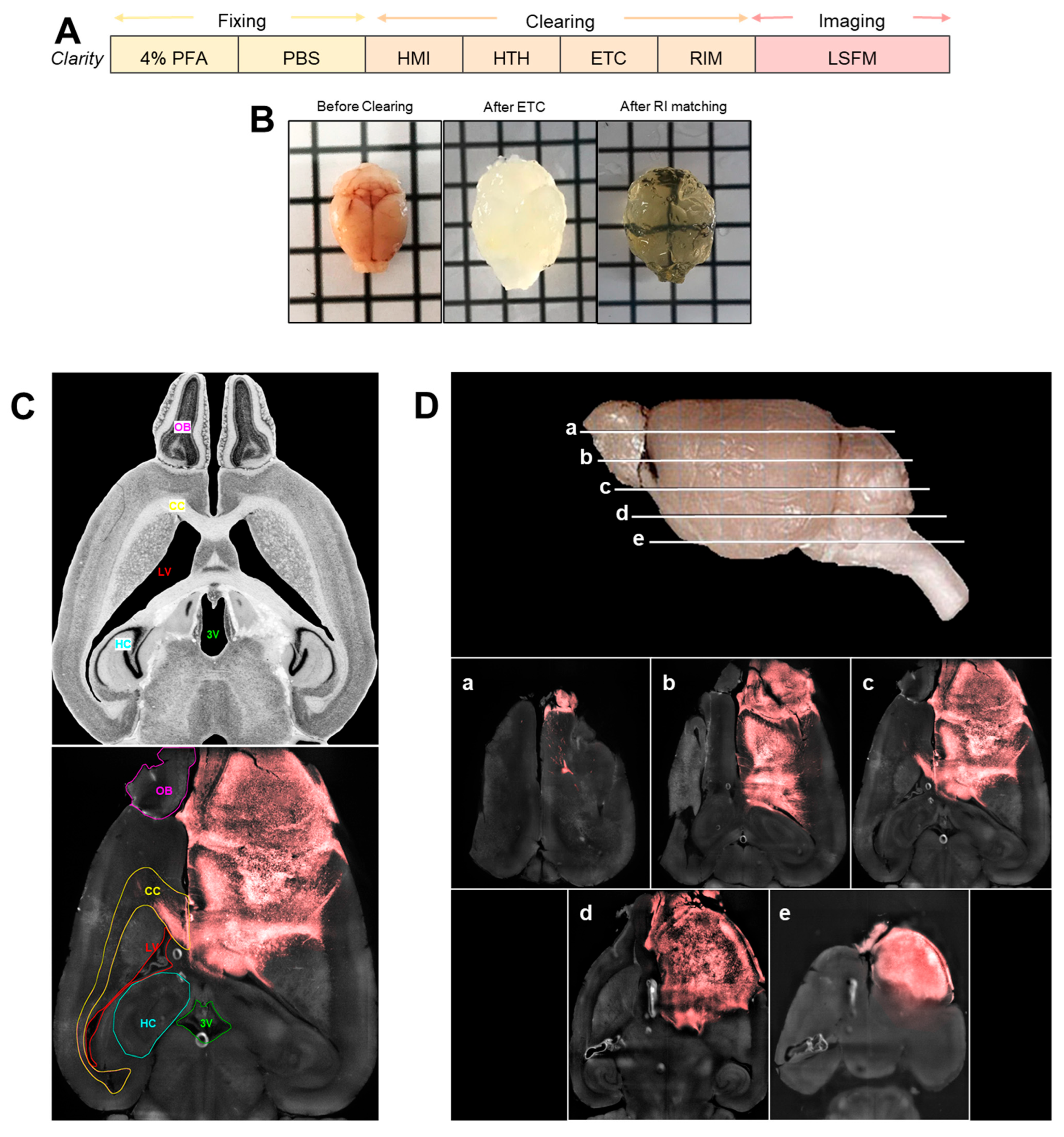
3.2. Complete Mouse Brain Clearing and Tumor Visualization in an Intact, Highly Transparent Brain
3.3. Multimodal Invasion Patterns Captured by 3D Imaging of Cleared Brains
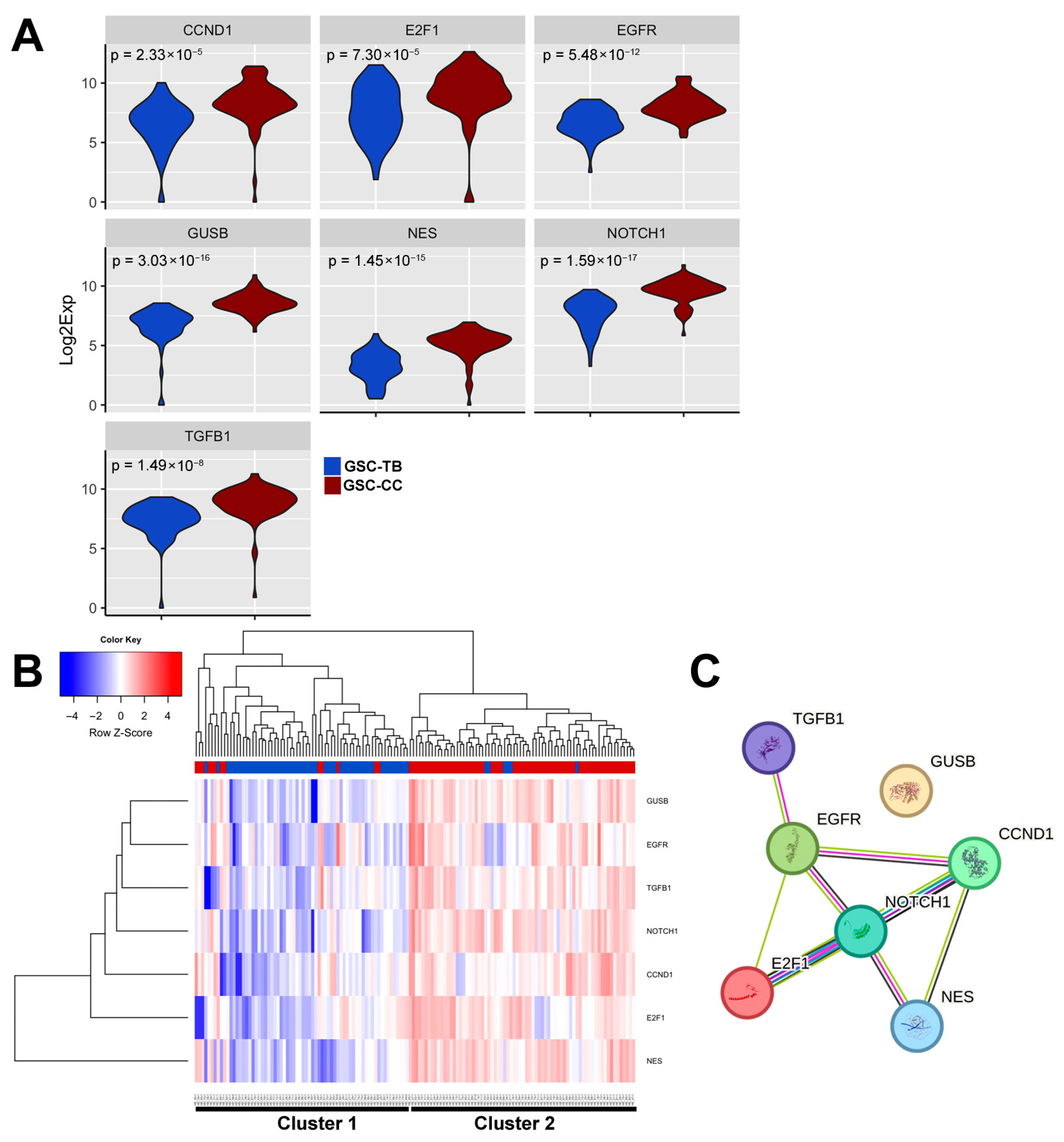
3.4. Spatially Resolved Single-Cell Gene Expression Reveals Distinct Transcriptional States in GSC Subpopulations
3.5. Differential Expression Genes (DEG) Reveal a Corpus Callosum Invasion Signature in Glioma Stem Cells
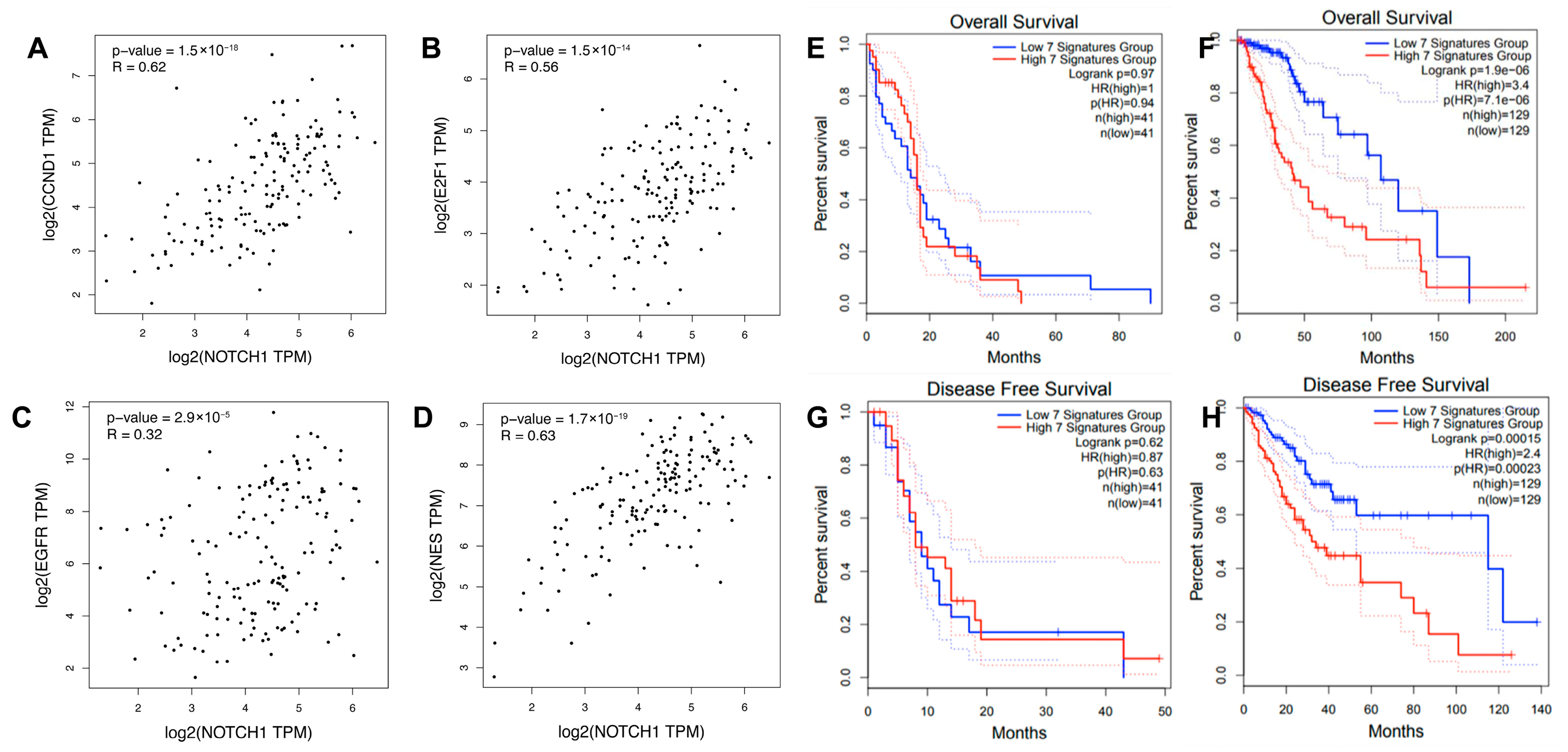
3.6. Confirmation of CC-Iv Signature Co-Expression and Clinical Relevance Using Public GBM Datasets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CC | Corpus Callosum |
| CC-Iv | Corpus Callosum invasion signature |
| CCND1 | Cyclin D1 |
| ECM | Extracellular Matrix |
| FP | red fluorescent protein TurboFP635 |
| GBM | Glioblastoma |
| GEPIA | Gene Expression Profiling Interactive Analysis |
| GSC | Glioma Stem-like Cell |
| HGG | High-Grade Glioma |
| ITH | intratumoral heterogeneity |
| LGG | Low-Grade Glioma |
| MMP | Matrix Metalloproteinase |
| NES | Nestin |
| scRNA-seq | Single-cell RNA sequencing |
| TB | tumor bulk |
| wkpi | weeks post-injection |
References
- Ichikawa, T.; Otani, Y.; Kurozumi, K.; Date, I. Phenotypic Transition as a Survival Strategy of Glioma. Neurol. Med. Chir. 2016, 56, 387–395. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Meyer, M.; Reimand, J.; Lan, X.; Head, R.; Zhu, X.; Kushida, M.; Bayani, J.; Pressey, J.C.; Lionel, A.C.; Clarke, I.D.; et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. USA 2015, 112, 851–856. [Google Scholar] [CrossRef]
