Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease
Abstract
:1. Introduction
1.1. Objective
1.2. Specific Aims
- To study modifications of the infants’ microbiome in relation to specific environmental factors, presence or absence of HLA DQ2 and/or DQ8 predisposing genes, and in relation to tolerance vs. immune response leading to the autoimmune intestinal insult typical of CD;
- To study the infants’ metabolomic phenotype variation in relation to tolerance vs. immune response leading to the autoimmune intestinal insult typical of CD; and
- To investigate the impact of specific bacteria-derived metabolites on gut mucosal molecular pathways leading to the early steps of CD pathogenesis.
2. Study Design
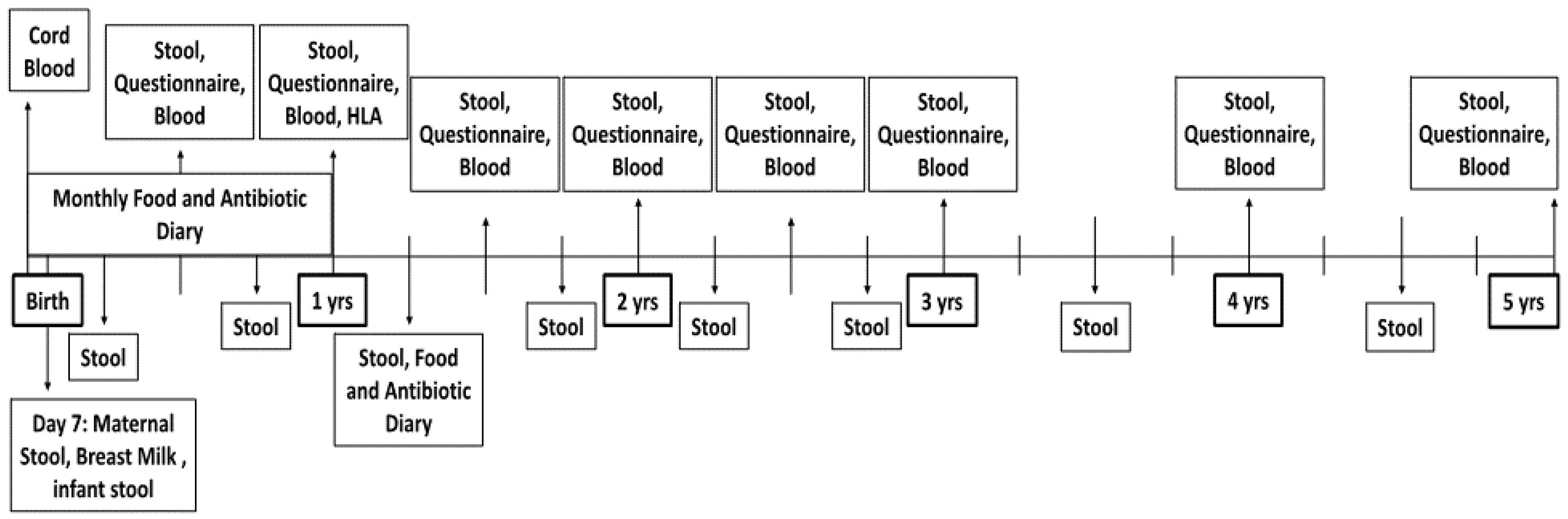
2.1. Participants
2.2. Data Collection
2.3. Parental and Child Questionnaires
2.4. Serological Markers
| Age in Months | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 15 | 18 | 21 | 24 | 27 | 30 | 33 | 36 | 42 | 48 | 54 | 60 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal Stool and Breast Milk Sample | X | ||||||||||||||||||||||||
| Cord Blood | X | ||||||||||||||||||||||||
| Blood Sample | X | X * | X | X | X | X | X | X | |||||||||||||||||
| Stool Sample | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||
| Food Diary | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||
| Antibiotic Diary | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||
| Maternal Diet | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||
| Anthropometrics | X | X | X | X | X | X | X | X | X | X | |||||||||||||||
| Medical History | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||||
| Parent and Child Demographics | X | X | X | X | X | X | |||||||||||||||||||
| Assessment of Sleep and Activity | X | X | X | X | X | X | X | X | X | X |
2.5. Whole Blood
2.6. Stool
