Trichothecenes in Cereal Grains – An Update
Abstract
:1. Introduction: Trichothecene-Producing Fungi and Their Impact on Food and Feed
2. Trichothecene Structure, Biosynthesis and Chemotype Distribution
2.1. Chemical Structure
2.2. Trichothecene Biosynthesis
2.3. Fusarium Species and Chemotype Distribution
3. Trichothecene Toxicity
3.1. Disruption of Eukaryotic Protein Synthesis
3.2. Structure-Activity Relationship: Importance of the 12,13-Epoxide
3.3. Structure-Activity Relationship: Influence of Substitution Patterns to the Trichothecene Core
4. Molecular Mechanisms of Trichothecene Resistance
4.1. Trichothecene Resistance at the Ribosome
4.2. Trichothecene Efflux
4.3. Enzymatic Detoxification of Trichothecenes
4.4. Other Trichothecene Resistance Genes
5. Fusarium Resistance to Limit Trichothecene Contamination of Food and Feed
5.1. Types and Forms of Fusarium Resistance
5.2. Screening for Fusarium and Trichothecene Resistance in Cereals
5.3. Trichothecences and FHB Pathogenicity
5.4. Breeding for Low Trichothecence Content in Wheat and Barley Grains
6. Food and Feed Safety
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Wilkins, K.; Nielsen, K.F.; Din, S.U. Patterns of volatile metabolites and nonvolatile trichothecenes produced by isolates of Stachybotrys, Fusarium, Trichoderma, Trichothecium and Memnoniella. Environ. Sci. Pollut. Res. 2003, 10, 162. [Google Scholar] [CrossRef]
- Abbas, H.K.; Johnson, B.B.; Shier, W.T.; Tak, H.; Jarvis, B.B.; Boyette, C.D. Phytotoxicity and mammalian cytotoxicity of macrocyclic trichothecene mycotoxins from Myrothecium verrucaria. Phytochem 2002, 59, 309–313. [Google Scholar] [CrossRef]
- Kang, Z.; Buchenauer, H. Immunocytochemical localization of fusarium toxins in infected wheat spikes by Fusarium culmorum. Physiol. Mol. Plant Pathol. 1999, 55, 275–288. [Google Scholar] [CrossRef]
- Geiser, D.M.; Aoki, T.; Bacon, C.W.; Baker, S.E.; Bhattacharyya, M.K.; Brandt, M.E.; Brown, D.W.; Burgess, L.W.; Chulze, S.; Coleman, J.J.; et al. One fungus, one name: Defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 2013, 103, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili Taheri, A.; Chatterton, S.; Foroud, N.A.; Gossen, B.D.; McLaren, D.L. Identification and community dynamics of fungi associated with root, crown, and foot rot of field pea in western Canada. Eur. J. Plant Pathol. 2017, 147, 489–500. [Google Scholar] [CrossRef]
- Saleh, A.A.; El_Komy, M.H.; Eranthodi, A.; Hamoud, A.S.; Molan, Y.Y. Variation in a molecular marker for resistance of Saudi date palm germplasm to Fusarium oxysporum f. sp. albedinis the causal agent of Bayoud disease. Eur. J. Plant Pathol. 2015, 143, 507–514. [Google Scholar] [CrossRef]
- Yang, J.-W.; Nam, S.-S.; Lee, H.-U.; Choi, K.-H.; Hwang, S.-G.; Paul, N.C. Fusarium root rot caused by Fusarium solani on sweet potato (Ipomoea batatas) in South Korea. Can. J. Plant Pathol. 2018, 40, 90–95. [Google Scholar] [CrossRef]
- Gao, M.L.; Luan, Y.S.; Yu, H.N.; Bao, Y.M. First report of tomato leaf spot caused by Fusarium proliferatum in China. Can. J. Plant Pathol. 2016, 38, 400–404. [Google Scholar] [CrossRef]
- Maryani, N.; Lombard, L.; Poerba, Y.S.; Subandiyah, S.; Crous, P.W.; Kema, G.H.J. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud. Mycol. 2019, 92, 155–194. [Google Scholar]
- O’Donnell, K.; Kistler, H.C.; Tacke, B.K.; Casper, H.H. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 2000, 97, 7905–7910. [Google Scholar] [CrossRef]
- O’Donnell, K.; Ward, T.J.; Aberra, D.; Kistler, H.C.; Aoki, T.; Orwig, N.; Kimura, M.; Bjørnstad, Å.; Klemsdal, S.S. Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genet. Biol. 2008, 45, 1514–1522. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Ward, T.J.; Geiser, D.M.; Corby Kistler, H.; Aoki, T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004, 41, 600–623. [Google Scholar] [CrossRef] [PubMed]
- Starkey, D.E.; Ward, T.J.; Aoki, T.; Gale, L.R.; Kistler, H.C.; Geiser, D.M.; Suga, H.; Toth, B.; Varga, J.; O’Donnell, K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007, 44, 1191–1204. [Google Scholar] [CrossRef] [PubMed]
- Yli-Mattila, T.; Gagkaeva, T.; Ward, T.J.; Aoki, T.; Kistler, H.C.; O’Donnell, K. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia 2009, 101, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Ndoye, M.; Zhang, J.B.; Li, H.P.; Liao, Y.C. Population structure and genetic diversity of the Fusarium graminearum species complex. Toxins 2011, 3, 1020–1037. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.C.; Clear, R.M.; O’Donnell, K.; McCormick, S.; Turkington, T.K.; Tekauz, A.; Gilbert, J.; Kistler, H.C.; Busman, M.; Ward, T.J. Diversity of Fusarium head blight populations and trichothecene toxin types reveals regional differences in pathogen composition and temporal dynamics. Fungal Genet. Biol. 2015, 82, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Ward, T.J.; Kistler, H.C.; O’Donnell, K. Systematics, phylogeny and trichothecene mycotoxin potential of Fusarium head blight cereal pathogens. Mycotoxins 2012, 62, 91–102. [Google Scholar] [CrossRef]
- van der Lee, T.; Zhang, H.; van Diepeningen, A.; Waalwijk, C. Biogeography of Fusarium graminearum species complex and chemotypes: A review. Food Addit. Contam. 2015, 32, 453–460. [Google Scholar] [CrossRef]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. MPMI Mol. Plant Microbe Interact. 1995, 8, 593–601. [Google Scholar] [CrossRef]
- Maier, F.J.; Miedaner, T.; Hadeler, B.; Felk, A.; Salomon, S.; Lemmens, M.; Kassner, H.; Schaefer, W. Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 2006, 7, 449–461. [Google Scholar] [CrossRef]
- Wegulo, S.N. Factors influencing deoxynivalenol accumulation in small grain cereals. Toxins 2012, 4, 1157–1180. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.; Koopmann, B.; Döll, K.; Karlovsky, P.; Kropf, U.; Schlüter, K.; von Tiedemann, A. Mechanisms regulating grain contamination with trichothecenes translocated from the stem base of Wheat (Triticum aestivum) infected with Fusarium culmorum. Phytopathology 2013, 103, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Mudge, A.M.; Dill-Macky, R.; Dong, Y.; Gardiner, D.M.; White, R.G.; Manners, J.M. A role for the mycotoxin deoxynivalenol in stem colonisation during crown rot disease of wheat caused by Fusarium graminearum and Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 2006, 69, 73–85. [Google Scholar] [CrossRef]
- Bucheli, T.D.; Wettstein, F.E.; Hartmann, N.; Erbs, M.; Vogelgsang, S.; Forrer, H.R.; Schwarzenbach, R.P. Fusarium mycotoxins: Overlooked aquatic micropollutants? J. Agric. Food Chem. 2008, 56, 1029–1034. [Google Scholar] [CrossRef]
- Kolpin, D.W.; Schenzel, J.; Meyer, M.T.; Phillips, P.J.; Hubbard, L.E.; Scott, T.M.; Bucheli, T.D. Mycotoxins: Diffuse and point source contributions of natural contaminants of emerging concern to streams. Sci. Total Environ. 2014, 470–471, 669–676. [Google Scholar] [CrossRef]
- Foroud, N.A.; Chatterton, S.; Reid, L.M.; Turkington, T.K.; Tittlemier, S.A.; Gräfenhan, T. Fusarium diseases of Canadian grain crops: Impact and disease management strategies. In Future Challenges in Crop Protection Against Fungal Pathogens; Goyal, A., Manoharachary, C., Eds.; Springer: New York, NY, USA, 2014; pp. 267–316. [Google Scholar]
- Mesterházy, Á.; Lemmens, M.; Reid, L.M. Breeding for resistance to ear rots caused by Fusarium spp. in maize—A review. Plant Breed. 2012, 131, 1–19. [Google Scholar] [CrossRef]
- Moretti, A.; Panzarini, G.; Somma, S.; Campagna, C.; Ravaglia, S.; Logrieco, A.F.; Solfrizzo, M. Systemic growth of F. graminearum in wheat plants and related accumulation of deoxynivalenol. Toxins 2014, 6, 1308–1324. [Google Scholar] [CrossRef]
- Backhouse, D.; Abubakar, A.A.; Burgess, L.W.; Dennis, J.I.; Hollaway, G.J.; Wildermuth, G.B.; Wallwork, H.; Henry, F.J. Survey of Fusarium species associated with crown rot of wheat and barley in eastern Australia. Australas. Plant Pathol. 2004, 33, 255–261. [Google Scholar] [CrossRef]
- Garreau de Loubresse, N.; Prokhorova, I.; Holtkamp, W.; Rodnina, M.V.; Yusupova, G.; Yusupov, M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014, 513, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Ueno, Y. Mode of action of trichothecenes. Pure Appl. Chem. 1977, 49, 1737–1745. [Google Scholar] [CrossRef]
- McLaughlin, C.S.; Vaughn, M.H.; Campbell, J.M.; Wei, C.M.; Stafford, M.E. Inhibition of protein synthesis by trichothecenes. In Mycotoxins in Human and Health; Rodricks, J.V., Hesseltine, C.W., Mehlman, M.A., Eds.; Pathotox Publishers: Park Forest South, IL, USA, 1977; pp. 263–275. [Google Scholar]
- Alexander, N.J.; McCormick, S.P.; Hohn, T.M. TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: Gene isolation and expression in yeast. Mol. Gen. Genet. 1999, 261, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J. Toxicological mechanisms and potential health effects of deoxynivalenol and nivalenol. World Mycotoxin J. 2010, 3, 323–347. [Google Scholar] [CrossRef]
- McCormick, S.P. Phytotoxicity of trichothecenes. In Mycotoxin Prevention and Control in Agriculture; Appell, M., Kendra, D.F., Trucksess, M.W., Eds.; American Chemical Society: Washington, DC, USA, 2009; pp. 143–155. [Google Scholar]
- Rocha, O.; Ansari, K.; Doohan, F.M. Effects of trichothecene mycotoxins on eukaryotic cells: A review. Food Addit. Contam. 2005, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Yagen, B.; Joffe, A.Z. Screening of toxic isolates of Fusarium poae and Fusarium sporotrichioides involved in causing alimentary toxic aleukia. Appl. Environ. Microbiol. 1976, 32, 423–427. [Google Scholar] [PubMed]
- Hendry, K.M.; Cole, E.C. A review of mycotoxins in indoor air. J. Toxicol. Environ. Health Sci. 1993, 38, 183–198. [Google Scholar] [CrossRef]
- Schoental, R. Mycotoxins in food and the plague of Athens. J. Nutr. Med. 1994, 4, 83–85. [Google Scholar] [CrossRef]
- Stack, R.W. History of Fusarium head blight with emphasis on North America. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2003; pp. 1–34. [Google Scholar]
- Mayer, C.F. Endemic panmyelotoxicosis in the Russian grain belt. I. The clinical aspects of alimentary toxic aleukia (ATA); a comprehensive review. Mil. Surg. 1953, 113, 173–189. [Google Scholar]
- Sarkisov, A.H. Mycotoxicoses (Fungal Poisonings); In State Publisher of Agricultural Literature: Moscow, Russia, 1954; p. 216. (In Russian) [Google Scholar]
- Shalak, A.V. To the evaluation of the scale of starvation 1946-1947. J. Econ. Hist. Hist. Econ. 2009, 10, 100–108. Available online: http://jhist.bgu.ru/reader/article.aspx?id=18942 (accessed on 22 January 2016).
