In Vitro Human Microbiota Response to Exposure to Silver Nanoparticles Biosynthesized with Mushroom Extract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Mushrooms, Extracts, and Obtaining NPs
2.3. Nanoparticle Characterization
2.4. In Vitro Colon-Simulation Tests
2.5. Antioxidant Activity
2.6. Metabolic Activities of the Microbiota Simulated in the In Vitro GIS1 System
2.7. Microbial Community Evolution by qPCR Analysis
2.8. The Antimicrobial Assays and Biofilm Inhibition of AgNPs
2.9. Statistical Analysis
3. Results
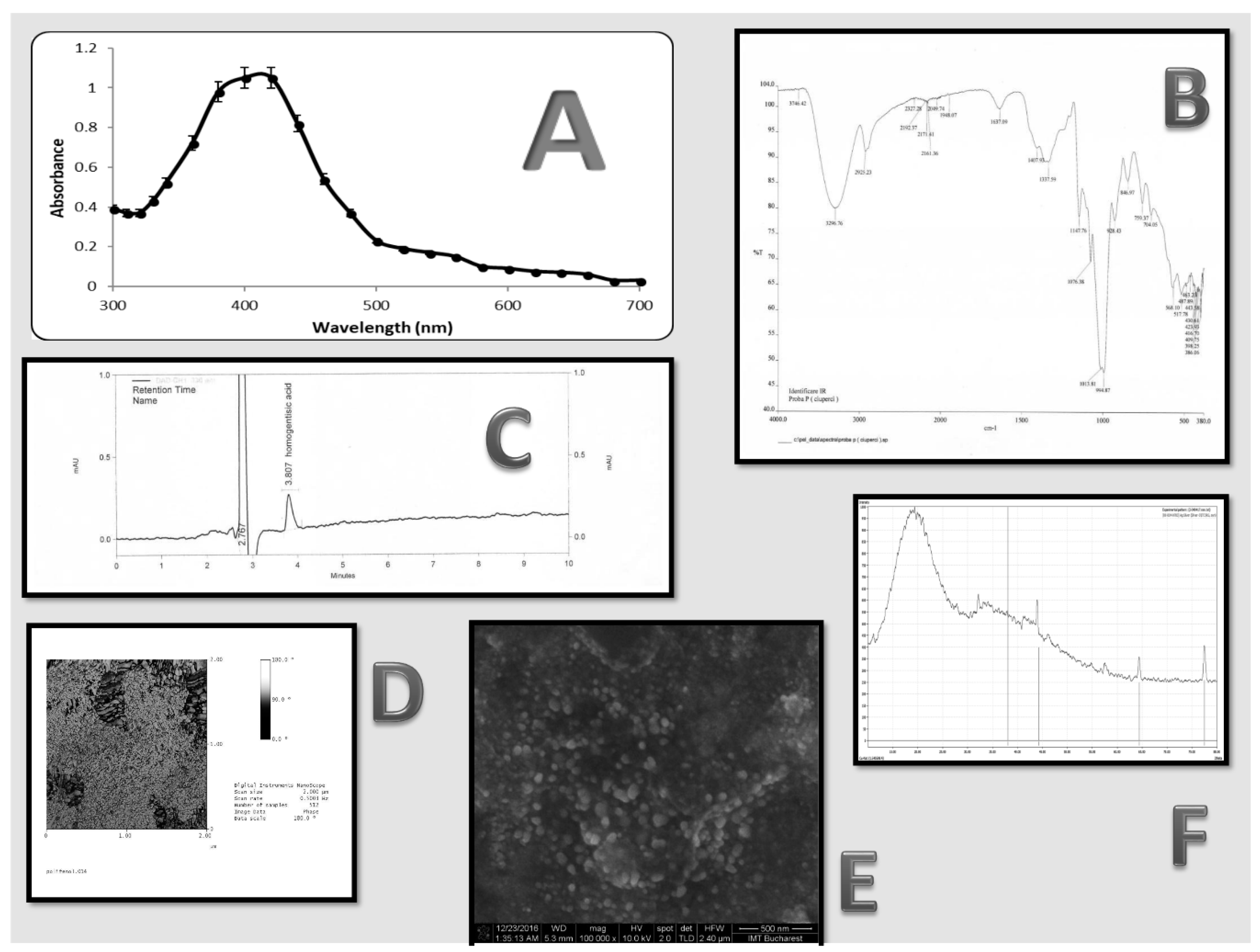
3.1. AgNP Characterization
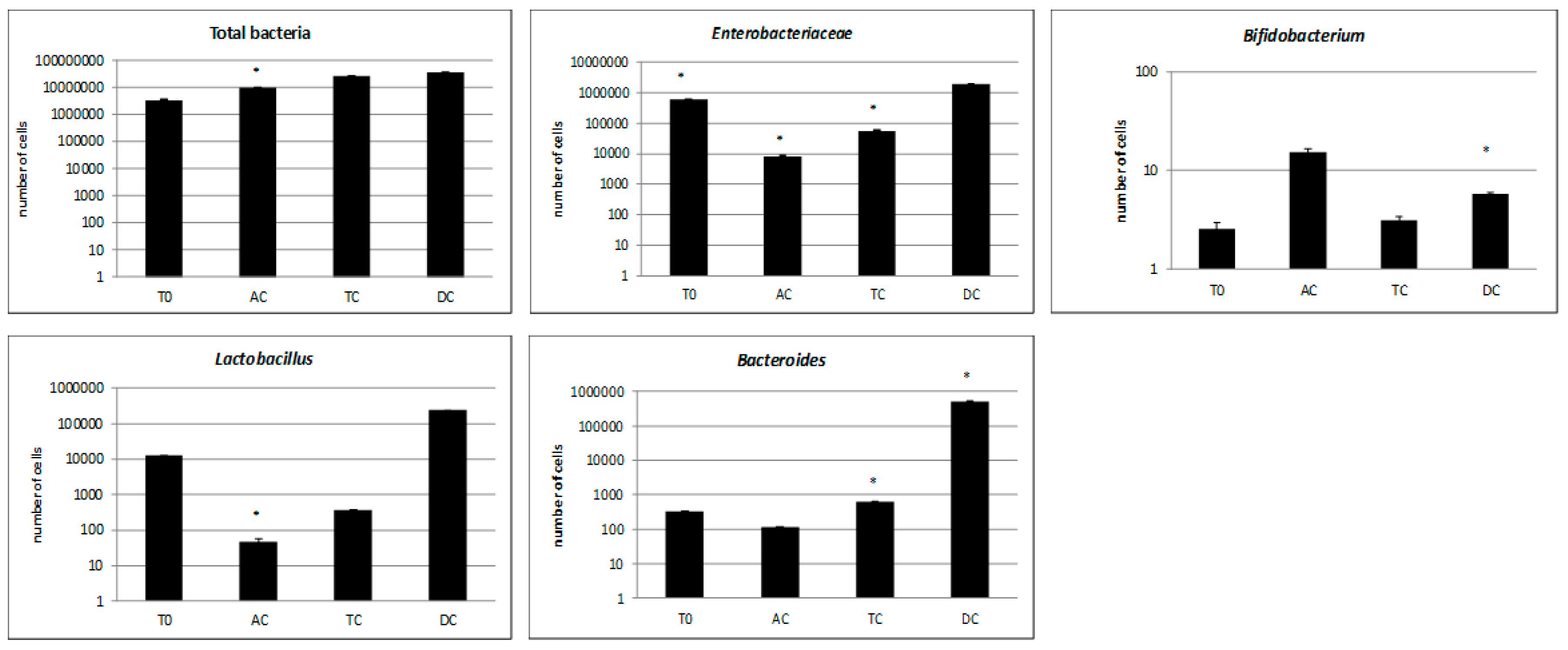
3.2. Microbiological Analysis of the Microbiota Composition. Approach of Antimicrobial Effect and Biofilm Inhibition
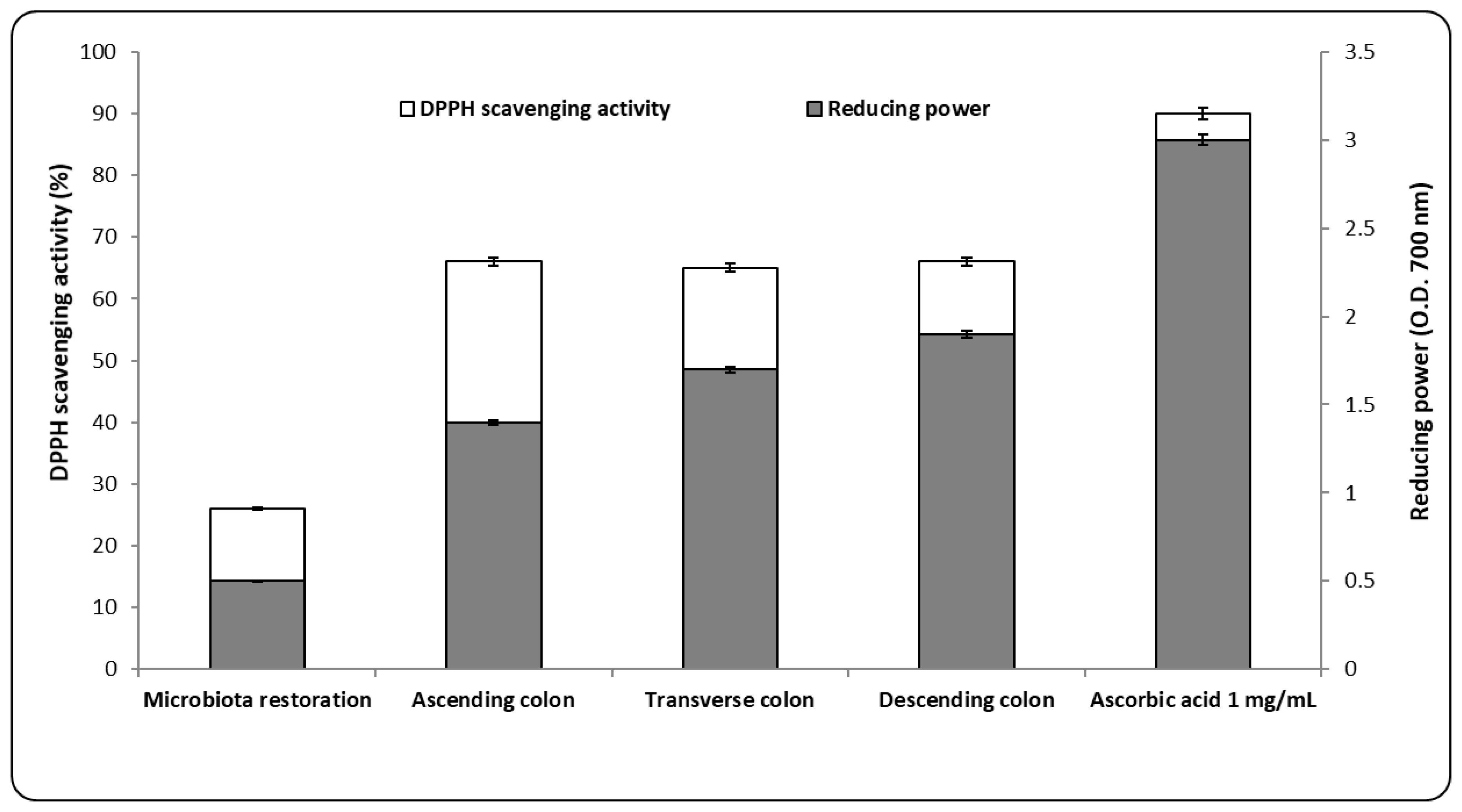