2.7. Maternal Samples
2.8. Diagnosis of Celiac Disease
3. Factors of Interest
3.1. Environmental
3.2. Genetic
3.3. Microbiome/Metabolome
4. Statistical Approach
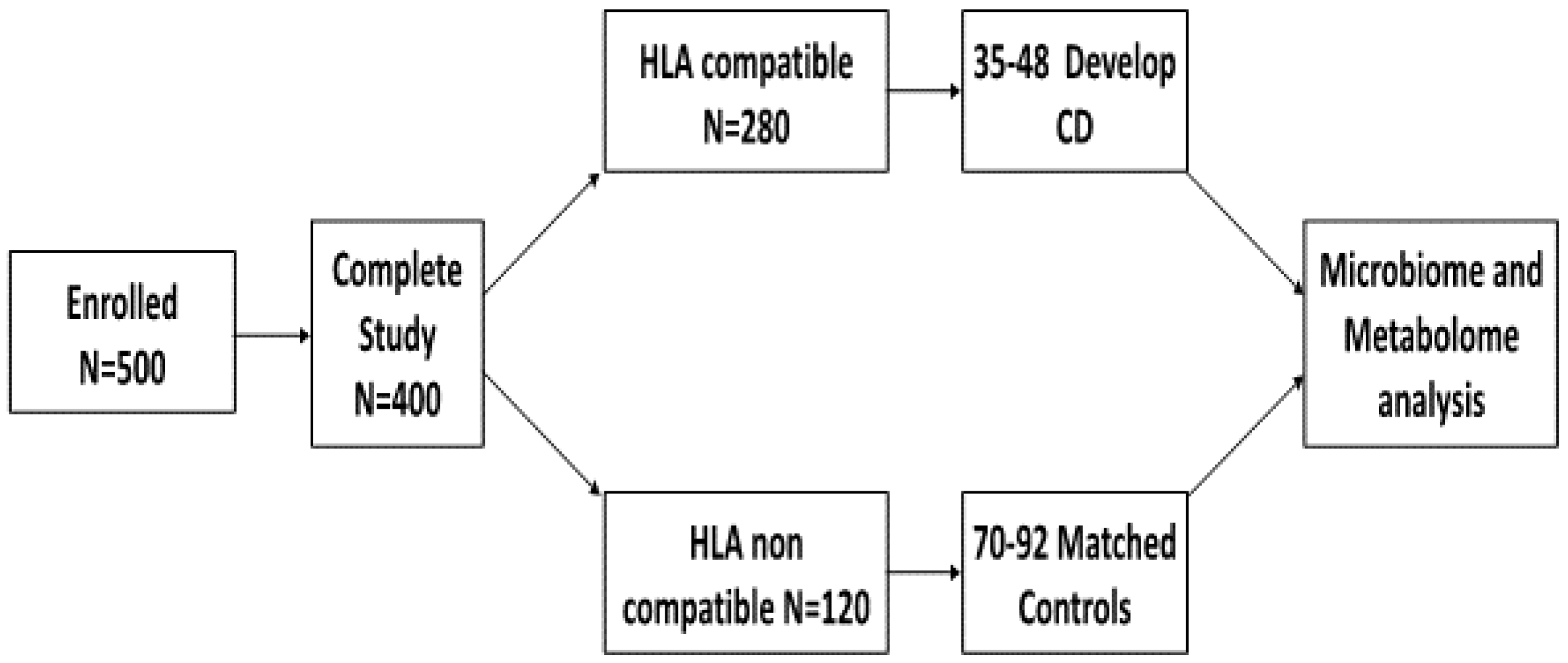
4.1. Power Analysis
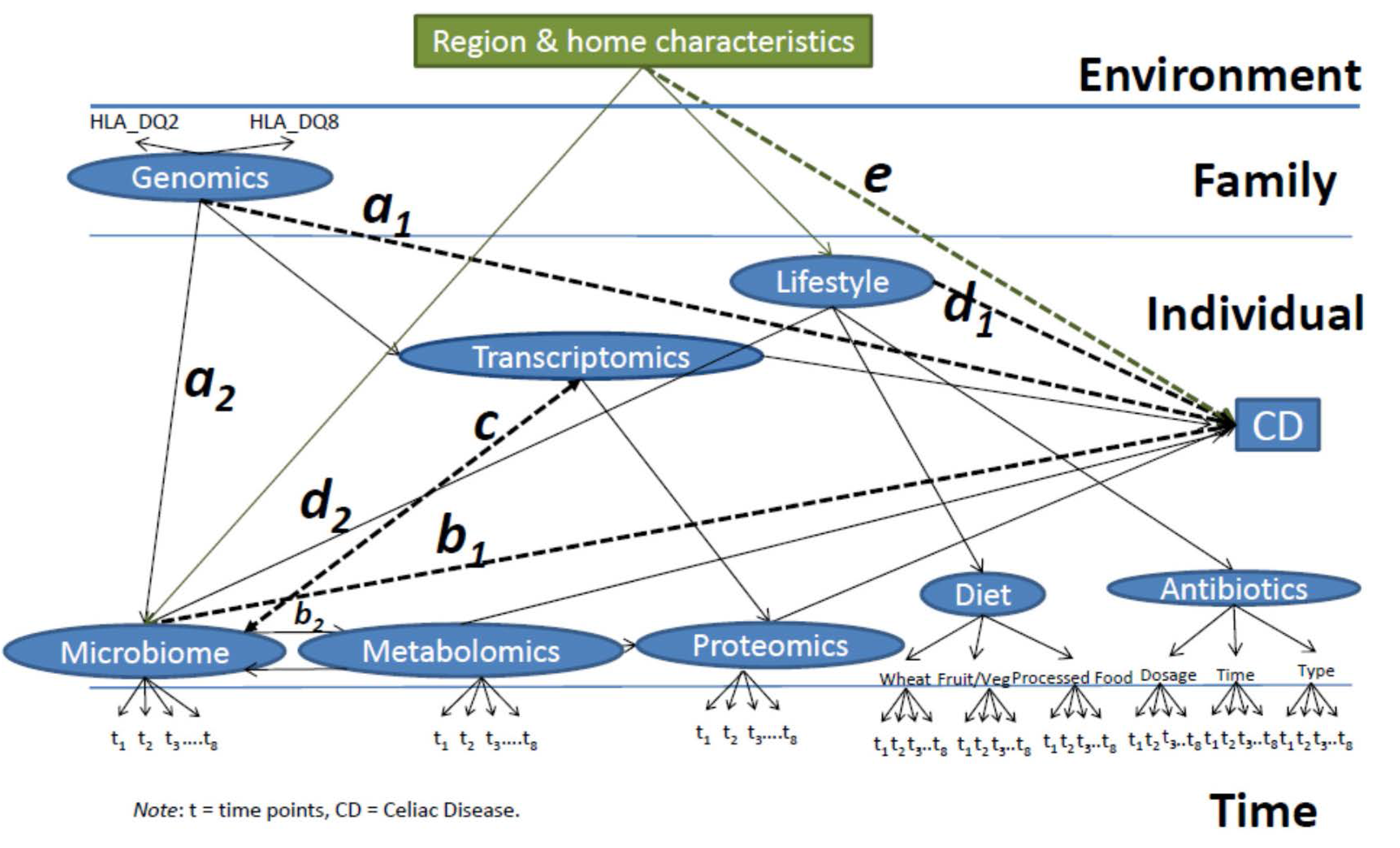
4.2. Developing an Integrative Multilevel Model to Predict Celiac Disease
- (1)
- Path a: a causal path between the genetics and the final effect on the onset of CD, with the microbiome as the mediator (a1 × a2), such that the intestinal microbiome will be altered in order for the genetics to ultimately influence the development of disease (microbiome-mediated epigenetic pressure);
- (2)
- Path b: a causal path of the microbiome’s effect on CD mediated (b1 × b2) by the metabolic activity of intestinal bacteria, which, through the production of metabolites, affects the intestinal transcriptome and proteome. Including a bidirectional path between microbiome and metabolomics to capture how metabolism could alter the microbial metabolism and ultimately the risk for CD;
- (3)
- Path c: A bidirectional path between the transcriptome and the microbiome as their causal relationship will depend on each other;
- (4)
- Path d: a direct relationship between lifestyle factors (here represented by dietary regimen and antibiotic intake) and the onset of the disease mediated (d1 × d2) by alterations in the microbiome;
- (5)
- Path e: The environment (including region characteristics, urban compared to rural, number of individuals living in the household, birth order, pets present) affecting the onset of the disease as a mediator and affecting the composition of the microbiome and resulting metabolome, in turn, influencing gene and protein expression;
- (6)
- Full Model: Time points will be clustered within individual characteristics, analyzing all the paths listed above (Figure 3), individuals will be clustered within families, and families within regions.