- Ueno, Y. Trichothecenes: Chemical, Biological and Toxicological Aspects; Elsevier Scientific Publishers: Amsterdam, The Netherlands, 1983. [Google Scholar]
- Rotter, B.A.; Prelusky, D.B.; Pestka, J.J. Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Environ. Health Sci. 1996, 48, 1–34. [Google Scholar] [CrossRef]
- Wu, W.; Flannery, B.M.; Sugita-Konishi, Y.; Watanabe, M.; Zhang, H.; Pestka, J.J. Comparison of murine anorectic responses to the 8-ketotrichothecenes 3-acetyldeoxynivalenol, 15-acetyldeoxynivalenol, fusarenon X and nivalenol. Food Chem. Toxicol. 2012, 50, 2056–2061. [Google Scholar] [CrossRef]
- Forsyth, D.M.; Yoshizawa, T.; Morooka, N.; Tuite, J. Emetic and refusal activity of deoxynivalenol to swine. Appl. Environ. Microbiol. 1977, 34, 547–552. [Google Scholar] [PubMed]
- Alm, H.; Greising, T.; Brüssow, K.P.; Torner, H.; Tiemann, U. The influence of the mycotoxins deoxynivalenol and zearalenol on in vitro maturation of pig oocytes and in vitro culture of pig zygotes. Toxicol. Vitr. 2002, 16, 643–648. [Google Scholar] [CrossRef]
- Tiemann, U.; Dänicke, S. In vivo and in vitro effects of the mycotoxins zearalenone and deoxynivalenol on different non-reproductive and reproductive organs in female pigs: A review. Food Addit. Contam. 2007, 24, 306–314. [Google Scholar] [CrossRef]
- Bonnet, M.S.; Roux, J.; Mounien, L.; Dallaporta, M.; Troadec, J.D. Advances in deoxynivalenol toxicity mechanisms: The brain as a target. Toxins 2012, 4, 1120–1138. [Google Scholar] [CrossRef] [PubMed]
- Naumov, N.A. Tempulent corn. In Observations under Some Species of Genus Fusarium; Bulletin of the Mycological and Plant Pathology Society: Petrograd, Russia, 1916. (In Russian) [Google Scholar]
- Voronin, M.C. About tempulent corn in the South-Ussuriiskij region. Bot. Notes 1890, 3, 13–21. (In Russian) [Google Scholar]
- Jaczewski, A.A. About tempulent corn. Sheet Inf. Dis. Control Russ. 1904, 11, 89–92. (In Russian) [Google Scholar]
- Morooka, N.; Uratsuji, N.; Yoshizawa, T.; Yamamoto, H. Studies on the toxic substances in barley Infected with Fusarium spp. Food Hyg. Saf. Sci. (Shokuhin Eiseigaku Zasshi) 1972, 13, 368–375. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Morooka, N. Deoxynivalenol and its monoacetate: New mycotoxins from Fusarium roseum and moldy barley. Agric. Biol. Chem. 1973, 37, 2933–2934. [Google Scholar] [CrossRef]
- Zouagui, Z.; Asrar, M.; Lakhdissi, H.; Abdennebi, E. Prevention of mycotoxin effects in dairy cows by adding an anti-mycotoxin product in feed. J. Mater. Environ. Sci. 2017, 8, 3766–3770. [Google Scholar]
- Foroud, N.A.; Eudes, F. Trichothecenes in cereal grains. Int. J. Mol. Sci. 2009, 10, 147–173. [Google Scholar] [CrossRef]
- Cole, R.J.; Cox, R.H. The trichothecenes. In Handbook of Toxic Fungal Metabolites; Cole, R.J., Cox, R.H., Eds.; Academic Press: Toronto, ON, Canada, 1981; pp. 152–263. [Google Scholar]
- Wei, C.M.; McLaughlin, C.S. Structure-function relationship in the 12, 13-epoxytrichothecenes novel inhibitors of protein synthesis. Biochem. Biophys. Res. Commun. 1974, 57, 838–844. [Google Scholar] [CrossRef]
- Cole, R.; Schweikert, M.; Jarvis, B. Handbook of Fungal Secondary Metabolites; Academic Press: San Diego, CA, USA, 2003. [Google Scholar]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From simple to complex mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Varga, E.; Wiesenberger, G.; Hametner, C.; Ward, T.J.; Dong, Y.; Schöfbeck, D.; McCormick, S.; Broz, K.; Stückler, R.; Schuhmacher, R.; et al. New tricks of an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015, 17, 2588–2600. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.M.; Xayamongkhon, H.; Broz, K.; Dong, Y.; McCormick, S.P.; Abramova, S.; Ward, T.J.; Ma, Z.H.; Kistler, H.C. Temporal dynamics and population genetic structure of Fusarium graminearum in the upper Midwestern United States. Fungal Genet. Biol. 2014, 73, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Malmierca, M.G.; Cardoza, R.E.; Alexander, N.J.; McCormick, S.P.; Collado, I.G.; Hermosa, R.; Monte, E.; Gutiérrez, S. Relevance of trichothecenes in fungal physiology: Disruption of tri5 in Trichoderma arundinaceum. Fungal Genet. Biol. 2013, 53, 22–33. [Google Scholar] [CrossRef]
- Semeiks, J.; Borek, D.; Otwinowski, Z.; Grishin, N.V. Comparative genome sequencing reveals chemotype-specific gene clusters in the toxigenic black mold Stachybotrys. BMC Genom. 2014, 15, 590. [Google Scholar] [CrossRef]
- Cardoza, R.E.; Malmierca, M.G.; Hermosa, M.R.; Alexander, N.J.; McCormick, S.P.; Proctor, R.H.; Tijerino, A.M.; Rumbero, A.; Monte, E.; Gutiérrez, S. Identification of loci and functional characterization of trichothecene biosynthesis genes in filamentous fungi of the genus Trichoderma. Appl. Environ. Microbiol. 2011, 77, 4867–4877. [Google Scholar] [CrossRef]
- Trapp, S.C.; Hohn, T.M.; McCormick, S.; Jarvis, B.B. Characterization of the gene cluster for biosynthesis of macrocyclic trichothecenes in Myrothecium roridum. Mol. Gen. Genet. 1998, 257, 421–432. [Google Scholar] [CrossRef]
- Brown, D.W.; Dyer, R.B.; McCormick, S.P.; Kendra, D.F.; Plattner, R.D. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 2004, 41, 454–462. [Google Scholar] [CrossRef]
- Brown, D.W.; Proctor, R.H.; Dyer, R.B.; Plattner, R.D. Characterization of a Fusarium 2-gene cluster involved in trichothecene C-8 modification. J. Agric. Food Chem. 2003, 51, 7936–7944. [Google Scholar] [CrossRef]
- Kimura, M.; Yamaguchi, I. The mystery of the trichothecene 3-O-acetyltransferase gene: Tri101 evolved independently of other trichothecene biosynthetic genes in the gene cluster. Pestic. Sci. 1999, 55, 372–374. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Larson, T.M.; Jurgenson, J.E. Expression of Tri15 in Fusarium sporotrichioides. Curr. Genet. 2004, 45, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Cane, D.E.; Ha, H.J.; Pargellis, C.; Waldmeier, F.; Swanson, S.; Murthy, P.P.N. Trichodiene biosynthesis and the stereochemistry of the enzymatic cyclization of farnesyl pyrophosphate. Bioorganic Chem. 1985, 13, 246–265. [Google Scholar] [CrossRef]
- Canevascini, S.; Caderas, D.; Mandel, T.; Fleming, A.J.; Dupuis, I.; Kuhlemeier, C. Tissue-specific expression and promoter analysis of the tobacco Itp1 gene. Plant Physiol. 1996, 112, 513–524. [Google Scholar] [CrossRef]
- Hohn, T.M.; Beremand, P.D. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 1989, 79, 131–138. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Hohn, T.M.; McCormick, S.P. Effect of gene disruption of trichodiene synthase on the virulence of Gibberella pulicaris. Mol. Plant Microbe Interact. 1992, 5, 214–222. [Google Scholar] [CrossRef]
- Hohn, T.M.; Desjardins, A.E.; McCormick, S.P. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 1995, 248, 95–102. [Google Scholar] [CrossRef]
- Tokai, T.; Koshino, H.; Takahashi, A.N.; Sato, M.; Fujimura, M.; Kimura, M. Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem. Biophys. Res. Commun. 2007, 353, 412–417. [Google Scholar] [CrossRef]
- McCormick, S.P.; Alexander, N.J.; Proctor, R.H. Fusarium Tri4 encodes a multifunctional oxygenase required for trichothecene biosynthesis. Can. J. Microbiol. 2006, 52, 636–642. [Google Scholar] [CrossRef]
- McCormick, S.P.; Taylor, S.L.; Plattner, R.D.; Beremand, M.N. Bioconversion of possible T-2 toxin precursors by a mutant strain of Fusarium sporotrichioides NRRL 3299. Appl. Environ. Microbiol. 1990, 56, 702–706. [Google Scholar]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- McCormick, S.P.; Harris, L.J.; Alexander, N.J.; Ouellet, T.; Saparno, A.; Allard, S.; Desjardins, A.E. Tri1 in Fusarium graminearum encodes a P450 oxygenase. Appl. Environ. Microbiol. 2004, 70, 2044–2051. [Google Scholar] [CrossRef] [PubMed]
- Meek, I.B.; Peplow, A.W.; Ake, C., Jr.; Phillips, T.D.; Beremand, M.N. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl. Environ. Microbiol. 2003, 69, 1607–1613. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P.; Hohn, T.M.; Desjardins, A.E. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 1996, 62, 353–359. [Google Scholar]
- Kimura, M.; Tokai, T.; Matsumoto, G.; Fujimura, M.; Hamamoto, H.; Yoneyama, K.; Shibata, T.; Yamaguchi, I. Trichothecene nonproducer Gibberella species have both functional and nonfunctional 3-O-acetyltransferase genes. Genetics 2003, 163, 677–684. [Google Scholar]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Restoration of wild-type virulence to Tri5 disruption mutants of Gibberella zeae via gene reversion and mutant complementation. Microbiology 1997, 143, 2583–2591. [Google Scholar] [CrossRef]
- Lee, T.; Han, Y.K.; Kim, K.H.; Yun, S.H.; Lee, Y.W. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 2002, 68, 2148–2154. [Google Scholar] [CrossRef]
- Brown, D.W.; McCormick, S.P.; Alexander, N.J.; Proctor, R.H.; Desjardins, A.E. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol. 2001, 32, 121–133. [Google Scholar] [CrossRef]
- McCormick, S.P.; Alexander, N.J. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 2002, 68, 2959–2964. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Waalwijk, C.; van der Lee, T.; Proctor, R.H. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet. Biol. 2011, 48, 485–495. [Google Scholar] [CrossRef]
- Alexander, N.J.; Hohn, T.M.; McCormick, S.P. The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 1998, 64, 221–225. [Google Scholar] [PubMed]
- Brown, D.W.; McCormick, S.P.; Alexander, N.J.; Proctor, R.H.; Desjardins, A.E. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol. 2002, 36, 224–233. [Google Scholar] [CrossRef]
- McCormick, S.P.; Alexander, N.J.; Trapp, S.E.; Hohn, T.M. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl. Environ. Microbiol. 1999, 65, 5252–5256. [Google Scholar] [PubMed]
- Kimura, M.; Shingu, Y.; Yoneyama, K.; Yamaguchi, I. Features of Tri101, the trichothecene 3-O-acetyltransferase gene, related to the self-defense mechanism in Fusarium graminearum. Biosci. Biotechnol. Biochem. 1998, 62, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P.; Desjardins, A.E. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 1995, 61, 1923–1930. [Google Scholar] [PubMed]
- Nasmith, C.G.; Walkowiak, S.; Wang, L.; Leung, W.W.Y.; Gong, Y.; Johnston, A.; Harris, L.J.; Guttman, D.S.; Subramaniam, R. Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum. PLoS Pathog. 2011, 7, e1002266. [Google Scholar] [CrossRef]
- Peplow, A.W.; Tag, A.G.; Garifullina, G.F.; Beremand, M.N. Identification of new genes positively regulated by Tri10 and a regulatory network for trichothecene mycotoxin production. Appl. Environ. Microbiol. 2003, 69, 2731–2736. [Google Scholar] [CrossRef]
- Tag, A.G.; Garifullina, G.F.; Peplow, A.W.; Ake, C., Jr.; Phillips, T.D.; Hohn, T.M.; Beremand, M.N. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 2001, 67, 5294–5302. [Google Scholar] [CrossRef]
- Menke, J.; Dong, Y.; Kistler, H.C. Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation. Mol. Plant Microbe Interact. 2012, 25, 1408–1418. [Google Scholar] [CrossRef]
- Nakajima, Y.; Koseki, N.; Sugiura, R.; Tominaga, N.; Maeda, K.; Tokai, T.; Izawa, M.; Kanamaru, K.; Kamakura, T.; Kobayashi, T.; et al. Effect of disrupting the trichothecene efflux pump encoded by FgTri12 in the nivalenol chemotype of Fusarium graminearum. J. Gen. Appl. Microbiol. 2015, 61, 93–96. [Google Scholar] [CrossRef]
- Dyer, R.B.; Plattner, R.D.; Kendra, D.F.; Brown, D.W. Fusarium graminearum TRI14 is required for high virulence and DON production on wheat but not for DON synthesis in vitro. J. Agric. Food Chem. 2005, 53, 9281–9287. [Google Scholar] [CrossRef] [PubMed]
- Wuchiyama, J.; Kimura, M.; Yamaguchi, I. A trichothecene efflux pump encoded by Tri102 in the biosynthetic gene cluster of Fusarium graminearum. J. Antibiot. 2000, 53, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, A.E. Natural product chemistry meets genetics: When is a genotype a chemotype? J. Agric. Food Chem. 2008, 56, 7587–7592. [Google Scholar] [CrossRef]
- Hohn, T.M.; Krishna, R.; Proctor, R.H. Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 1999, 26, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Ravensdale, M.; Rocheleau, H.; Wang, L.; Nasmith, C.; Ouellet, T.; Subramaniam, R. Components of priming-induced resistance to Fusarium head blight in wheat revealed by two distinct mutants of Fusarium graminearum. Mol. Plant Pathol. 2014, 15, 948–956. [Google Scholar] [CrossRef]
- Scherm, B.; Orrù, M.; Balmas, V.; Spanu, F.; Azara, E.; Delogu, G.; Hammond, T.M.; Keller, N.P.; Migheli, Q. Altered trichothecene biosynthesis in TRI6-silenced transformants of Fusarium culmorum influences the severity of crown and foot rot on durum wheat seedlings. Mol. Plant Pathol. 2011, 12, 759–771. [Google Scholar] [CrossRef]