- Yeo, A.T.; Rawal, S.; Delcuze, B.; Christofides, A.; Atayde, A.; Strauss, L.; Balaj, L.; Rogers, V.A.; Uhlmann, E.J.; Varma, H.; et al. Single-cell RNA sequencing reveals evolution of immune landscape during glioblastoma progression. Nat. Immunol. 2022, 23, 971–984. [Google Scholar] [CrossRef]
- Rosenberg, S.; Verreault, M.; Schmitt, C.; Guegan, J.; Guehennec, J.; Levasseur, C.; Marie, Y.; Bielle, F.; Mokhtari, K.; Hoang-Xuan, K.; et al. Multi-omics analysis of primary glioblastoma cell lines shows recapitulation of pivotal molecular features of parental tumors. Neuro Oncol. 2017, 19, 219–228. [Google Scholar] [CrossRef]
- Darmanis, S.; Sloan, S.A.; Croote, D.; Mignardi, M.; Chernikova, S.; Samghababi, P.; Zhang, Y.; Neff, N.; Kowarsky, M.; Caneda, C.; et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017, 21, 1399–1410. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Jill, P.; Alexe, G.; et al. Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Shcherbo, D.; Merzlyak, E.M.; Chepurnykh, T.V.; Fradkov, A.F.; Ermakova, G.V.; Solovieva, E.A.; Lukyanov, K.; Bogdanova, E.A.; Zaraisky, A.G.; Lukyanov, S.; et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 2007, 4, 741–746. [Google Scholar] [CrossRef]
- Jandrey, E.H.F.; Barnabé, G.F.; Maldaun, M.; Asprino, P.F.; Dos Santos, N.C.; Inoue, L.T.; Rozanski, A.; Galante, P.A.F.; Marie, S.K.N.; Oba-Shinjo, S.M.; et al. A novel program of infiltrative control in astrocytomas: ADAM23 depletion promotes cell invasion by activating γ-secretase complex. Neuro-Oncol. Adv. 2023, 5, vdad147. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.; Wallace, J.; Kim, S.