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Green, P.H.R.; Cellier, C. Celiac disease. N. Engl. J. Med. 2007, 357, 1731–1743. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Kryszak, D.; Bhatti, B.; Sturgeon, C.; Helzlsouer, K.; Clipp, S.L.; Gelfond, D.; Puppa, E.; Sferruzza, A.; Fasano, A. Natural history of celiac disease autoimmunity in a USA cohort followed since 1974. Ann. Med. 2010, 42, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Sellitto, M.; Bai, G.; Serena, G.; Fricke, W.F.; Sturgeon, C.; Gajer, P.; White, J.R.; Koenig, S.S.; Sakamoto, J.; Boothe, D.; et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE 2012, 7, e33387. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, E.; Castellaneta, S.; Francavilla, R.; Pulvirenti, A.; Tonutti, E.; Amarri, S.; Barbato, M.; Barbera, C.; Barera, G.; Bellantoni, A.; et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 2014, 371, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Vriezinga, S.L.; Auricchio, R.; Bravi, E.; Castillejo, G.; Chmielewska, A.; Crespo Escobar, P.; Kolacek, S.; Koletzko, S.; Korponay-Szabo, I.R.; Mummert, E.; et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 2014, 371, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Decker, E.; Engelmann, G.; Findeisen, A.; Gerner, P.; Laass, M.; Ney, D.; Posovszky, C.; Hoy, L.; Hornef, M.W. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatrics 2010, 125, e1433–e1440. [Google Scholar] [CrossRef] [PubMed]
- Emilsson, L.; Magnus, M.L.; Stordal, K. Perinatal risk factors for development of celiac disease in children, base on the propsective norwegian mother and child cohort study. Clin. Gastroenterol. Hepataol. 