- Seong, K.Y.; Pasquali, M.; Zhou, X.; Song, J.; Hilburn, K.; McCormick, S.; Dong, Y.; Xu, J.R.; Kistler, H.C. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 2009, 72, 354–367. [Google Scholar] [CrossRef]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef]
- Brown, N.A.; Bass, C.; Baldwin, T.K.; Chen, H.; Massot, F.; Carion, P.W.C.; Urban, M.; van de Meene, A.M.L.; Hammond-Kosack, K.E. Characterisation of the Fusarium graminearum-wheat floral interaction. J. Pathog. 2011, 2011, 9. [Google Scholar] [CrossRef]
- Harris, L.J.; Alexander, N.J.; Saparno, A.; Blackwell, B.; McCormick, S.P.; Desjardins, A.E.; Robert, L.S.; Tinker, N.; Hattori, J.; Piche, C.; et al. A novel gene cluster in Fusarium graminearum contains a gene that contributes to butenolide synthesis. Fungal Genet. Biol. 2007, 44, 293–306. [Google Scholar] [CrossRef]
- Lysøe, E.; Bone, K.R.; Klemsdal, S.S. Real-time quantitative expression studies of the zearalenone biosynthetic gene cluster in Fusarium graminearum. Phytopathology 2009, 99, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Mirocha, C.J.; Xie, W.; Filho, E.R. Chemistry and detection of Fusarium mycotoxins. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2003; pp. 144–164. [Google Scholar]
- Schoental, R. Trichothecenes, zearalenone, and other carcinogenic metabolites of Fusarium and related microfungi. In Advances in Cancer Research; Klein, G., Sidney, W., Eds.; Academic Press: Cambridge, MA, USA, 1985; Volume 45, pp. 217–290. [Google Scholar]
- Yerkovich, N.; Palazzini, J.M.; Sulyok, M.; Chulze, S.N. Trichothecene genotypes, chemotypes and zearalenone production by Fusarium graminearum species complex strains causing Fusarium head blight in Argentina during an epidemic and non-epidemic season. Trop. Plant Pathol. 2017, 42, 190–196. [Google Scholar] [CrossRef]
- Jestoi, M. Emerging Fusarium-mycotoxins fusaproliferin, beauvericin, enniatins, and moniliformin—A review. Crit. Rev. Food Sci. Nutr. 2008, 48, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Bottalico, A.; Perrone, G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur. J. Plant Pathol. 2002, 108, 611–624. [Google Scholar] [CrossRef]
- Tittlemier, S.A.; Roscoe, M.; Trelka, M.; Gaba, D.; Chan, J.; Patrick, S.; Sulyok, M.; Krska, R.; McKendry, T.; Gräfenhan, T. Fusarium damage in cereal grains from western Canada. 2. Occurrence of mycotoxins and their source organisms in Fusarium damaged durum wheat harvested in 2010. J. Agric. Food Chem. 2013, 61, 5438–5448. [Google Scholar] [CrossRef]
- Reid, L.M.; Nicol, R.W.; Ouellet, T.; Savard, M.; Miller, J.D.; Young, J.C.; Stewart, D.W.; Schaafsma, A.W. Interaction of Fusarium graminearum and F. moniliforme in maize ears: Disease progress, fungal biomass, and mycotoxin accumulation. Phytopathology 1999, 89, 1028–1037. [Google Scholar] [CrossRef]
- Tóth, B.; Mesterházy, Á.; Horvath, Z.; Bartók, T.; Varga, M.; Varga, J. Genetic variability of central European isolates of the Fusarium graminearum species complex. Eur. J. Plant Pathol. 2005, 113, 35–45. [Google Scholar] [CrossRef]
- Ward, T.J.; Bielawski, J.P.; Kistler, H.C.; Sullivan, E.; O’Donnell, K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 2002, 99, 9278–9283. [Google Scholar] [CrossRef]
- McMullen, M.P.; Bergstrom, G.C.; De Wolf, E.; Dill-Macky, R.; Hershman, D.E.; Shaner, G.; Van Sanford, D.A. A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis. 2012, 96, 1712–1728. [Google Scholar] [CrossRef]
- Stępień, Ł.; Chełkowski, J. Fusarium head blight of wheat: Pathogenic species and their mycotoxins. World Mycotoxin J. 2010, 3, 107–119. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Gagkaeva, T. Molecular chemotyping of Fusarium graminearum, F. culmorum, and F. cerealis isolates from Finland and Russia. In Molecular Identification of Fungi; Gherbawy, Y., Voigt, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 159–177. [Google Scholar]
- Doohan, F.M.; Brennan, J.; Cooke, B.M. Influence of climatic factors on Fusarium species pathogenic to cereals. Eur. J. Plant Pathol. 2003, 109, 755–768. [Google Scholar] [CrossRef]
- Miller, J.D.; Greenhalgh, R.; Wang, Y.; Lu, M. Trichothecene chemotypes of three Fusarium species. Mycologia 1991, 83, 121–130. [Google Scholar] [CrossRef]
- Mirocha, C.J.; Abbas, H.K.; Windels, C.E.; Xie, W. Variation in deoxynivalenol, 15-acetyldeoxynivalenol, 3-acetyldeoxynivalenol, and zearalenone production by Fusarium graminearum isolates. Appl. Environ. Microbiol. 1989, 55, 1315–1316. [Google Scholar] [PubMed]
- Yoshizawa, T.; Jin, Y.Z. Natural occurrence of acetylated derivatives of deoxynivalenol and nivalenol in wheat and barley in Japan. Food Addit. Contam. 1995, 12, 689–694. [Google Scholar] [CrossRef]
- Pasquali, M.; Beyer, M.; Logrieco, A.; Audenaert, K.; Balmas, V.; Basler, R.; Boutigny, A.L.; Chrpová, J.; Czembor, E.; Gagkaeva, T.; et al. A European database of Fusarium graminearum and F. culmorum trichothecene genotypes. Front. Microbiol. 2016, 7, 406. [Google Scholar] [CrossRef]
- Talas, F.; Parzies, H.K.; Miedaner, T. Diversity in genetic structure and chemotype composition of Fusarium graminearum sensu stricto populations causing wheat head blight in individual fields in Germany. Eur. J. Plant Pathol. 2011, 131, 39–48. [Google Scholar] [CrossRef]
- Bilska, K.; Jurczak, S.; Kulik, T.; Ropelewska, E.; Olszewski, J.; Żelechowski, M.; Zapotoczny, P. Species composition and trichothecene genotype profiling of Fusarium field isolates recovered from wheat in Poland. Toxins 2018, 10, 325. [Google Scholar] [CrossRef]
- Boutigny, A.L.; Ward, T.J.; Ballois, N.; Iancu, G.; Ioos, R. Diversity of the Fusarium graminearum species complex on French cereals. Eur. J. Plant Pathol. 2014, 138, 133–148. [Google Scholar] [CrossRef]
- Fredlund, E.; Gidlund, A.; Sulyok, M.; Borjesson, T.; Krska, R.; Olsen, M.; Lindblad, M. Deoxynivalenol and other selected Fusarium toxins in Swedish oats - Occurrence and correlation to specific Fusarium species. Int. J. Food Microbiol. 2013, 167, 276–283. [Google Scholar] [CrossRef]
- Aamot, H.U.; Ward, T.J.; Brodal, G.; Vrålstad, T.; Larsen, G.B.; Klemsdal, S.S.; Elameen, A.; Uhlig, S.; Hofgaard, I.S. Genetic and phenotypic diversity within the Fusarium graminearum species complex in Norway. Eur. J. Plant Pathol. 2015, 142, 501–519. [Google Scholar] [CrossRef]
- Nielsen, L.K.; Jensen, J.D.; Rodriguez, A.; Jorgensen, L.N.; Justesen, A.F. TRI12 based quantitative real-time PCR assays reveal the distribution of trichothecene genotypes of F. graminearum and F. culmorum isolates in Danish small grain cereals. Int. J. Food Microbiol. 2012, 157, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.J.; Clear, R.M.; Rooney, A.P.; O’Donnell, K.; Gaba, D.; Patrick, S.; Starkey, D.E.; Gilbert, J.; Geiser, D.M.; Nowicki, T.W. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 2008, 45, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Spolti, P.; Del Ponte, E.M.; Cummings, J.A.; Dong, Y.; Bergstrom, G.C. Fitness attributes of Fusarium graminearum isolates from wheat in New York possessing a 3-ADON or 15-ADON trichothecene genotype. Phytopathology 2013, 104, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.; Brûlé-Babel, A.; Guerrieri, A.T.; Clear, R.M.; Patrick, S.; Slusarenko, K.; Wolfe, C. Ratio of 3-ADON and 15-ADON isolates of Fusarium graminearum recovered from wheat kernels in Manitoba from 2008 to 2012. Can. J. Plant Pathol. 2014, 36, 54–63. [Google Scholar] [CrossRef]
- von der Ohe, C.; Gauthier, V.; Tamburic-Ilincic, L.; Brule-Babel, A.; Fernando, W.G.D.; Clear, R.; Ward, T.J.; Miedaner, T. A comparison of aggressiveness and deoxynivalenol production between Canadian Fusarium graminearum isolates with 3-acetyl and 15-acetyldeoxynivalenol chemotypes in field-grown spring wheat. Eur. J. Plant Pathol. 2010, 127, 407–417. [Google Scholar] [CrossRef]
- Gilbert, J.; Clear, R.M.; Ward, T.J.; Gaba, D.; Tekauz, A.; Turkington, T.K.; Woods, S.M.; Nowicki, T.; O’Donnell, K. Relative aggressiveness and production of 3- or 15-acetyl deoxynivalenol and deoxynivalenol by Fusarium graminearum in spring wheat. Can. J. Plant Pathol. 2010, 32, 146–152. [Google Scholar] [CrossRef]
- Foroud, N.A.; McCormick, S.P.; MacMillan, T.; Badea, A.; Kendra, D.F.; Ellis, B.E.; Eudes, F. Greenhouse studies reveal increased aggressiveness of emergent Canadian Fusarium graminearum chemotypes in wheat. Plant Dis. 2012, 96, 1271–1279. [Google Scholar] [CrossRef]
- Clear, R.M.; Tucker, J.R.; Gaba, D.; Patrick, S.K.; Lee, S.J.; Demeke, T.; Tittlemier, S.A.; Legge, W.G.; Gräfenhan, T. Deoxynivalenol levels and chemotype frequency in barley cultivars inoculated with two chemotypes of Fusarium graminearum. Can. J. Plant Pathol. 2013, 35, 37–45. [Google Scholar] [CrossRef]
- Puri, K.D.; Zhong, S. The 3ADON population of Fusarium graminearum found in North Dakota is more aggressive and produces a higher level of DON than the prevalent 15ADON population in spring wheat. Phytopathology 2010, 100, 1007–1014. [Google Scholar] [CrossRef]
- Serajazari, M.; Hudson, K.; Kaviani, M.; Navabi, A. Fusarium graminearum chemotype–spring wheat genotype interaction effects in type I and II resistance response assays. Phytopathology 2019, 109, 643–649. [Google Scholar] [CrossRef]
- Walkowiak, S.; Bonner, C.T.; Wang, L.; Blackwell, B.; Rowland, O.; Subramaniam, R. Intraspecies interaction of Fusarium graminearum contributes to reduced toxin production and virulence. Mol. Plant Microbe Interact. 2015, 28, 1256–1267. [Google Scholar] [CrossRef] [PubMed]
- Walkowiak, S.; Rowland, O.; Rodrigue, N.; Subramaniam, R. Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genom. 2016, 17, 1014. [Google Scholar] [CrossRef] [PubMed]
- Gang, G.; Miedaner, T.; Schuhmacher, U.; Schollenberger, M.; Geiger, H.H. Deoxynivalenol and nivalenol production by Fusarium culmorum isolates differing in aggressiveness toward winter rye. Phytopathology 1998, 88, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Gale, L.R.; Harrison, S.A.; Ward, T.J.; O’Donnell, K.; Milus, E.A.; Gale, S.W.; Kistler, H.C. Nivalenol-type populations of Fusarium graminearum and F. asiaticum are prevalent on wheat in southern Louisiana. Phytopathology 2011, 101, 124–134. [Google Scholar] [CrossRef]
- Miedaner, T.; Reinbrecht, C. Trichothecene content of rye and wheat genotypes inoculated with a deoxynivalenol- and a nivalenol-producing isolate of Fusarium culmorum. J. Phytopathol. 2001, 149, 245–251. [Google Scholar] [CrossRef]
- Malihipour, A.; Gilbert, J.; Piercey-Normore, M.; Cloutier, S. Molecular phylogenetic analysis, trichothecene chemotype patterns, and variation in aggressiveness of Fusarium isolates causing head blight in wheat. Plant Dis. 2012, 96, 1016–1025. [Google Scholar] [CrossRef]
- Davari, M.; Wei, C.M.; Babay-Ahari, A.; Arzanlou, M.; Waalwijk, C.; van der Lee, T.A.J.; Zare, R.; Gerrits van den Ende, A.H.G.; de Hoog, G.S.; van Diepeningen, A.D. Geographic differences in trichothecene chemotypes of Fusarium graminearum in the Northwest and North of Iran. World Mycotoxin J. 2013, 6, 137–150. [Google Scholar] [CrossRef]
- Kelly, A.C.; Ward, T.J. Population genomics of Fusarium graminearum reveals signatures of divergent evolution within a major cereal pathogen. PLoS ONE 2018, 13, e0194616. [Google Scholar] [CrossRef]
- Gale, L.R.; Ward, T.J.; Kistler, H.C. A Subset of the Newly Discovered Northland Population of Fusarium Graminearum from the U.S. Does Not Produce the B-Type Trichothecenes DON, 15ADON, 3ADON or NIV. In Proceedings of the National Fusarium Head Blight Forum, Milwaukee, WI, USA, 7–9 December 2010; Canty, S., Clark, A., Anderson-Scully, A., Ellis, D., Van Sanford, D., Eds.; University of Kentucky: Lexington, KY, USA, 2010; pp. 48–49. [Google Scholar]
- Kelly, A.; Proctor, R.H.; Belzile, F.; Chulze, S.N.; Clear, R.M.; Cowger, C.; Elmer, W.; Lee, T.; Obanor, F.; Waalwijk, C.; et al. The geographic distribution and complex evolutionary history of the NX-2 trichothecene chemotype from Fusarium graminearum. Fungal Genet. Biol. 2016, 95, 39–48. [Google Scholar] [CrossRef]
- Lofgren, L.; Riddle, J.; Dong, Y.; Kuhnem, P.R.; Cummings, J.A.; Del Ponte, E.M.; Bergstrom, G.C.; Kistler, H.C. A high proportion of NX-2 genotype strains are found among Fusarium graminearum isolates from northeastern New York State. Eur. J. Plant Pathol. 2018, 150, 791–796. [Google Scholar] [CrossRef]
- Liang, J.; Lofgren, L.; Ma, Z.; Ward, T.J.; Kistler, H.C. Population subdivision of Fusarium graminearum from barley and wheat in the upper Midwestern United States at the turn of the century. Phytopathology 2015, 105, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Brennan, J.M.; Fagan, B.; van Maanen, A.; Cooke, B.M.; Doohan, F.M. Studies on in vitro growth and pathogenicity of European Fusarium fungi. Eur. J. Plant Pathol. 2003, 109, 577–587. [Google Scholar] [CrossRef]