-Y.; Kalyanasundaram, S.; Andalman, A.S.; Davidson, T.J.; Mirzabekov, J.J.; Zalocusky, K.A.; Mattis, J.; Denisin, A.K.; et al. Structural and molecular interrogation of intact biological systems. Nature 2013, 497, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Voigt, F.F.; Kirschenbaum, D.; Platonova, E.; Pagès, S.; Campbell, R.A.A.; Kastli, R.; Schaettin, M.; Egolf, L.; van der Bourg, A.; Bethge, P.; et al. The mesoSPIM initiative: Open-source light-sheet microscopes for imaging cleared tissue. Nat. Methods 2019, 16, 1105–1108. [Google Scholar] [CrossRef]
- Hörl, D.; Rusak, F.R.; Preusser, F.; Tillberg, P.; Randel, N.; Chhetri, R.K.; Cardona, A.; Keller, P.J.; Harz, H.; Leonhardt, H.; et al. BigStitcher: Reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 2019, 16, 870–874. [Google Scholar] [CrossRef]
- Collins, S.C.; Wagner, C.; Gagliardi, L.; Kretz, P.F.; Fischer, M.; Kessler, P.; Kannan, M.; Yalcin, B. A Method for Parasagittal Sectioning for Neuroanatomical Quantification of Brain Structures in the Adult Mouse. Curr. Protoc. Mouse Biol. 2018, 8, e48. [Google Scholar] [CrossRef]
- DeLaughter, D.M. The Use of the Fluidigm C1 for RNA Expression Analyses of Single Cells. Curr. Protoc. Mol. Biol. 2018, 122, e55. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- De Bonis, P.; Anile, C.; Pompucci, A.; Fiorentino, A.; Balducci, M.; Chiesa, S.; Lauriola, L.; Maira, G.; Mangiola, A. The influence of surgery on recurrence pattern of glioblastoma. Clin. Neurol. Neurosurg. 2013, 115, 37–43. [Google Scholar] [CrossRef]
- Mahdi, A.; Aittaleb, M.; Tissir, F. Targeting Glioma Stem Cells: Therapeutic Opportunities and Challenges. Cells 2025, 14, 675. [Google Scholar] [CrossRef]
- Inoue, A.; Takahashi, H.; Harada, H.; Kohno, S.; Ohue, S.; Kobayashi, K.; Yano, H.; Tanaka, J.; Ohnishi, T. Cancer stem-like cells of glioblastoma characteristically express MMP-13 and display highly invasive activity. Int. J. Oncol. 