2015, 13, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Szajewska, H.; Shamir, R.; Chmielewska, A.; Piescik-Lech, M.; Auricchio, R.; Ivarsson, A.; Kolacek, S.; Koletzko, S.; Korponay-Szabo, I.; Mearin, M.L.; et al. Systematic review with meta-analysis: Early infant feeding and coeliac disease—Update 2015. Aliment. Pharmacol. Ther. 2015, 41, 1038–1054. [Google Scholar] [CrossRef] [PubMed]
- Canova, C.; Zabeo, V.; Pitter, G.; Romor, P.; Baldovin, T.; Zanotti, R.; Simonato, L. Association of maternal education, early infections, and antibiotic use with celiac disease: A population-based birth cohort study in northeastern Italy. Am. J. Epidemiol. 2014, 180. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Lee, H.S.; Aronsson, C.A.; Hagopian, W.A.; Koletzko, S.; Rewers, M.J.; Eisenbarth, G.S.; Bingley, P.J.; Bonifacio, E.; Simell, V.; et al. Risk of pediatric celiac disease according to HLA haplotype and country. N. Engl. J. Med. 2014, 371, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Neef, A.; Castillejo, G.; Palma, G.D.; Varea, V.; Capilla, A.; Palau, F.; Nova, E.; Marcos, A.; Polanco, I.; et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut 2015, 64, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Feigelson, H.S.; Bischoff, K.; Ardini, M.E.; Ravel, J.; Gail, M.H.; Flores, R.; Goedert, J.J. Feasibility of self-collection of fecal specimens by randomly sampled women for health-related studies of the gut microbiome. BMC Res. Notes 2014, 7, 204. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Fasano, A. Celiac disease diagnosis: Simple rules are better than complicated algorithms. Am. J. Med. 2010, 123, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Division of Nutrition, Physical Activity, and Obesity, National Center for Chronic Disease Prevention and Health Promotion. Infant Feeding Practices II. Available online: http://www.cdc.gov/nccdphp/dnpao/index.html (accessed on 14 July 2015).