- Knutsen, A.K.; Torp, M.; Holst-Jensen, A. Phylogenetic analyses of the Fusarium poae, Fusarium sporotrichioides and Fusarium langsethiae species complex based on partial sequences of the translation elongation factor-1 alpha gene. Int. J. Food Microbiol. 2004, 95, 287–295. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Gagkaeva, T.Y. Fusarium toxins in cereals in northern Europe and Asia. In Fungi: Applications and Management Strategies; Deshmukh, S.K., Misra, J.K., Tewari, J.P., Papp, T., Eds.; CRC Press, Taylor & Francis Group: Borarton, FL, USA, 2016; pp. 293–317. [Google Scholar]
- Langseth, W.; Rundberget, T. The occurrence of HT-2 toxin and other trichothecenes in Norwegian cereals. Mycopathologia 1999, 147, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez-Vea, M.; Lizarraga, E.; González-Peñas, E.; López de Cerain, A. Co-occurrence of type-A and type-B trichothecenes in barley from a northern region of Spain. Food Control 2012, 25, 81–88. [Google Scholar] [CrossRef]
- Scudamore, K.A.; Patel, S.; Edwards, S.G. HT-2 toxin and T-2 toxin in commercial cereal processing in the United Kingdom, 2004-2007. World Mycotoxin J. 2009, 2, 357–365. [Google Scholar] [CrossRef]
- Fredlund, E.; Gidlund, A.; Pettersson, H.; Olsen, M.; Börjesson, T. Real-time PCR detection of Fusarium species in Swedish oats and correlation to T-2 and HT-2 toxin content. World Mycotoxin J. 2010, 3, 77–88. [Google Scholar] [CrossRef]
- Hietaniemi, V.; Rämö, S.; Yli-Mattila, T.; Jestoi, M.; Peltonen, S.; Kartio, M.; Sieviläinen, E.; Koivisto, T.; Parikka, P. Updated survey of Fusarium species and toxins in Finnish cereal grains. Food Addit Contam 2016, 33, 831–848. [Google Scholar] [CrossRef]
- Edwards, S.G. Fusarium mycotoxin content of UK organic and conventional oats. Food Addit. Contam. 2009, 26, 1063–1069. [Google Scholar] [CrossRef] [Green Version]
- Orlando, B.; Barrier-Guillot, B.; Gourdain, E.; Maumené, C. Identification of agronomic factors that influence the levels of T-2 and HT-2 toxins in barley grown in France. World Mycotoxin J. 2010, 3, 169–174. [Google Scholar] [CrossRef]
- Pettersson, H.; Börjesson, T.; Persson, L.; Lerenius, C.; Berg, G.; Gustafsson, G. T-2 and HT-2 toxins in oats grown in Northern Europe. Cereal Res. Commun. 2008, 36, 591–592. [Google Scholar]
- Edwards, S.G. Fusarium mycotoxin content of UK organic and conventional wheat. Food Addit. Contam. 2009, 26, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.G.; Barrier-Guillot, B.; Clasen, P.E.; Hietaniemi, V.; Pettersson, H. Emerging issues of HT-2 and T-2 toxins in European cereal production. World Mycotoxin J. 2009, 2, 173–179. [Google Scholar] [CrossRef]
- Schöneberg, T.; Jenny, E.; Wettstein, F.E.; Bucheli, T.D.; Mascher, F.; Bertossa, M.; Musa, T.; Seifert, K.; Gräfenhan, T.; Keller, B.; et al. Occurrence of Fusarium species and mycotoxins in Swiss oats—Impact of cropping factors. Eur. J. Agron. 2018, 92, 123–132. [Google Scholar] [CrossRef]
- Martin, C.; Schöneberg, T.; Vogelgsang, S.; Mendes Ferreira, C.S.; Morisoli, R.; Bertossa, M.; Bucheli, T.D.; Mauch-Mani, B.; Mascher, F. Responses of oat grains to Fusarium poae and F. langsethiae infections and mycotoxin contaminations. Toxins 2018, 10, 47. [Google Scholar] [CrossRef]
- van der Fels-Klerx, H.; Stratakou, I. T-2 toxin and HT-2 toxin in grain and grain-based commodities in Europe: Occurrence, factors affecting occurrence, co-occurrence and toxicological effects. World Mycotoxin J. 2010, 3, 349–367. [Google Scholar] [CrossRef]
- Torp, M.; Langseth, W. Production of T-2 toxin by a Fusarium resembling Fusarium poae. Mycopathologia 1999, 147, 89–96. [Google Scholar] [CrossRef]
- Imathiu, S.M.; Edwards, S.G.; Ray, R.V.; Back, M.A. Fusarium langsethiae—A HT-2 and T-2 toxins producer that needs more attention. J. Phytopathol. 2013, 161, 1–10. [Google Scholar] [CrossRef]
- Imathiu, S.M.; Ray, R.V.; Back, M.; Hare, M.C.; Edwards, S.G. Fusarium langsethiae pathogenicity and aggressiveness towards oats and wheat in wounded and unwounded in vitro detached leaf assays. Eur. J. Plant Pathol. 2009, 124, 117–126. [Google Scholar] [CrossRef]
- Hofgaard, I.S.; Aamot, H.U.; Torp, T.; Jestoi, M.; Lattanzio, V.M.T.; Klemsdal, S.S.; Waalwijk, C.; Lee, T.; Brodal, G. Associations between Fusarium species and mycotoxins in oats and spring wheat from farmers’ fields in Norway over a six-year period. World Mycotoxin J. 2016, 9, 365–378. [Google Scholar] [CrossRef]
- Edwards, S.G.; Imathiu, S.M.; Ray, R.V.; Back, M.; Hare, M.C. Molecular studies to identify the Fusarium species responsible for HT-2 and T-2 mycotoxins in UK oats. Int. J. Food Microbiol. 2012, 156, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Bočarov-Stančić, A.S.; Lević, J.; Stanković, S.Ž.; Tančić, S.L.; Krnjaja, V.; Salma, N. Toxigenic potential of Fusarium langsethiae isolates from Serbian wheat kernels. Cereal Res. Commun. 2008, 36, 345–346. [Google Scholar]
- Infantino, A.; Pucci, N.; Conca, G.; Santori, A. First report of Fusarium langsethiae on durum wheat kernels in Italy. Plant Dis. 2007, 91, 1362. [Google Scholar] [CrossRef] [PubMed]
- Lukanowski, A.; Sadowski, C. Fusarium langsethiae on kernels of winter wheat in Poland—Occurrence and mycotoxigenic abilities. Cereal Res. Commun. 2008, 36, 453–457. [Google Scholar] [CrossRef]
- Suproniene, S.; Justesen, A.F.; Nicolaisen, M.; Mankeviciene, A.; Dabkevicius, Z.; Semaskiene, R.; Leistrumaite, A. Distribution of trichothecene and zearalenone producing Fusarium species in grain of different cereal species and cultivars grown under organic farming conditions in Lithuania. Ann. Agric. Environ. Med. 2010, 17, 79–86. [Google Scholar]
- Hudec, K.; Roháčik, T. The occurrence and predominance of Fusarium species on barley kernels in Slovakia. Cereal Res. Commun. 2009, 37, 101–109. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Gavrilova, O.; Hussien, T.; Gagkaeva, T. Identification of the first Fusarium sibiricum isolate in Iran and Fusarium langsethiae isolate in Siberia by morphology and species-specific primers. J. Plant Pathol. 2015, 97, 183–187. [Google Scholar]
- Wilcoxson, R.D.; Kommedahl, T.; Ozmon, E.A.; Windels, C.E. Occurrence of Fusarium species in scabby wheat from Minnesota and their pathogenicity to wheat. Phytopathology 1988, 78, 586–589. [Google Scholar] [CrossRef]
- Vigier, B.; Reid, L.M.; Seifert, K.A.; Stewart, D.W.; Hamilton, R.I. Distribution and prediction of Fusarium species associated with maize ear rot in Ontario. Can. J. Plant Pathol. 1997, 19, 60–65. [Google Scholar] [CrossRef]
- Ueno, Y.; Hosoya, M.; Morita, Y.; Ueno, I.; Tatsuno, T. Inhibition of the protein synthesis in rabbit reticulocyte by nivalenol, a toxic principle isolated from Fusarium nivale-growing rice. J. Biochem. 1968, 64, 479–485. [Google Scholar] [CrossRef]
- Arunachalam, C.; Doohan, F.M. Trichothecene toxicity in eukaryotes: Cellular and molecular mechanisms in plants and animals. Toxicol. Lett. 2013, 217, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Cundliffe, E.; Cannon, M.; Davies, J. Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins. Proc. Natl. Acad. Sci. USA 1974, 71, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Schindler, D. Two classes of inhibitors of peptidyl transferase activity in eukaryotes. Nature 1974, 249, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Stafford, M.E.; McLaughlin, C.S. Trichodermin, a possible inhibitor of the termination process of protein synthesis. J. Cell. Physiol. 1973, 82, 121–128. [Google Scholar] [CrossRef]
- Tate, W.P.; Caskey, C.T. Peptidyltransferase inhibition by trichodermin. J. Biol. Chem. 1973, 248, 7970–7972. [Google Scholar]
- Wei, C.M.; Hansen, B.S.; Vaughan, M.H.; McLaughlin, C.S. Mechanism of action of the mycotoxin trichodermin, a 12,13-epoxytrichothecene. Proc. Natl. Acad. Sci. USA 1974, 71, 713–717. [Google Scholar] [CrossRef]
- Carrasco, L.; Barbacid, M.; Vazquez, D. The trichodermin group of antibiotics, inhibitors of peptide bond formation by eukaryotic ribosomes. Biochim. Biophys. Acta 1973, 312, 368–376. [Google Scholar] [CrossRef]
- Carter, C.J.; Cannon, M.; Smith, K.E. Inhibition of protein synthesis in reticulocyte lysates by trichodermin. Biochem. J. 1976, 154, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.M.; Campbell, I.M.; McLaughlin, C.S.; Vaughan, M.H. Binding of trichodermin to mammalian ribosomes and its inhibition by other 12,13-epoxytrichothecenes. Mol. Cell. Biochem. 1974, 3, 215–219. [Google Scholar] [CrossRef]
- Ehrlich, K.C.; Daigle, K.W. Protein synthesis inhibition by 8 oxo-12,13-epoxytrichothecenes. Biochim. Biophys. Acta 1987, 923, 206–213. [Google Scholar] [CrossRef]
- Mayer, E.; Novak, B.; Springler, A.; Schwartz-Zimmermann, H.E.; Nagl, V.; Reisinger, N.; Hessenberger, S.; Schatzmayr, G. Effects of deoxynivalenol (DON) and its microbial biotransformation product deepoxy-deoxynivalenol (DOM-1) on a trout, pig, mouse, and human cell line. Mycotoxin Res. 2017, 33, 297–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foroud, N.A.; Shank, R.A.; Kiss, D.; Eudes, F.; Hazendonk, P. Solvent and water mediated structural variations in deoxynivalenol and their potential implications on the disruption of ribosomal function. Front. Microbiol. 2016, 7, 1239. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Shank, R.A.; Montina, T.; Goettel, J.T.; Foroud, N.A.; Hazendonk, P.; Eudes, F. Hydrogen-bonding interactions in T-2 toxin studied using solution and solid-state NMR. Toxins 2011, 3, 1310–1331. [Google Scholar] [CrossRef] [PubMed]
- Shank, R.A.; Foroud, N.A.; Hazendonk, P.; Eudes, F.; Blackwell, B.A. Current and future experimental strategies for structural analysis of trichothecene mycotoxins—A prospectus. Toxins 2011, 3, 1518–1553. [Google Scholar] [CrossRef] [PubMed]
- Nierhaus, K.H. Mg2+, K+, and the Ribosome. J. Bacteriol. 2014, 196, 3817–3819. [Google Scholar] [CrossRef]
- Kimura, M.; Kaneko, I.; Komiyama, M.; Takatsuki, A.; Koshino, H.; Yoneyama, K.; Yamaguchi, I. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. J. Biol. Chem. 1998, 273, 1654–1661. [Google Scholar] [CrossRef]
- Alexander, N.J.; McCormick, S.P.; Ziegenhorn, S.L. Phytotoxicity of selected trichothecenes using Chlamydomonas reinhardtii as a model system. Nat. Toxins 1999, 7, 265–269. [Google Scholar] [CrossRef]
- Eriksen, G.S.; Pettersson, H.; Lundh, T. Comparative cytotoxicity of deoxynivalenol, nivalenol, their acetylated derivatives and de-epoxy metabolites. Food Chem. Toxicol. 2004, 42, 619–624. [Google Scholar] [CrossRef]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glossl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef]
- Thompson, W.L.; Wannemacher, R.W., Jr. Structure-function relationships of 12,13-epoxytrichothecene mycotoxins in cell culture: Comparison to whole animal lethality. Toxicon 1986, 24, 985–994. [Google Scholar] [CrossRef]
- Desjardins, A.E.; McCormick, S.P.; Appell, M. Structure—Activity relationships of trichothecene toxins in an Arabidopsis thaliana leaf assay. J. Agric. Food Chem. 2007, 55, 6487–6492. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.; Tekauz, A. Effects of delayed harvest on Manitoba wheat. In Proceedings of the 1993 Regional Scab Forum, Moorhead, MN, USA, 3 November 1993. [Google Scholar]
- Eudes, F.; Comeau, A.; Rioux, S.; Collin, J. Phytotoxicité de huit mycotoxines associées à la fusariose de l’épi chez le blé. Can. J. Plant Pathol. 2000, 22, 286–292. [Google Scholar] [CrossRef]
- Minervini, F.; Fornelli, F.; Flynn, K.M. Toxicity and apoptosis induced by the mycotoxins nivalenol, deoxynivalenol and fumonisin B1 in a human erythroleukemia cell line. Toxicol. Vitr. 2004, 18, 21–28. [Google Scholar] [CrossRef]
- Ueno, Y. Toxicological features of T-2 toxin and related trichothecenes. Toxicol. Sci. 1984, 4, 124–132. [Google Scholar] [CrossRef]
- Mitterbauer, R.; Poppenberger, B.; Raditschnig, A.; Lucyshyn, D.; Lemmens, M.; Glössl, J.; Adam, G. Toxin-dependent utilization of engineered ribosomal protein L3 limits trichothecene resistance in transgenic plants. Plant Biotechnol. J. 2004, 2, 329–340. [Google Scholar] [CrossRef]
- Jimenez, A.; Sanchez, L.; Vazquez, D. Simultaneous ribosomal resistance to trichodermin and anisomycin in Saccharomyces cerevisiae mutants. Biochim. Biophys. Acta 1975, 383, 427–434. [Google Scholar] [CrossRef]
- Meskauskas, A.; Dinman, J.D. Ribosomal protein L3 functions as a ‘rocker switch’ to aid in coordinating of large subunit-associated functions in eukaryotes and Archaea. Nucleic Acids Res. 2008, 36, 6175–6186. [Google Scholar] [CrossRef]
- Meskauskas, A.; Dinman, J.D. Ribosomal protein L3: Gatekeeper to the A-Site. Mol. Cell 2007, 25, 877–888. [Google Scholar] [CrossRef]
- Miller, J.D.; Ewen, M.A. Toxic effects of deoxynivalenol on ribosomes and tissues of the spring wheat cultivars Frontana and Casavant. Nat. Toxins 1997, 5, 234–237. [Google Scholar] [CrossRef]