2012, 40, 1461–1468. [Google Scholar] [CrossRef]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Kohanbash, G.; Liu, S.J.; Alvarado, B.; Carrera, D.; Bhaduri, A.; Watchmaker, P.B.; Yagnik, G.; Di Lullo, E.; Malatesta, M.; et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation. Nat. Genet. 2020, 49, 595–605. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, H.; Hu, J.; Fu, H.; Qu, Y.; Yang, Y.; Cai, K.Q.; Efimov, A.; Wu, M.; Yen, T.; et al. Nestin Is Required for Spindle Assembly and Cell-Cycle Progression in Glioblastoma Cells. Mol. Cancer Res. 2021, 19, 1651–1665. [Google Scholar] [CrossRef]
- Tian, R.; Wang, J.; Yan, H.; Wu, J.; Xu, Q.; Zhan, X.; Gui, Z.; Ding, M.; He, J. Differential expression of miR16 in glioblastoma and glioblastoma stem cells: Their correlation with proliferation, differentiation, metastasis and prognosis. Oncogene 2017, 36, 5861–5873. [Google Scholar] [CrossRef]
- Xie, H.; Lv, S.; Wang, Z.; Yuan, X. E2F transcription factor 1 elevates cyclin D1 expression by suppressing transcription of microRNA-107 to augment progression of glioma. Brain Behav. 2021, 11, e2399. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Guo, Y.; Ning, W.; Wang, X.; Li, S.; Chen, Y.; Ma, L.; Qu, Y.; Song, Y.; Zhang, H. Comprehensive analyses of glucose metabolism in glioma reveal the glioma-promoting effect of GALM. Front. Cell Dev. Biol. 2022, 9, 717182. [Google Scholar] [CrossRef]
- Wang, J.; Wakeman, T.P.; Lathia, J.D.; Hjelmeland, A.B.; Wang, X.F.; White, R.R.; Rich, J.N.; Sullenger, B.A. Notch promotes radioresistance of glioma stem cells. Stem Cells 2010, 28, 17–28. [Google Scholar] [CrossRef]
- Colonna, M.; Konopka, G.; Liddelow, S.A.; Nowakowski, T.; Awatramani, R.; Bateup, H.S.; Cadwell, C.R.; Caglayan, E.; Chen, J.L.; Gillis, J.; et al. Implementation and validation of single-cell genomics experiments in neuroscience. Nat. Neurosci. 2024, 27, 2310–2325. [Google Scholar] [CrossRef]