- Gorzelak, M.A.; Gill, S.K.; Tasnim, N.; Ahmadi-Vand, Z.; Jay, M.; Gibson, D.L. Methods for Improving Human Gut Microbiome Data by Reducing Variability through Sample Processing and Storage of Stool. PLoS ONE 2015, 10, e0134802. [Google Scholar] [CrossRef] [PubMed]
- Magnus, P.; Igrens, L.M.; Haug, K.; Nystad, W.; Skjaerven, R.; Stoltenberg, C.; MoBa Study Group. Cohort profile: The norwegian mother and child cohort study (NoBa). Int. J. Epidemiol. 2006, 35, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Ivarsson, A.; Persson, L.A.; Nystrom, L.; Ascher, H.; Cavell, B.; Danielsson, L.; Dannaeus, A.; Lindberg, T.; Lindquist, B.; Stenhammar, L.; et al. Epidemic of coeliac disease in Swedish children. Acta Paediatr. 2000, 89, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Marlid, K.; Weimin, Y.; Lebwohl, B.; Green, P.H.R.; Blaser, M.J.; Card, T.; Ludvigsson, J.F. Antibiotic exposure and the development of coeliac disease: A nationwide case-control study. BMC Gastroenterol. 2013, 13. [Google Scholar] [CrossRef]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Fairchild, A.J.; MacKinnon, D.P. A general model for testing mediation and moderation effects. Prev. Sci. 2009, 10, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Blood, E.A.; Cheng, D.M. The use of mixed models for the analysis of mediated data with time-dependent predictors. J. Environ. Public Health 2011, 2011, 435078:1–435078:12. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.M.; Kenny, D.A. The moderator-mediator variable distinction in social psychological research: Conceptual, strategic, and statistical considerations. J. Pers. Soc. Psychol. 1986, 51, 1173–1182. [Google Scholar] [PubMed]
- Kenny, D.A.; Korchmaros, J.D.; Bolger, N. Lower level mediation in multilevel models. Psychol. Methods 2003, 8, 115–128. [Google Scholar] [CrossRef] [PubMed]
- US Department of Health and Human Services, National Institutes of Health National Institute of Allergy and Infectious Diseases. Progress in Autoimmune Diseases Research; 2005. Available online: http://www.niaid.nih.gov/topics/autoimmune/Documents/adccfinal.pdf (accessed on 16 June 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonard, M.M.; Camhi, S.; Huedo-Medina, T.B.; Fasano, A. Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease. Nutrients 2015, 7, 9325-9336. https://doi.org/10.3390/nu7115470
Leonard MM, Camhi S, Huedo-Medina TB, Fasano A. Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease. Nutrients. 2015; 7(11):9325-9336. https://doi.org/10.3390/nu7115470
Chicago/Turabian StyleLeonard, Maureen M., Stephanie Camhi, Tania B. Huedo-Medina, and Alessio Fasano. 2015. "Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease" Nutrients 7, no. 11: 9325-9336. https://doi.org/10.3390/nu7115470