- Harris, L.J.; Gleddie, S.C. A modified Rpl3 gene from rice confers tolerance of the Fusarium graminearum mycotoxin deoxynivalenol to transgenic tobacco. Physiol. Mol. Plant Pathol. 2001, 58, 173–181. [Google Scholar] [CrossRef]
- Kant, P.; Gulati, A.; Harris, L.; Gleddie, S.; Singh, J.; Pauls, K.P. Transgenic corn plants with modified ribosomal protein L3 show decreased ear rot disease after inoculation with Fusarium graminearum. Aust. J. Crop Sci. 2012, 6, 1598–1605. [Google Scholar]
- Di, R.; Blechl, A.; Dill-Macky, R.; Tortora, A.; Tumer, N.E. Expression of a truncated form of yeast ribosomal protein L3 in transgenic wheat improves resistance to Fusarium head blight. Plant Sci. 2010, 178, 374–380. [Google Scholar] [CrossRef]
- Di, R.; Tumer, N.E. Expression of a truncated form of ribosomal protein L3 confers resistance to pokeweed antiviral protein and the Fusarium mycotoxin deoxynivalenol. Mol. Plant Microbe Interact. 2005, 18, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Foroud, N.A.; Ouellet, T.; Laroche, A.; Oosterveen, B.; Jordan, M.C.; Ellis, B.E.; Eudes, F. Differential transcriptome analyses of three wheat genotypes reveal different host response pathways associated with Fusarium head blight and trichothecene resistance. Plant Pathol. 2012, 61, 296–314. [Google Scholar] [CrossRef]
- Hall, H.C.; Samuel, M.A.; Ellis, B.E. SIPK conditions transcriptional responses unique to either bacterial or oomycete elicitation in tobacco. Mol. Plant Pathol. 2007, 8, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Mauro, V.P.; Edelman, G.M. The ribosome filter redux. Cell Cycle 2007, 6, 2246–2251. [Google Scholar] [CrossRef]
- Gerst, J.E. Pimp my ribosome: Ribosomal protein paralogs specify translational control. Trends Genet. 2018, 34, 832–845. [Google Scholar] [CrossRef]
- Del Sorbo, G.; Schoonbeek, H.J.; De Waard, M.A. Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet. Biol. 2000, 30, 1–15. [Google Scholar] [CrossRef]
- Perlin, M.H.; Andrews, J.; San Toh, S. Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. In Advances in Genetics; Friedmann, T., Dunlap, J.C., Goodwin, S.F., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 85, pp. 201–253. [Google Scholar]
- Adam, G.; Mitterbauer, R.; Raditschnig, A.; Poppenberger, B.; Karl, T.; Goritschnig, S.; Weindorfer, H.; Glössl, J. Molecular mechanisms of deoxynivalenol resistance in the yeast Saccharomyces cerevisiae. Mycotoxin Res. 2001, 17, 19–23. [Google Scholar] [CrossRef]
- Demissie, Z.A.; Foote, S.J.; Tan, Y.; Loewen, M.C. Profiling of the transcriptomic responses of Clonostachys rosea upon treatment with Fusarium graminearum secretome. Front. Microbiol. 2018, 9, 1061. [Google Scholar] [CrossRef]
- Hue, A.G.; Voldeng, H.D.; Savard, M.E.; Fedak, G.; Tian, X.; Hsiang, T. Biological control of fusarium head blight of wheat with Clonostachys rosea strain ACM941. Can. J. Plant Pathol. 2009, 31, 169–179. [Google Scholar] [CrossRef]
- Xue, A.G.; Chen, Y.; Voldeng, H.D.; Fedak, G.; Savard, M.E.; Längle, T.; Zhang, J.; Harman, G.E. Concentration and cultivar effects on efficacy of CLO-1 biofungicide in controlling Fusarium head blight of wheat. Biol. Control 2014, 73, 2–7. [Google Scholar] [CrossRef]
- Biselli, C.; Bagnaresi, P.; Faccioli, P.; Hu, X.; Balcerzak, M.; Mattera, M.G.; Yan, Z.; Ouellet, T.; Cattivelli, L.; Valè, G. Comparative transcriptome profiles of near-isogenic hexaploid wheat lines differing for effective alleles at the 2DL FHB resistance QTL. Front. Plant Sci. 2018, 9, 37. [Google Scholar] [CrossRef]
- Boddu, J.; Cho, S.; Kruger, W.M.; Muehlbauer, G.J. Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol. Plant Microbe Interact. 2006, 19, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Boddu, J.; Cho, S.; Muehlbauer, G.J. Transcriptome analysis of trichothecene-induced gene expression in barley. Mol. Plant Microbe Interact. 2007, 20, 1364–1375. [Google Scholar] [CrossRef]
- Handa, H.; Namiki, N.; Xu, D.; Ban, T. Dissecting of the FHB resistance QTL on the short arm of wheat chromosome 2D using a comparative genomic approach: From QTL to candidate gene. Mol. Breed. 2008, 22, 71–84. [Google Scholar] [CrossRef]
- Kosaka, A.; Manickavelu, A.; Kajihara, D.; Nakagawa, H.; Ban, T. Altered gene expression profiles of wheat genotypes against Fusarium head blight. Toxins 2015, 7, 604–620. [Google Scholar] [CrossRef]
- Liu, J.; Li, L.; Foroud, N.A.; Li, C.; Gong, X.; Li, T. Proteomics of bulked rachides combined with documented QTLs uncovers genotype non-specific players of the Fusarium head blight responses in wheat. Phytopathology 2019, 109, 111–119. [Google Scholar] [CrossRef]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef] [Green Version]
- Walter, S.; Brennan, J.M.; Arunachalam, C.; Ansari, K.I.; Hu, X.; Khan, M.R.; Trognitz, F.; Trognitz, B.; Leonard, G.; Egan, D.; et al. Components of the gene network associated with genotype-dependent response of wheat to the Fusarium mycotoxin deoxynivalenol. Funct. Integr. Genom. 2008, 8, 421–427. [Google Scholar] [CrossRef]
- Chetouhi, C.; Bonhomme, L.; Lasserre-Zuber, P.; Cambon, F.; Pelletier, S.; Renou, J.P.; Langin, T. Transcriptome dynamics of a susceptible wheat upon Fusarium head blight reveals that molecular responses to Fusarium graminearum infection fit over the grain development processes. Funct. Integr. Genom. 2016, 16, 183–201. [Google Scholar] [CrossRef] [PubMed]
- Boutigny, A.L.; Richard-Forget, F.; Barreau, C. Natural mechanisms for cereal resistance to the accumulation of Fusarium trichothecenes. Eur. J. Plant Pathol. 2008, 121, 411–423. [Google Scholar] [CrossRef]
- Powell, J.J.; Carere, J.; Fitzgerald, T.L.; Stiller, J.; Covarelli, L.; Xu, Q.; Gubler, F.; Colgrave, M.L.; Gardiner, D.M.; Manners, J.M.; et al. The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum L.). Ann. Bot. 2017, 119, 853–867. [Google Scholar]
- Steiner, B.; Kurz, H.; Lemmens, M.; Buerstmayr, H. Differential gene expression of related wheat lines with contrasting levels of head blight resistance after Fusarium graminearum inoculation. Theor. Appl. Genet. 2009, 118, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, S.A.; Boddu, J.; Berthiller, F.; Hametner, C.; Stupar, R.M.; Adam, G.; Muehlbauer, G.J. Transcriptome analysis of the barley-deoxynivalenol interaction: Evidence for a role of glutathione in deoxynivalenol detoxification. Mol. Plant Microbe Interact. 2010, 23, 962–976. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shin, S.; Heinen, S.; Dill-Macky, R.; Berthiller, F.; Nersesian, N.; Clemente, T.; McCormick, S.; Muehlbauer, G.J. Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum. Mol. Plant Microbe Interact. 2015, 28, 1237–1246. [Google Scholar] [CrossRef]
- Xing, L.; Gao, L.; Chen, Q.; Pei, H.; Di, Z.; Xiao, J.; Wang, H.; Ma, L.; Chen, P.; Cao, A.; et al. Over-expressing a UDP-glucosyltransferase gene (Ta-UGT3) enhances Fusarium Head Blight resistance of wheat. Plant Growth Regul. 2018, 84, 561–571. [Google Scholar] [CrossRef]
- Gatti, M.; Cambon, F.; Tassy, C.; Macadre, C.; Guerard, F.; Langin, T.; Dufresne, M. The Brachypodium distachyon UGT Bradi5gUGT03300 confers type II fusarium head blight resistance in wheat. Plant Pathol. 2019, 68, 334–343. [Google Scholar] [CrossRef]
- Shin, S.; Torres-Acosta, J.A.; Heinen, S.J.; McCormick, S.; Lemmens, M.; Paris, M.P.K.; Berthiller, F.; Adam, G.; Muehlbauer, G.J. Transgenic Arabidopsis thaliana expressing a barley UDP-glucosyltransferase exhibit resistance to the mycotoxin deoxynivalenol. J. Exp. Bot. 2012, 63, 4731–4740. [Google Scholar] [CrossRef]
- Li, X.; Muehlbauer, G.J.; Adam, G.; Wiesenberger, G.; Michlmayr, H.; Schweiger, W.; Malachova, A.; Berthiller, F.; Shin, S.; Huang, Y.; et al. A barley UDP-glucosyltransferase inactivates nivalenol and provides Fusarium Head Blight resistance in transgenic wheat. J. Exp. Bot. 2017, 68, 2187–2197. [Google Scholar] [CrossRef] [PubMed]
- Mandalà, G.; Tundo, S.; Francesconi, S.; Gevi, F.; Zolla, L.; Ceoloni, C.; D’Ovidio, R. Deoxynivalenol detoxification in transgenic wheat confers resistance to Fusarium head blight and crown rot diseases. Mol. Plant Microbe Interact. 2019, 32, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Tokai, T.; Fujimura, M.; Inoue, H.; Aoki, T.; Ohta, K.; Shibata, T.; Yamaguchi, I.; Kimura, M. Concordant evolution of trichothecene 3-O-acetyltransferase and an rDNA species phylogeny of trichothecene-producing and non-producing fusaria and other ascomycetous fungi. Microbiology 2005, 151, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Khatibi, P.A.; Newmister, S.A.; Rayment, I.; McCormick, S.P.; Alexander, N.J.; Schmale, D.G. Bioprospecting for trichothecene 3-O-acetyltransferases in the fungal genus Fusarium yields functional enzymes with different abilities to modify the mycotoxin deoxynivalenol. Appl. Environ. Microbiol. 2011, 77, 1162–1170. [Google Scholar] [CrossRef]
- Okubara, P.A.; Blechl, A.E.; McCormick, S.P.; Alexander, N.J.; Dill-Macky, R.; Hohn, T.M. Engineering deoxynivalenol metabolism in wheat through the expression of a fungal trichothecene acetyltransferase gene. Theor. Appl. Genet. 2002, 106, 74–83. [Google Scholar] [CrossRef]
- Ohsato, S.; Ochiai-Fukuda, T.; Nishiuchi, T.; Takahashi-Ando, N.; Koizumi, S.; Hamamoto, H.; Kudo, T.; Yamaguchi, I.; Kimura, M. Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep. 2007, 26, 531–538. [Google Scholar] [CrossRef]
- Manoharan, M.; Dahleen, L.S.; Hohn, T.M.; Neate, S.M.; Yu, X.H.; Alexander, N.J.; McCormick, S.P.; Bregitzer, P.; Schwarz, P.B.; Horsley, R.D. Expression of 3-OH trichothecene acetyltransferase in barley (Hordeum vulgare L.) and effects on deoxynivalenol. Plant Sci. 2006, 171, 699–706. [Google Scholar] [CrossRef]
- Alexander, N.J. The TRI101 story: Engineering wheat and barley to resist Fusarium head blight. World Mycotoxin J. 2008, 1, 31–37. [Google Scholar] [CrossRef]
- Garvey, G.S.; McCormick, S.P.; Rayment, I. Structural and functional characterization of the TRI101 trichothecene 3-O-acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: Kinetic insights into combating Fusarium head blight. J. Biol. Chem. 2008, 283, 1660–1669. [Google Scholar] [CrossRef]
- Wu, L.; Wang, B. Evaluation on levels and conversion profiles of DON, 3-ADON, and 15-ADON during bread making process. Food Chem. 2015, 185, 509–516. [Google Scholar] [CrossRef]
- He, W.J.; Yuan, Q.S.; Zhang, Y.B.; Guo, M.W.; Gong, A.D.; Zhang, J.B.; Wu, A.B.; Huang, T.; Qu, B.; Li, H.P.; et al. Aerobic de-epoxydation of trichothecene mycotoxins by a soil bacterial consortium isolated using in situ soil enrichment. Toxins 2016, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; He, J.; Young, J.C.; Zhu, H.; Li, X.Z.; Ji, C.; Zhou, T. Transformation of trichothecene mycotoxins by microorganisms from fish digesta. Aquaculture 2009, 290, 290–295. [Google Scholar] [CrossRef]
- Eriksen, G.S.; Pettersson, H.; Johnsen, K.; Lindberg, J.E. Transformation of trichothecenes in ileal digesta and faeces from pigs. Arch. Anim. Nutr. 2002, 56, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Swanson, S.P.; Helaszek, C.; Buck, W.B.; Rood, H.D.; Haschek, W.M. The role of intestinal microflora in the metabolism of trichothecene mycotoxins. Food Chem. Toxicol. 1988, 26, 823–829. [Google Scholar] [CrossRef]
- Gratz, S.W.; Duncan, G.; Richardson, A.J. The human fecal microbiota metabolizes deoxynivalenol and deoxynivalenol-3-glucoside and may be responsible for urinary deepoxy-deoxynivalenol. Appl. Environ. Microbiol. 2013, 79, 1821–1825. [Google Scholar] [CrossRef]
- Turner, P.C.; Hopton, R.P.; Lecluse, Y.; White, K.L.M.; Fisher, J.; Lebailly, P. Determinants of urinary deoxynivalenol and de-epoxy deoxynivalenol in male farmers from Normandy, France. J. Agric. Food Chem. 2010, 58, 5206–5212. [Google Scholar] [CrossRef]
- Eriksen, G.S.; Pettersson, H. Lack of de-epoxidation of type B trichothecenes in incubates with human faeces. Food Addit. Contam. 2003, 20, 579–582. [Google Scholar] [CrossRef]
- Papageorgiou, M.; Wells, L.; Williams, C.; White, K.; De Santis, B.; Liu, Y.; Debegnach, F.; Miano, B.; Moretti, G.; Greetham, S.; et al. Assessment of urinary deoxynivalenol biomarkers in UK children and adolescents. Toxins 2018, 10, 50. [Google Scholar] [CrossRef]
- Ito, M.; Sato, I.; Ishizaka, M.; Yoshida, S.I.; Koitabashi, M.; Yoshida, S.; Tsushima, S. Bacterial cytochrome P450 system catabolizing the Fusarium toxin deoxynivalenol. Appl. Environ. Microbiol. 2013, 79, 1619–1628. [Google Scholar] [CrossRef]
- Gunupuru, L.R.; Arunachalam, C.; Malla, K.B.; Kahla, A.; Perochon, A.; Jia, J.; Thapa, G.; Doohan, F.M. A wheat cytochrome P450 enhances both resistance to deoxynivalenol and grain yield. PLoS ONE 2018, 13, e0204992. [Google Scholar] [CrossRef]