- Baisiwala, S.; Hall, R.R., III; Saathoff, M.R.; Shireman, J.; Park, C.; Budhiraja, S.; Goel, C.; Warnke, L.; Hardiman, C.; Wang, J.Y.; et al. LNX1 Modulates Notch1 Signaling to Promote Expansion of the Glioma Stem Cell Population during Temozolomide Therapy in Glioblastoma. Cancers 2020, 12, 3505. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tong, F.; Zhao, J.; Cui, X.; Wang, Y.; Wang, G.; Kang, C.; Liu, X.; Wang, Q. Identification of the E2F1-RAD51AP1 axis as a key factor in MGMT-methylated GBM TMZ resistance. Cancer Biol. Med. 2023, 20, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Yin, Y.; Chen, C.; Wan, Q.; Xia, D.; Wang, M.; Pu, Z.; Zhang, B.; Zou, J. Characterization of EGFR-reprogrammable temozolomide-resistant cells in a model of glioblastoma. Cell Death Discov. 2022, 8, 438. [Google Scholar] [CrossRef]
- Namestnikova, D.D.; Gubskiy, I.L.; Cherkashova, E.A.; Sukhinich, K.K.; Melnikov, P.A.; Gabashvili, A.N.; Kurilo, V.V.; Chekhonin, V.P.; Gubsky, L.V.; Yarygin, K.N. Therapeutic Efficacy and Migration of Mesenchymal Stem Cells after Intracerebral Transplantation in Rats. with Experimental Ischemic Stroke. Bull. Exp. Biol. Med. 2023, 175, 116–125. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | Gene Name | HGNC ID | Fold-Change | FDR |
|---|---|---|---|---|
| NES | Nestin | HGNC:7757 | 1.6 | 1.02 × 10−14 |
| CCND1 | Cyclin D1 | HGNC:1582 | 1.3 | 1.70 × 10−7 |
| GUSB | Glucuronidase Beta | HGNC:4696 | 1.3 | 2.12 × 10−15 |
| NOTCH1 | Notch Receptor 1 | HGNC:7881 | 1.25 | 1.11 × 10−16 |
| E2F1 | E2F Transcription Factor 1 | HGNC:3113 | 1.21 | 5.11 × 10−4 |
| EGFR | Epidermal Growth Factor Receptor | HGNC:3236 | 1.2 | 3.84 × 10−11 |