- Gunupuru, L.R.; Perochon, A.; Doohan, F.M. Deoxynivalenol resistance as a component of FHB resistance. Trop. Plant Pathol. 2017, 42, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Walter, S.; Doohan, F. Transcript profiling of the phytotoxic response of wheat to the Fusarium mycotoxin deoxynivalenol. Mycotoxin Res. 2011, 27, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Masuda, D.; Ishida, M.; Yamaguchi, K.; Nishiuchi, T.; Yamaguchi, I.; Kimura, M. Phytotoxic effects of trichothecenes on the growth and morphology of Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Pritsch, C.; Vance, C.P.; Bushnell, W.R.; Somers, D.A.; Hohn, T.M.; Muehlbauer, G.J. Systemic expression of defense response genes in wheat spikes as a response to Fusarium graminearum infection. Physiol. Mol. Plant Pathil. 2001, 58, 1–12. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, Z.; Rocheleau, H.; Fauteux, F.; Wang, Y.; McCartney, C.; Ouellet, T. Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genom. 2018, 19, 642. [Google Scholar] [CrossRef]
- Buhrow, L.M.; Cram, D.; Tulpan, D.; Foroud, N.A.; Loewen, M.C. Exogenous abscisic acid and gibberellic acid elicit opposing effects on Fusarium graminearum infection in wheat. Phytopathology 2016, 106, 986–996. [Google Scholar] [CrossRef]
- Qi, P.F.; Balcerzak, M.; Rocheleau, H.; Leung, W.; Wei, Y.M.; Zheng, Y.L.; Ouellet, T. Jasmonic acid and abscisic acid play important roles in host–pathogen interaction between Fusarium graminearum and wheat during the early stages of fusarium head blight. Physiol. Mol. Plant Pathol. 2016, 93, 39–48. [Google Scholar] [CrossRef]
- Gottwald, S.; Samans, B.; Luck, S.; Friedt, W. Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: Two essential mechanisms of Fusarium head blight resistance in wheat? BMC Genom. 2012, 13, 369. [Google Scholar] [CrossRef]
- Makandar, R.; Nalam, V.J.; Lee, H.; Trick, H.N.; Dong, Y.; Shah, J. Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol. Plant Microbe Interact. 2012, 25, 431–439. [Google Scholar] [CrossRef]
- Li, G.; Yen, Y. Jasmonate and ethylene signaling pathway may mediate Fusarium head blight resistance in wheat. Crop Sci. 2008, 48, 1888–1896. [Google Scholar] [CrossRef]
- Foroud, N.A.; Pordel, R.; Goyal, R.K.; Ryabova, D.; Chatterton, S.; Kovalchuk, I. Chemical activation of the ethylene signalling pathway promotes wheat resistance to Fusarium graminearum. Phytopathology 2019, 109, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Steed, A.; Travella, S.; Keller, B.; Nicholson, P. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 2009, 182, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Gunupuru, L.R.; Kumar, G.B.S.; Khan, M.; Scofield, S.; Nicholson, P.; Doohan, F.M. Plant disease resistance is augmented in uzu barley lines modified in the brassinosteroid receptor BRI1. BMC Plant Biol. 2014, 14, 227. [Google Scholar] [CrossRef]
- Desmond, O.J.; Edgar, C.I.; Manners, J.M.; Maclean, D.J.; Schenk, P.M.; Kazan, K. Methyl jasmonate induced gene expression in wheat delays symptom development by the crown rot pathogen Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 2005, 67, 171–179. [Google Scholar] [CrossRef]
- Miedaner, T.; Herter, C.P.; Ebmeyer, E.; Kollers, S.; Korzun, V. Use of non-adapted quantitative trait loci for increasing Fusarium head blight resistance for breeding semi-dwarf wheat. Plant Breed. 2019, 138, 140–147. [Google Scholar] [CrossRef]
- Brauer, E.K.; Rocheleau, H.; Balcerzak, M.; Pan, Y.; Fauteux, F.; Liu, Z.; Wang, L.; Zheng, W.; Ouellet, T. Transcriptional and hormonal profiling of Fusarium graminearum-infected wheat reveals an association between auxin and susceptibility. Physiol. Mol. Plant Pathol. 2019, 107, 33–39. [Google Scholar] [CrossRef]
- Wang, L.; Li, Q.; Liu, Z.; Surendra, A.; Pan, Y.; Li, Y.; Zaharia, L.I.; Ouellet, T.; Fobert, P.R. Integrated transcriptome and hormone profiling highlight the role of multiple phytohormone pathways in wheat resistance against fusarium head blight. PLoS ONE 2018, 13, e0207036. [Google Scholar] [CrossRef]
- Perochon, A.; Jianguang, J.; Kahla, A.; Arunachalam, C.; Scofield, S.R.; Bowden, S.; Wallington, E.; Doohan, F.M. TaFROG encodes a Pooideae orphan protein that interacts with SnRK1 and enhances resistance to the mycotoxigenic fungus Fusarium graminearum. Plant Physiol. 2015, 169, 2895–2906. [Google Scholar] [CrossRef]
- Jia, H.; Cho, S.; Muehlbauer, G.J. Transcriptome analysis of a wheat near-isogenic line pair carrying Fusarium head blight-resistant and -susceptible alleles. Mol. Plant Microbe Interact. 2009, 22, 1366–1378. [Google Scholar] [CrossRef]
- Perochon, A.; Kahla, A.; Vranić, M.; Jia, J.; Malla, K.B.; Craze, M.; Wallington, E.; Doohan, F.M. A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium Head Blight disease. Plant Biotechnol. J. 2019, 17, 1892–1904. [Google Scholar] [CrossRef]
- Becher, R.; Miedaner, T.; Wirsel, S.R. Biology, diversity, and management of FHB-causing Fusarium species in small-grain cereals. In Agricultural Applications; Kempken, F., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; Volume 11, pp. 199–241. [Google Scholar]
- Gilbert, J.; Tekauz, A. Strategies for management of fusarium head blight (FHB) in cereals. Prairie Soils Crops J. 2011, 4, 97–104. [Google Scholar]
- Beres, B.L.; Brûlé-Babel, A.L.; Ye, Z.; Graf, R.J.; Turkington, T.K.; Harding, M.W.; Kutcher, H.R.; Hooker, D.C. Exploring Genotype × Environment × Management synergies to manage fusarium head blight in wheat. Can. J. Plant Pathol. 2018, 40, 179–188. [Google Scholar] [CrossRef]
- Ye, Z.; Brûlé-Babel, A.L.; Graf, R.J.; Mohr, R.; Beres, B.L. The role of genetics, growth habit, and cultural practices in the mitigation of Fusarium head blight. Can. J. Plant Sci. 2017, 97, 316–328. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Loewen, M.; Rajagopalan, N.; Surendra, A. Perspectives on the specific targeting of Fusarium graminearum for the development of alternative head blight treatment approaches. Plant Pathol. 2017, 66, 1391–1403. [Google Scholar] [CrossRef]
- Rojas, E.C.; Jørgensen, H.J.; Jensen, B.; Collinge, D.B. Fusarium diseases: Biology and management perspectives. In Integrated Disease Management of Wheat and Barley; Burleigh Dodds Series in Agricultural Science; Burleigh Dodds Science Publishing Limited: Cambridge, UK, 2018; Volume 19. [Google Scholar]
- Mesterházy, Á. Breeding wheat for Fusarium head blight resistance in Europe. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2003; pp. 211–240. [Google Scholar]
- Schroeder, H.W.; Christensen, J.J. Factors affecting resistance of wheat to scab by Gibberella zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Wang, Y.Z.; Miller, J.D. Effects of Fusarium graminearum metabolites on wheat tissue in relation to Fusarium Head Blight resistance. J. Phytopathol. 1988, 122, 118–125. [Google Scholar] [CrossRef]
- Miller, J.D.; Young, J.C.; Sampson, D.R. Deoxynivalenol and Fusarium head blight resistance in spring cereals. J. Phytopathol. 1985, 113, 359–367. [Google Scholar] [CrossRef]
- Miller, D.J.; Arnison, P.G. Degradation of deoxynivalenol by suspension cultures of the Fusarium head blight resistant wheat cultivar Frontana. Can. J. Plant Pathol. 1986, 8, 147–150. [Google Scholar] [CrossRef]
- Shaner, G. Resistance in hexaploid wheat to Fusarium head blight. In Proceedings of the National Fusarium Head Blight Forum, Erlanger, KY, USA, 7–9 December 2002. [Google Scholar]
- McCallum, B.D.; Tekauz, A. Influence of inoculation method and growth stage on fusarium head blight in barley. Can. J. Plant Pathol. 2002, 24, 77–80. [Google Scholar] [CrossRef]
- Dill-Macky, R. Inoculation methods and evaluation of Fusarium head blight. In Fusarium Head Blight of Wheat and Barley; Leonard, K.J., Bushnell, W.R., Eds.; The American Phytopathological Society: St. Paul, MN, USA, 2003; pp. 184–210. [Google Scholar]
- Geddes, J.; Eudes, F.; Tucker, J.R.; Legge, W.G.; Selinger, L.B. Evaluation of inoculation methods on infection and deoxynivalenol production by Fusarium graminearum on barley. Can. J. Plant Pathol. 2008, 30, 66–73. [Google Scholar] [CrossRef]
- Miedaner, T.; Moldovan, M.; Ittu, M. Comparison of spray and point inoculation to assess resistance to Fusarium head blight in a multienvironment wheat trial. Phytopathology 2003, 93, 1068–1072. [Google Scholar] [CrossRef] [PubMed]
- Hautsalo, J.; Jalli, M.; Manninen, O.; Veteläinen, M. Evaluation of resistance to Fusarium graminearum in oats. Euphytica 2018, 214, 139. [Google Scholar] [CrossRef]
- Wang, Q.; Gottwald, S. Wheat root-dip inoculation with Fusarium graminearum and assessment of root rot disease severity. Bio Protoc. 2017, 7, e2189. [Google Scholar] [CrossRef]
- Mesterhazy, A. Selection of head blight resistant wheats through improved seedling resistance. Plant Breed. 1987, 98, 25–36. [Google Scholar] [CrossRef]
- Miedaner, T. Breeding wheat and rye for resistance to Fusarium diseases. Plant Breed. 1997, 116, 201–220. [Google Scholar] [CrossRef]
- Wang, Q.; Shao, B.; Shaikh, F.I.; Friedt, W.; Gottwald, S. Wheat resistances to Fusarium root rot and head blight are both associated with deoxynivalenol and jasmonate related gene expression. Phytopathology 2018, 108, 602–616. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef]
- Yoshida, M.; Kawada, N.; Nakajima, T. Effect of infection timing on Fusarium head blight and mycotoxin accumulation in open- and closed-flowering barley. Phytopathology 2007, 97, 1054–1062. [Google Scholar] [CrossRef]
- Clear, R.M.; Patrick, S.K.; Platford, R.G.; Desjardins, M. Occurrence and distribution of Fusarium species in barley and oat seed from Manitoba in 1993 and 1994. Can. J. Plant Pathol. 1996, 18, 409–414. [Google Scholar] [CrossRef]
- He, X.; Osman, M.; Helm, J.; Capettini, F.; Singh, P.K. Evaluation of Canadian barley breeding lines for Fusarium head blight resistance. Can. J. Plant Sci. 2015, 95, 923–929. [Google Scholar] [CrossRef] [Green Version]
- Del Ponte, E.M.; Fernandes, J.M.C.; Bergstrom, G.C. Influence of growth stage on Fusarium head blight and deoxynivalenol production in wheat. J. Phytopathol. 2007, 155, 577–581. [Google Scholar] [CrossRef]
- Tittlemier, S.A.; Gaba, D.; Chan, J.M. Monitoring of Fusarium trichothecenes in Canadian cereal grain shipments from 2010 to 2012. J. Agric. Food Chem. 2013, 61, 7412–7418. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Brera, C.; Iha, M.H.; Krska, R.; Lattanzio, V.M.T.; MacDonald, S.; Malone, R.J.; Maragos, C.; Solfrizzo, M.; Stranska-Zachariasova, M.; et al. Developments in mycotoxin analysis: An update for 2015–2016. World Mycotoxin J. 2017, 10, 5–29. [Google Scholar] [CrossRef]
- Casale, W.L.; Pestka, J.J.; Hart, L.P. Enzyme-linked immunosorbent assay employing monoclonal antibody specific for deoxynivalenol (vomitoxin) and several analogues. J. Agric. Food Chem. 1988, 36, 663–668. [Google Scholar] [CrossRef]
- Sinha, R.C.; Savard, M.E.; Lau, R. Production of monoclonal antibodies for the specific detection of deoxynivalenol and 15-acetyldeoxynivalenol by ELISA. J. Agric. Food Chem. 1995, 43, 1740–1744. [Google Scholar] [CrossRef]
- Bai, G.H.; Desjardins, A.E.; Plattner, R.D. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 2002, 153, 91–98. [Google Scholar] [CrossRef]
- Langevin, F.; Eudes, F.; Comeau, A. Effect of trichothecenes produced by Fusarium graminearum during Fusarium head blight development in six cereal species. Eur. J. Plant Pathol. 2004, 110, 735–746. [Google Scholar] [CrossRef]
- Jansen, C.; von Wettstein, D.; Schaefer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef]
- Eudes, F.; Comeau, A.; Rioux, S.; Collin, J. Impact of trichothecenes on Fusarium head blight (Fusarium graminearum) development in spring wheat (Triticum aestivum). Can. J. Plant Pathol. 2001, 23, 318–322. [Google Scholar] [CrossRef]
- Bürstmayr, H.; Ban, T.; Anderson, A.J. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Paul, P.A.; Lipps, P.E.; Madden, L.V. Relationship between visual estimates of Fusarium head blight intensity and deoxynivalenol accumulation in harvested wheat grain: A meta-analysis. Phytopathology 2005, 95, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Buerstmayr, H.; Lemmens, M. Breeding healthy cereals: Genetic improvement of Fusarium resistance and consequences for mycotoxins. World Mycotoxin J. 2015, 8, 591–602. [Google Scholar] [CrossRef]
- van Eeuwijk, F.A.; Mesterhazy, A.; Kling, C.I.; Ruckenbauer, P.; Saur, L.; Bürstmayr, H.; Lemmens, M.; Keizer, L.C.P.; Maurin, N.; Snijders, C.H.A. Assessing non-specificity of resistance in wheat to head blight caused by inoculation with European strains of Fusarium culmorum, F. graminearum and F. nivale using a multiplicative model for interaction. Theor. Appl. Genet. 1995, 90, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Lemmens, M.; Steiner, B.; Sulyok, M.; Nicholson, P.; Mesterhazy, A.; Buerstmayr, H. Masked mycotoxins: Does breeding for enhanced Fusarium head blight resistance result in more deoxynivalenol-3-glucoside in new wheat varieties? World Mycotoxin J. 2016, 9, 741–754. [Google Scholar] [CrossRef]
- Audenaert, K.; De Boevre, M.; Vanheule, A.; Callewaert, J.; Bekaert, B.; Höfte, M.; De Saeger, S.; Haesaert, G. Mycotoxin glucosylation in commercial wheat varieties: Impact on resistance to Fusarium graminearum under laboratory and field conditions. Food Control 2013, 34, 756–762. [Google Scholar] [CrossRef]
- Dall’Asta, C.; Dall’Erta, A.; Mantovani, P.; Massi, A.; Galaverna, G. Occurrence of deoxynivalenol and deoxynivalenol-3-glucoside in durum wheat. World Mycotoxin J. 2013, 6, 83–91. [Google Scholar] [CrossRef]
- Tucker, J.R.; Badea, A.; Blagden, R.; Pleskach, K.; Tittlemier, S.A.; Fernando, W.G.D. Deoxynivalenol-3-glucoside content is highly associated with deoxynivalenol levels in two-row barley genotypes of importance to Canadian barley breeding programs. Toxins 2019, 11, 319. [Google Scholar] [CrossRef]
- Waldron, B.L.; Moreno Sevilla, B.; Anderson, J.A.; Stack, R.W.; Frohberg, R.C. RFLP mapping of QTL for fusarium head blight resistance in wheat. Crop Sci. 1999, 39, 805–811. [Google Scholar] [CrossRef]
- Kolb, F.L.; Bai, G.H.; Muehlbauer, G.J.; Anderson, J.A.; Smith, K.P.; Fedak, G. Host plant resistance genes for Fusarium head blight: Mapping and manipulation with molecular markers. Crop Sci. 2001, 41, 611–619. [Google Scholar] [CrossRef]
- Massman, J.; Cooper, B.; Horsley, R.; Neate, S.; Dill-Macky, R.; Chao, S.; Dong, Y.; Schwarz, P.; Muehlbauer, G.J.; Smith, K.P. Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol. Breed. 2011, 27, 439–454. [Google Scholar] [CrossRef]
- Islam, M.S.; Brown-Guedira, G.; Van Sanford, D.; Ohm, H.; Dong, Y.; McKendry, A.L. Novel QTL associated with the Fusarium head blight resistance in Truman soft red winter wheat. Euphytica 2016, 207, 571–592. [Google Scholar] [CrossRef]
- Jiang, G.L.; Shi, J.; Ward, R.W. QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. I. Resistance to fungal spread. Theor. Appl. Genet. 2007, 116, 3–13. [Google Scholar] [CrossRef]
- Jiang, G.L.; Dong, Y.; Shi, J.; Ward, R.W. QTL analysis of resistance to Fusarium head blight in the novel wheat germplasm CJ 9306. II. Resistance to deoxynivalenol accumulation and grain yield loss. Theor. Appl. Genet. 2007, 115, 1043–1052. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.J.; Fedak, G.; Clarke, J.; Cao, W. Mapping of FHB resistance QTLs in tetraploid wheat. Genome 2006, 49, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Lemmens, M.; Scholz, U.; Berthiller, F.; Dall’Asta, C.; Koutnik, A.; Schuhmacher, R.; Adam, G.; Bürstmayr, H.; Mesterhazy, A.; Krska, R.; et al. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol. Plant Microbe Interact. 2005, 18, 1318–1324. [Google Scholar] [CrossRef]
- Ma, H.X.; Zhang, K.M.; Gao, L.; Bai, G.H.; Chen, H.G.; Cai, Z.X.; Lu, W.Z. Quantitative trait loci for resistance to Fusarium head blight and deoxynivalenol accumulation in Wangshuibai wheat under field conditions. Plant Pathol. 2006, 55, 739–745. [Google Scholar] [CrossRef]
- Yu, J.B.; Bai, G.H.; Zhou, W.C.; Dong, Y.H.; Kolb, F.L. Quantitative trait loci for Fusarium head blight resistance in a recombinant inbred population of Wangshuibai/Wheaton. Phytopathology 2008, 98, 87–94. [Google Scholar] [CrossRef]
- Jayatilake, D.V.; Bai, G.H.; Dong, Y.H. A novel quantitative trait locus for Fusarium head blight resistance in chromosome 7A of wheat. Theor. Appl. Genet. 2011, 122, 1189–1198. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, G.; Leng, Y.; Wanjugi, H.; Xi, P.; Grosz, M.D.; Mergoum, M.; Zhong, S. Molecular mapping of Fusarium head blight resistance in the spring wheat line ND2710. Phytopathology 2018, 108, 972–979. [Google Scholar] [CrossRef]
- He, X.; Singh, P.K.; Dreisigacker, S.; Singh, S.; Lillemo, M.; Duveiller, E. Dwarfing genes Rht-B1b and Rht-D1b are associated with both type I FHB susceptibility and low anther extrusion in two bread wheat populations. PLoS ONE 2016, 11, e0162499. [Google Scholar] [CrossRef]
- Draeger, R.; Gosman, N.; Steed, A.; Chandler, E.; Thomsett, M.; Schondelmaier, J.; Bürstmayr, H.; Lemmens, M.; Schmolke, M.; Mesterházy, Á.; et al. Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina. Theor. Appl. Genet. 2007, 115, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Griffey, C.A.; Hall, M.D.; McKendry, A.L.; Chen, J.; Brooks, W.S.; Brown-Guedira, G.; Van Sanford, D.; Schmale, D.G. Molecular characterization of field resistance to Fusarium head blight in two US soft red winter wheat cultivars. Theor. Appl. Genet. 2013, 126, 2485–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, S.; Lyerly, J.H.; McKendry, A.L.; Islam, M.S.; Brown-Guedira, G.; Cowger, C.; Dong, Y.; Murphy, J.P. Validation of Fusarium head blight resistance QTL in US winter wheat. Crop Sci. 2017, 57, 1–12. [Google Scholar] [CrossRef]
- He, X.; Lillemo, M.; Shi, J.; Wu, J.; Bjørnstad, Å.; Belova, T.; Dreisigacker, S.; Duveiller, E.; Singh, P. QTL characterization of Fusarium head blight resistance in CIMMYT bread wheat line Soru#1. PLoS ONE 2016, 11, e0158052. [Google Scholar]
- Chu, C.; Niu, Z.; Zhong, S.; Chao, S.; Friesen, T.L.; Halley, S.; Elias, E.M.; Dong, Y.; Faris, J.D.; Xu, S.S. Identification and molecular mapping of two QTLs with major effects for resistance to Fusarium head blight in wheat. Theor. Appl. Genet. 2011, 123, 1107. [Google Scholar] [CrossRef]
- Zhao, M.; Leng, Y.; Chao, S.; Xu, S.S.; Zhong, S. Molecular mapping of QTL for Fusarium head blight resistance introgressed into durum wheat. Theor. Appl. Genet. 2018, 131, 1939–1951. [Google Scholar] [CrossRef]
- Schweiger, W.; Steiner, B.; Vautrin, S.; Nussbaumer, T.; Siegwart, G.; Zamini, M.; Jungreithmeier, F.; Gratl, V.; Lemmens, M.; Mayer, K.F.X.; et al. Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 2016, 129, 1607–1623. [Google Scholar] [CrossRef]
- Rawat, N.; Pumphrey, M.O.; Liu, S.; Zhang, X.; Tiwari, V.K.; Ando, K.; Trick, H.N.; Bockus, W.W.; Akhunov, E.; Anderson, J.A.; et al. Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 2016, 48, 1576. [Google Scholar] [CrossRef]
- Su, Z.; Bernardo, A.; Tian, B.; Chen, H.; Wang, S.; Ma, H.; Cai, S.; Liu, D.; Zhang, D.; Li, T.; et al. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 2019, 51, 1099–1105. [Google Scholar] [CrossRef]
- Li, G.; Zhou, J.; Jia, H.; Gao, Z.; Fan, M.; Luo, Y.; Zhao, P.; Xue, S.; Li, N.; Yuan, Y.; et al. Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 2019, 51, 1106–1112. [Google Scholar] [CrossRef]
- Lagudah, E.S.; Krattinger, S.G. A new player contributing to durable Fusarium resistance. Nat. Genet. 2019, 51, 1070–1071. [Google Scholar] [CrossRef] [PubMed]
- Miedaner, T.; Wilde, F.; Steiner, B.; Bürstmayr, H.; Korzun, V.; Ebmeyer, E. Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxynivalenol (DON) content and disease severity. Theor. Appl. Genet. 2006, 112, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Lillemo, M.; Skinnes, H.; He, X.; Shi, J.; Ji, F.; Dong, Y.; Bjørnstad, Å. Anther extrusion and plant height are associated with Type I resistance to Fusarium head blight in bread wheat line ‘Shanghai-3/Catbird’. Theor. Appl. Genet. 2013, 126, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.J.; Fedak, G.; Savard, M. Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome 2003, 46, 555–564. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Dreisigacker, S.; Singh, R.P.; Singh, P.K. Genetics for low correlation between Fusarium head blight disease and deoxynivalenol (DON) content in a bread wheat mapping population. Theor. Appl. Genet. 2019, 132, 2401–2411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semagn, K.; Skinnes, H.; Bjørnstad, Å.; Marøy, A.G.; Tarkegne, Y. Quantitative trait loci controlling Fusarium head blight resistance and low deoxynivalenol content in hexaploid wheat population from ‘Arina’ and NK93604. Crop Sci. 2007, 47, 294–303. [Google Scholar] [CrossRef]
- Ágnes, S.H.; Szabolcs, L.K.; Mónika, V.; László, P.; János, P.; Csaba, L.; Ákos, M. Differential influence of QTL linked to Fusarium head blight, Fusarium-damaged kernel, deoxynivalenol contents and associated morphological traits in a Frontana-derived wheat population. Euphytica 2014, 200, 9–26. [Google Scholar] [CrossRef]
- Venske, E.; dos Santos, R.S.; Farias, D.D.; Rother, V.; da Maia, L.C.; Pegoraro, C.; Costa de Oliveira, A. Meta-analysis of the QTLome of Fusarium head blight resistance in bread wheat: Refining the current puzzle. Front. Plant Sci. 2019, 10, 727. [Google Scholar] [CrossRef]
- Wilde, F.; Korzun, V.; Ebmeyer, E.; Geiger, H.H.; Miedaner, T. Comparison of phenotypic and marker-based selection for Fusarium head blight resistance and DON content in spring wheat. Mol. Breed. 2007, 19, 357–370. [Google Scholar] [CrossRef]
- Brar, G.S.; Brûlé-Babel, A.L.; Ruan, Y.; Henriquez, M.A.; Pozniak, C.J.; Kutcher, H.R.; Hucl, P.J. Genetic factors affecting Fusarium head blight resistance improvement from introgression of exotic Sumai 3 alleles (including Fhb1, Fhb2, and Fhb5) in hard red spring wheat. BMC Plant Biol. 2019, 19, 179. [Google Scholar] [CrossRef]
- McCartney, C.A.; Somers, D.J.; Fedak, G.; DePauw, R.M.; Thomas, J.; Fox, S.L.; Humphreys, D.G.; Lukow, O.; Savard, M.E.; McCallum, B.D.; et al. The evaluation of FHB resistance QTLs introgressed into elite Canadian spring wheat germplasm. Mol. Breed. 2007, 20, 209–221. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [PubMed]
- Rutkoski, J.; Benson, J.; Jia, Y.; Brown-Guedira, G.; Jannink, J.L.; Sorrells, M. Evaluation of genomic prediction methods for Fusarium head blight resistance in wheat. Plant Genome 2012, 5, 51–61. [Google Scholar] [CrossRef]
- Arruda, M.P.; Lipka, A.E.; Brown, P.J.; Krill, A.M.; Thurber, C.; Brown-Guedira, G.; Dong, Y.; Foresman, B.J.; Kolb, F.L. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 84. [Google Scholar] [CrossRef]
- Yu, G.T.; Franckowiak, J.D.; Neate, S.M.; Zhang, B.; Horsley, R.D. A native QTL for Fusarium head blight resistance in North American barley (Hordeum vulgare L.) independent of height, maturity, and spike type loci. Genome 2010, 53, 111–118. [Google Scholar] [CrossRef]
- de la Pena, R.C.; Smith, K.P.; Capettini, F.; Muehlbauer, G.J.; Gallo, M.M.; Dill, M.R.; Somers, D.A.; Rasmusson, D.C. Quantitative trait loci associated with resistance to Fusarium head blight and kernel discoloration in barley. Theor. Appl. Genet. 1999, 99, 561–569. [Google Scholar] [CrossRef]
- Ma, Z.; Steffenson, B.J.; Prom, L.K.; Lapitan, N.L. Mapping of quantitative trait loci for fusarium head blight resistance in barley. Phytopathology 2000, 90, 1079–1088. [Google Scholar] [CrossRef]
- Horsley, R.D.; Schmierer, D.; Maier, C.; Kudrna, D.; Urrea, C.A.; Steffenson, B.J.; Schwarz, P.B.; Franckowiak, J.D.; Green, M.J.; Zhang, B.; et al. Identification of QTLs associated with Fusarium head blight resistance in barley accession CIho 4196. Crop Sci. 2006, 46, 145–156. [Google Scholar] [CrossRef]
- Mesfin, A.; Smith, K.P.; Dill-Macky, R.; Evans, C.K.; Waugh, R.; Gustus, C.D.; Muehlbauer, G.J. Quantitative trait loci for Fusarium head blight resistance in barley detected in a two-rowed by six-rowed population. Crop Sci. 2003, 43, 307–318. [Google Scholar] [CrossRef]
- Huang, Y.; Haas, M.; Heinen, S.; Steffenson, B.J.; Smith, K.P.; Muehlbauer, G.J. QTL mapping of Fusarium head blight and correlated agromorphological traits in an elite barley cultivar Rasmusson. Front. Plant Sci. 2018, 9, 1260. [Google Scholar] [CrossRef]
- Canci, P.C.; Nduulu, L.M.; Muehlbauer, G.J.; Dill-Macky, R.; Rasmusson, D.C.; Smith, K.P. Validation of quantitative trait loci for Fusarium head blight and kernel discoloration in barley. Mol. Breed. 2004, 14, 91–104. [Google Scholar] [CrossRef]
- Dahleen, L.; Agrama, H.; Horsley, R.; Steffenson, B.; Schwarz, P.; Mesfin, A.; Franckowiak, J. Identification of QTLs associated with Fusarium head blight resistance in Zhedar 2 barley. Theor. Appl. Genet. 2003, 108, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Gilchrist, L.; Hayes, P.; Kleinhofs, A.; Kudrna, D.; Liu, Z.; Prom, L.; Steffenson, B.; Toojinda, T.; Vivar, H. Does function follow form? Principal QTLs for Fusarium head blight (FHB) resistance are coincident with QTLs for inflorescence traits and plant height in a doubled-haploid population of barley. Theor. Appl. Genet. 1999, 99, 1221–1232. [Google Scholar] [CrossRef]
- Kleinhofs, A.; Kilian, A.; Saghai Maroof, M.A.; Biyashev, R.M.; Hayes, P.; Chen, F.Q.; Lapitan, N.; Fenwick, A.; Blake, T.K.; Kanazin, V.; et al. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor. Appl. Genet. 1993, 86, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Millett, B.P.; Beaubien, K.A.; Dahl, S.K.; Steffenson, B.J.; Smith, K.P.; Muehlbauer, G.J. Haplotype diversity and population structure in cultivated and wild barley evaluated for Fusarium head blight responses. Theor. Appl. Genet. 2013, 126, 619–636. [Google Scholar] [CrossRef]