| TGFB1 | Transforming Growth Factor Beta 1 | HGNC:11766 | 1.2 | 1.04 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, N.; Aquino, A.; Preußer, F.; Rusak, F.R.; Jandrey, E.H.F.; Uno, M.; Furuya, T.K.; Lancellotti, C.L.P.; Maldaun, M.V.C.; Chammas, R.; et al. Innovative In Vivo Imaging and Single Cell Expression from Tumor Bulk and Corpus Callosum Reveal Glioma Stem Cells with Unique Regulatory Programs. Cancers 2025, 17, 3851. https://doi.org/10.3390/cancers17233851
dos Santos N, Aquino A, Preußer F, Rusak FR, Jandrey EHF, Uno M, Furuya TK, Lancellotti CLP, Maldaun MVC, Chammas R, et al. Innovative In Vivo Imaging and Single Cell Expression from Tumor Bulk and Corpus Callosum Reveal Glioma Stem Cells with Unique Regulatory Programs. Cancers. 2025; 17(23):3851. https://doi.org/10.3390/cancers17233851
Chicago/Turabian Styledos Santos, Natalia, Aline Aquino, Friedrich Preußer, Fabio Rojas Rusak, Elisa Helena Farias Jandrey, Miyuki Uno, Tatiane Katsue Furuya, Carmen Lucia Penteado Lancellotti, Marcos Vinicius Calfat Maldaun, Roger Chammas, and et al. 2025. "Innovative In Vivo Imaging and Single Cell Expression from Tumor Bulk and Corpus Callosum Reveal Glioma Stem Cells with Unique Regulatory Programs" Cancers 17, no. 23: 3851. https://doi.org/10.3390/cancers17233851
APA Styledos Santos, N., Aquino, A., Preußer, F., Rusak, F. R., Jandrey, E. H. F., Uno, M., Furuya, T. K., Lancellotti, C. L. P., Maldaun, M. V. C., Chammas, R., Preibisch, S., Camargo, A. A., Masotti, C., & Costa, E. T. (2025). Innovative In Vivo Imaging and Single Cell Expression from Tumor Bulk and Corpus Callosum Reveal Glioma Stem Cells with Unique Regulatory Programs. Cancers, 17(23), 3851. https://doi.org/10.3390/cancers17233851