- Mamo, B.E.; Steffenson, B.J. Genome-wide association mapping of Fusarium head blight resistance and agromorphological traits in barley landraces from Ethiopia and Eritrea. Crop Sci. 2015, 55, 1494–1512. [Google Scholar] [CrossRef]
- Sallam, A.H.; Endelman, J.B.; Jannink, J.L.; Smith, K.P. Assessing genomic selection prediction accuracy in a dynamic barley breeding population. Plant Genome 2015, 8. [Google Scholar] [CrossRef]
- Tiede, T.; Smith, K.P. Evaluation and retrospective optimization of genomic selection for yield and disease resistance in spring barley. Mol. Breed. 2018, 38, 55. [Google Scholar] [CrossRef]
- Fadel, F.; Wenzel, G. In vitro selection for tolerance to Fusarium in F1 microspore populations of wheat. Plant Breed. 1993, 110, 89–95. [Google Scholar] [CrossRef]
- Eudes, F.; Comeau, A.; Rioux, S.; Collin, J. Trichothecene-mediated in vitro selection in wheat for reduced mycotoxin accumulation caused by Fusarium graminearum. Can. J. Plant Sci. 2008, 88, 1115–1125. [Google Scholar] [CrossRef]
- Legge, W.G.; Tucker, J.R.; Bizimungu, B.; Tekauz, A.; Noll, J.S.; Fetch, T.G.; Menzies, J.G.; Haber, S.; Savard, M.E.; Vigier, B.J.; et al. Norman barley. Can. J. Plant Sci. 2011, 91, 1105–1113. [Google Scholar] [CrossRef]
- Legge, W.G.; Tucker, J.R.; Bizimungu, B.; Tekauz, A.; Fetch, T.G.; Haber, S.; Menzies, J.G.; Noll, J.S.; Turkington, T.K.; Martin, R.A.; et al. Taylor barley. Can. J. Plant Sci. 2013, 93, 969–977. [Google Scholar] [CrossRef]
- Marchand, S.; Clermont, I.; Belzile, F. In vitro selection of barley microspores for increased deoxynivalenol tolerance. In Proceedings of the 7th Canadian Workshop of Fusarium Head Blight, Ottawa, ON, Canada, 20–22 November 2016; p. 35. [Google Scholar]
- Jiang, F.; Ryabova, D.; Diedhiou, J.; Hucl, P.; Randhawa, H.; Marillia, E.F.; Foroud, N.A.; Eudes, F.; Kathiria, P. Trichostatin A increases embryo and green plant regeneration in wheat. Plant Cell Rep. 2017, 36, 1701–1706. [Google Scholar] [CrossRef] [PubMed]
- Dänicke, S.; Brezina, U. Kinetics and metabolism of the Fusarium toxin deoxynivalenol in farm animals: Consequences for diagnosis of exposure and intoxication and carry over. Food Chem. Toxicol. 2013, 60, 58–75. [Google Scholar] [CrossRef]
- Yiannikouris, A.; Jouany, J.P. Mycotoxins in feeds and their fate in animals: A review. Anim. Res. 2002, 51, 81–99. [Google Scholar] [CrossRef]
- Schoental, R. Chronic, including teratogenic and carcinogenic effects of trichothecenes: A short review. Vet. Res. Commun. 1983, 7, 165–170. [Google Scholar] [CrossRef]
- Doi, K.; Ishigami, N.; Sehata, S. T-2 toxin-induced toxicity in pregnant mice and rats. Int. J. Mol. Sci. 2008, 9, 2146–2158. [Google Scholar] [CrossRef]
- Cope, R.B. Chapter 75—Trichothecenes. In Veterinary Toxicology, 3rd ed.; Gupta, R.C., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 1043–1053. [Google Scholar]
- Berthiller, F.; Dall’Asta, C.; Schuhmacher, R.; Lemmens, M.; Adam, G.; Krska, R. Masked mycotoxins: Determination of a deoxynivalenol glucoside in artificially and naturally contaminated wheat by liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 2005, 53, 3421–3425. [Google Scholar] [CrossRef]
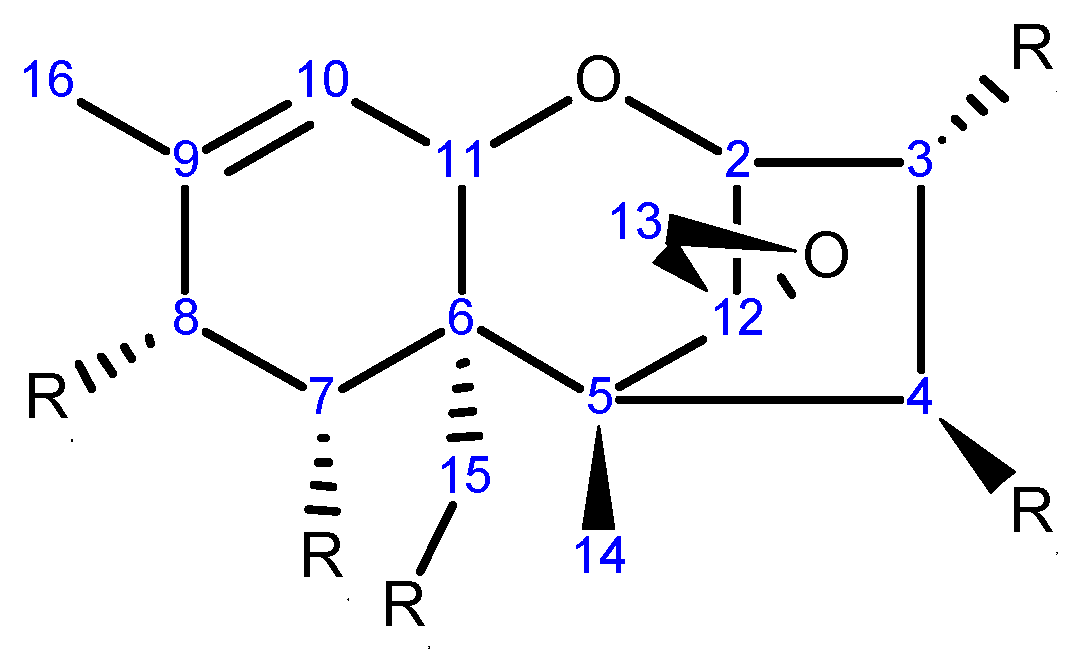

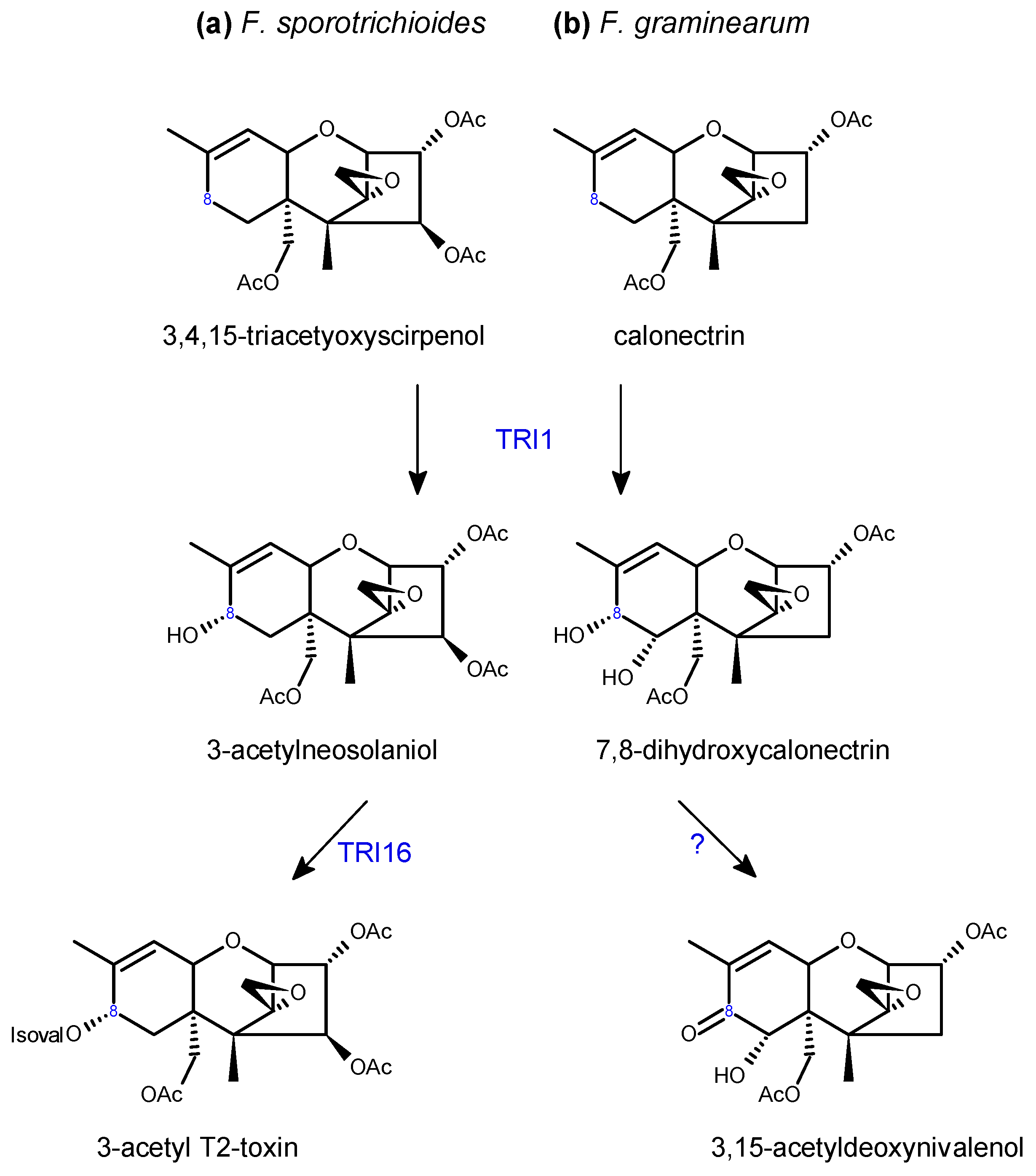
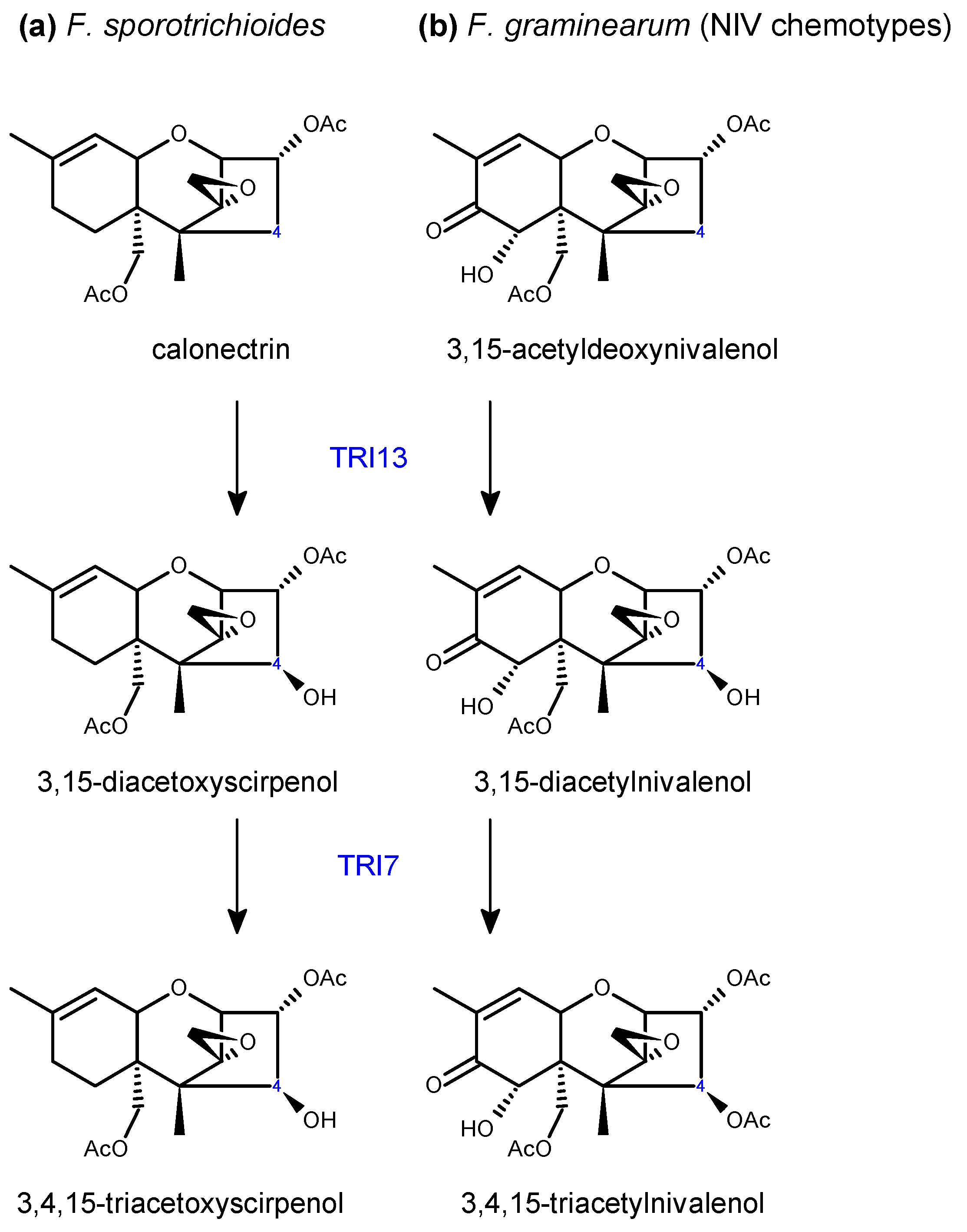
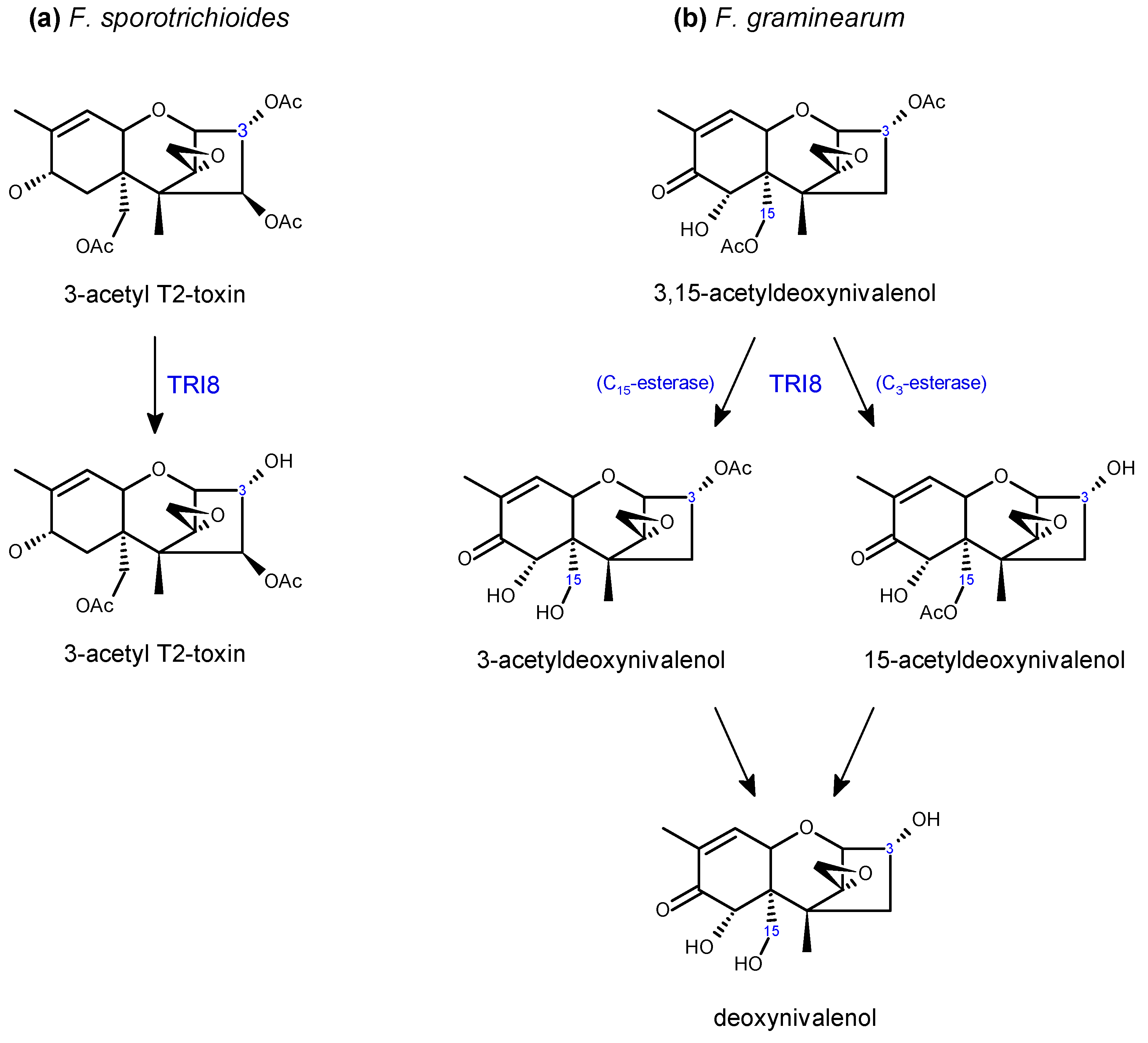

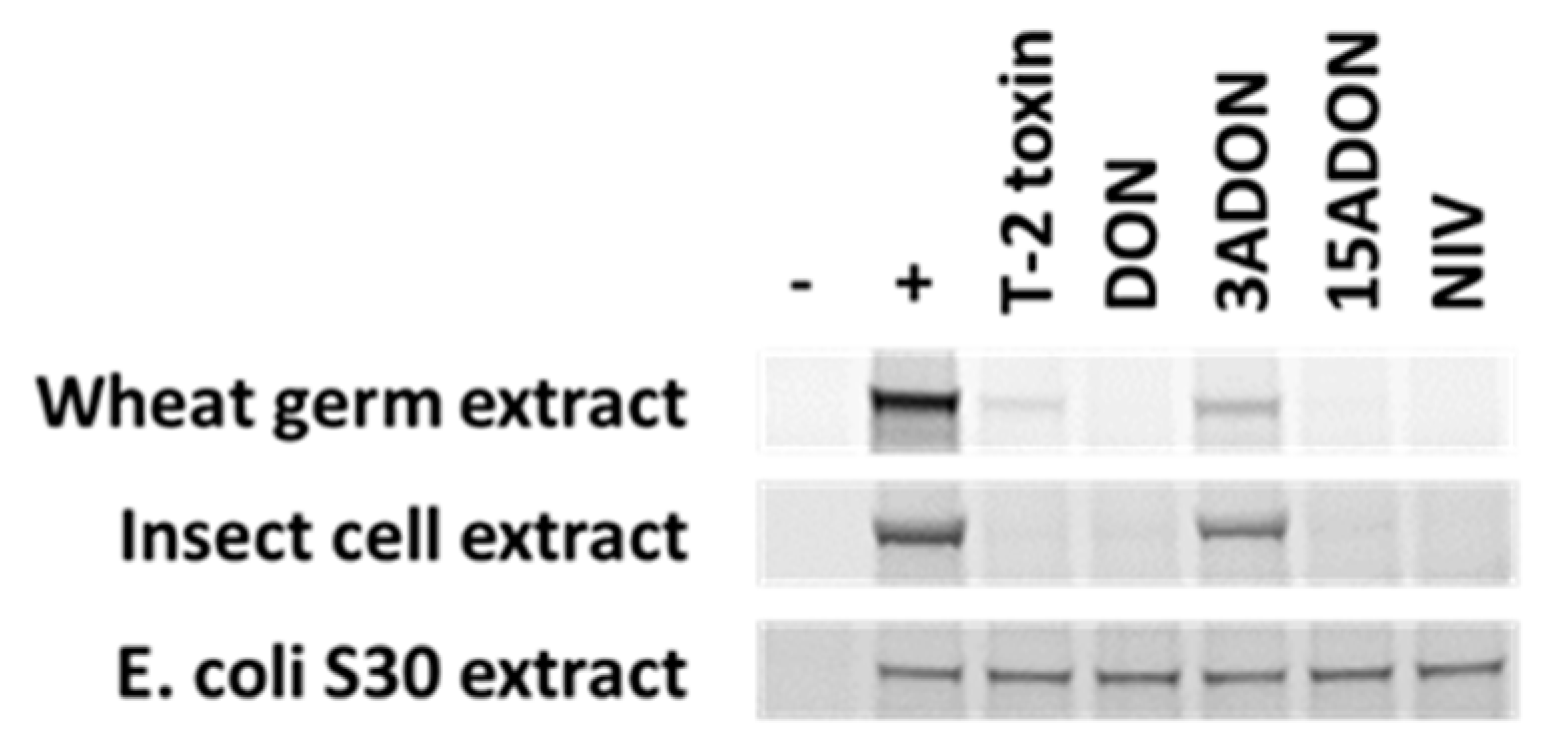

| C3 | C4 | C7 | C8 | C15 | |
|---|---|---|---|---|---|
| Type A | |||||
| diacetoxyscirpenol (DAS) | -OH | -OAc | -H | -H | -OAc |
| trichodermin | -H | -OAc | -H | -H | -H |
| trichodermol | -H | -OH | -H | -H | -H |
| T-2 toxin | -OH | -OAc | -H | -OIsoval | -OAc |
| HT-2 toxin | -OH | -OH | -H | -OIsoval | -OAc |
| NX-2 | -OAc | -H | -OH | -H | -OH |
| NX-3 | -OH | -H | -OH | -H | -OH |
| Type B | |||||
| nivalenol (NIV) | -OH | -OH | -OH | =O | -OH |
| 4-O-acetyl-NIV (4ANIV) | -OH | -OAc | -OH | =O | -OH |
| 4-deoxy-nivalenol (DON) | -OH | -H | -OH | =O | -OH |
| 3-O-acetyl-DON (3-ADON) | -OAc | -H | -OH | =O | -OH |
| 15-O-acetyl-DON (15-ADON) | -OH | -H | -OH | =O | -OAc |
| trichothecin | -H | -OIsoval | -H | =O | -H |
| Gene | Cluster | Gene Description | Activity |
|---|---|---|---|
| Biosynthesis | |||
| TRI1 | Tri1-Tri16 | Cytochrome p450 monooxygenase | C7 hydroxylation [81] C8 hydroxylation [69,81,82] |
| TRI3 | Core Tri | Acetyltransferase | C15 acetylation [83,84] |
| TRI4 | Core Tri | Cytochrome p450 monooxygenase | C2 hydroxylation [76] C3 hydroxylation [77,78] C11 hydroxylation [77,78] C12,13 epoxidation [77,78] |
| TRI5 | Core Tri | Sesquiterpene cyclase | Cyclization of farnesyl pyrophosphate; TRICHODIENE SYNTHASE [19,74,85] |
| TRI7 | Core Tri | Acetyltransferase | C4 acetylation [86,87] |
| TRI8 | Core Tri | Esterase | C3 deacetylation [88,89] C15 deacetylation [89] |
| TRI11 | Core Tri | Cytochrome p450 monooxygenase | C15 hydroxylation [90] |
| TRI13 | Core Tri | Cytochrome p450 monooxygenase | C4 hydroxylation [86,91] |
| TRI16 | Tri1-Tri16 | Acetyltransferase | C8 acetylation [69] |
| TRI101 | None | Acetyltransferase | C3 acetylation [70,92,93] |
| Transcription Factors | |||
| TRI6 | Core Tri | Zinc-finger DNA binding protein | Regulation of TRI gene expression [94,95] |
| TRI10 | Core Tri | Transcription factor | Regulation of TRI gene expression [96,97] |
| TRI15 | None | Zinc-finger DNA binding protein | Negative regulation of trichothecene biosynthesis [71] |
| Toxin Efflux | |||
| TRI12* | Core Tri | MFS transporter | Toxin efflux [33,98,99] |
| Unknown | |||
| TRI9 | Core Tri | ||
| TRI14 | Core Tri | Role in pathogenesis and trichothecene production [100] | |
| Trichothecene | Translation Inhibition Type |
|---|---|
| DAS | I-type [186] |
| HT-2 toxin | I-type [186,187] |
| trichothecin | E-type [187], E-type [187] |
| T-2 toxin | I-type [186,187], T-type [189] |
| trichodermin | I-type [187], E-type [186,187,189,192], T-type [186,187,188,189,190,193] |
| trichodermol | E-type [187], T-type [187] |
| NIV | I-type [186,187] |
| Type | Description |
|---|---|
| FHB resistance | |
| Type I | Resistance to initial infection [294] |
| Type II | Resistance to disease spread from one spikelet to another [294] |
| Type III | Resistance to kernel infection [293] |
| Type IV | Tolerance against FHB and trichothecenes [295] |
| Type V Class 1 Class 2 | Resistance to trichothecenes [296] by Degradation or detoxification [238] by Preventing their accumulation [238] |
| FEB resistance | |
| Silk resistance | Resistance to silk penetration [27] |
| Kernel resistance | Resistance to cob penetration, preventing spread from one kernel to another [27] |
| Reference | Chromosomal Allocation of QTL | Designated Gene/QTL | QTL of FHB Resistance Traits * | Coinciding Morpho/Phenological QTL | Source of Resistance Allele |
|---|---|---|---|---|---|
| Islam et al. [332] | 2DS | Ppd-D1 | type2, FHB sev, FDK, DON | flowering time, plant height | Truman |
| Jiang et al. [333,334] | 2DL | Qfhs.nau-2DL | type2, DON | CJ 9306 | |
| Somers et al., Lemmens et al. Ma et al., Jiang et al., Yu et al., Jayatilake et al., and Zhao et al. [333,335,336,337,338,339,340] | 3BS | Fhb1 | type1, type2, FDK, DON, DON resistance | Sumai-3, CM-82036, Nyu Bai, Wangshiubai, CJ 9306, ND2710 | |
| He et al. [341] | 4BS | Rht-B1 | FHB sev, FDK, DON | plant height, anther extrusion | Ocoroni F86 |
| Draeger et al., Liu et al., and Petersen et al. [342,343,344] | 4DS | Rht-D1 | type1, FHB sev, FHB index, AUDPC, FDK, DON | plant height | Arina, Ernie, Bess |
| Ma et al., and Jiang et al. [333,334,337] | 5AS | Fhb5 | type2, DON | Wangshuibai, CJ 9306 | |
| He et al. [341,345] | 5AL | Vrn-A1 | type2, FHB sev, FHB index, FDK, DON | heading date, plant height | Soru#1 synthetic hexaploid wheat, Ivan/Soru#2 |
| Chu et al., and Zhao et al. [346,347] | 5AL | q | type2, FHB sev, FDK, DON | plant height, spelt type | PI277012 |
| Zhao et al. [340] | 6B | Fhb2 | type2, DON | ND2710 | |
| Jayatilake et al. [339] | 7A | Fhb7AC | type2, FDK, DON | Sumai-3 |
| Reference | Chromosomal Allocation of QTL* | Designated QTL | QTL of FHB Resistance Traits** | Coinciding Morpho/Phenological QTL | Source of Resistance Allele |
|---|---|---|---|---|---|
| de la Penna et al., and Ma et al. [367,368] | 1H | NA | DON, FHB sev^, KD; DON | heading date, plant height; number of nodes per centimeter of rachis | Chevron |
| Canci et al., Dahleen et al., de la Penna et al., Horsley et al., Ma et al., Mesfin et al., and Zhu et al. [367,368,369,370,372,373,374] | 2H (bin 8 to 10) | Qrgz-2H-8, Qrgz-2H-10 | DON, FHB sev, KD | lateral floret size, heading date, plant height, rachis node number, seeds per inflorescence, spike type | CI 4196, Chevron, CMB643 (Azafran), Fredrickson, Zhedar 2 |
| Horsley et al., and Zhu et al. [369,374] | 4H (bin 2 and 4) | QDON-4H-2; NA | DON; DON, FHB sev | NA; seeds per inflorescence | CI 4196; Gobernadora (Zhenmai-1) |
| Dahleen et al. [373] | 6H | NA | DON, FHB sev | heading date | Zhedar 2 |
| Ma et al. [368] | 7H | NA | DON, FHB sev | heading date, plant height, | Chevron |
| Food products | |
| Finished wheat products for consumption by humans | 1000 ppb (USA) |
| Uncleaned soft wheat for human consumption | 2000 ppb (Canada) |
| Unprocessed cereals (excluding durum wheat, oats, and maize) | 1250 ppb (Europe) |
| Unprocessed durum wheat and oats | 1750 ppb (Europe) |
| Unprocessed maize | 1750 ppb (Europe) |
| Cereal flour, maize flour, maize, grits, and maize meal | 750 ppb (Europe) |
| Bread, biscuits, pastries, cereal snacks and breakfast cereals | 500 ppb (Europe) |
| Dry pasta | 750 ppb (Europe) |
| Processed cereal based baby and infant food | 200 ppb (Europe) |
| Feed | |
| Grains and grain by-products destined for ruminating beef and feedlot cattle older than 4 months and for chickens | 10 ppm (Europe) |
| Grains and grain by-products destined for ruminating beef and feedlot cattle older than 4 months and chickens (not exceeding 50% of the cattle or chicken total diet) | 10 ppm (USA) |
| Grains and grain by-products destined for ruminating beef and feedlot cattle older than 4 months and chickens (not exceeding 50% of the cattle or chicken total diet) | 5 ppm (Canada) |
| Grain and grain by-products destined for swine | 5 ppm (Europe) |
| Grain and grain by-products destined for swine | 5 ppm (USA) |
| Grain and grain by-products destined for swine | 1 ppm (Canada) |
| Grain and grain by-products destined for other animals | 5 ppm (Europe) |
| Grains and grain by-products (not exceeding 40% of the diet) for young calves, & lactating dairy animals | 1 ppm (Canada) |
| Grains and grain by-products (not exceeding 40% of the diet) for young calves, & lactating dairy animals | 5 ppm (USA) |
| Cereals and cereal products with the exception of maize by-products | 8 ppm (Europe) |
| Maize by-products | 12 ppm (Europe) |
| Complementary and complete feeding stuffs | 5 ppm (Europe) |
| Complementary and complete feeding stuffs for pigs | 0.9 ppm (Europe) |
| Complementary and complete feeding stuffs for calves (<4 months), lambs, kids and dogs | 2 ppm (Europe) |
| Unprocessed cereals | |
| Barley (including malting barley) and maize | 200 ppb |
| Oats (with husk) | 1000 ppb |
| Wheat, rye and other cereals | 100 ppb |
| Cereal grains for direct human consumption | |
| Oats | 200 ppb |
| Maize | 100 ppb |
| Other Cereals | 50 ppb |
| Cereal products for human consumption | |
| Oat bran and flaked oats | 200 ppb |
| Cereal bran except oat bran, oat milling products other than oat bran and flaked oats and maize milling products | 100 ppb |
| Other cereal milling products | 50 ppb |
| Breakfast cereals including formed cereal flakes | 75 ppb |
| Cereal products for feed and compound feed | |
| Oat milling products (husks) | 2000 ppb (Europe) |
| Other cereal products | 500 ppb (Europe) |
| Compound feed, with the exception of feed for cats | 250 ppb (Europe) |
| Compound feed for cats | 50 ppb (Europe) |
| Diets for cattle and poultry | 0.1 ppm (Canada) |
| Diets for dairy animals | 0.025 ppm (Canada) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in Cereal Grains – An Update. Toxins 2019, 11, 634. https://doi.org/10.3390/toxins11110634
Foroud NA, Baines D, Gagkaeva TY, Thakor N, Badea A, Steiner B, Bürstmayr M, Bürstmayr H. Trichothecenes in Cereal Grains – An Update. Toxins. 2019; 11(11):634. https://doi.org/10.3390/toxins11110634
Chicago/Turabian StyleForoud, Nora A., Danica Baines, Tatiana Y. Gagkaeva, Nehal Thakor, Ana Badea, Barbara Steiner, Maria Bürstmayr, and Hermann Bürstmayr. 2019. "Trichothecenes in Cereal Grains – An Update" Toxins 11, no. 11: 634. https://doi.org/10.3390/toxins11110634