3.3. Antioxidant and Metabolic Capacity after AgNP Administration
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Vasile, O.R.; Serdaru, I.; Andronescu, E.; Trusca, R.; Surdu, V.A.; Vasile, B.S.; Oprea, O.; Ilie, A. Influence of the size and the morphology of ZnO nanoparticles on cell viability. C. R. Chim. 2015, 18, 1335–1343. [Google Scholar] [CrossRef]
- Sivaranjani, T.; Asha, A.; Thirunavukkarasu, P.; Asha, S. Silver nanoparticle synthesis from plant extracts and its application—A review. Int. J. Adv. Res. Phys. Sci. 2016, 3, 5–8. [Google Scholar]
- Carrillo-López, L.M.; Soto-Hernández, R.M.; Zavaleta-Mancera, H.A.; Vilchis-Néstor, A.R. Study of the performance of the organic extracts of Chenopodium ambrosioides for Ag nanoparticle synthesis. J. Nanomater. 2016, 2016, 4714162. [Google Scholar] [CrossRef]
- Anthony, K.J.P.; Murugan, M.; Jeyaraj, M.; Rathinam, N.K.; Sangiliyandi, G. Synthesis of silver nanoparticles using pine mushroom extract: A potential antimicrobial agent against E. coli and B. subtilis. J. Ind. Eng. Chem. 2014, 20, 2325–2331. [Google Scholar] [CrossRef]
- Omidi, B.; Hashemi, S.J.; Bayat, M.; Larijani, K. Biosynthesis of silver nanoparticles by Lactobacillus fermentum. Bull. Environ. Pharmacol. Life Sci. 2014, 3, 186–192. [Google Scholar]
- Al-Bahrani, R.; Raman, J.; Lakshmanan, H.; Hassan, A.A.; Sabaratnam, V. Green synthesis of silver nanoparticles using tree oyster mushroom Pleurotus ostreatus and its inhibitory activity against pathogenic bacteria. Mater. Lett. 2017, 186, 21–25. [Google Scholar] [CrossRef]
- Bhat, R.; Sharanabasava, V.G.; Deshpande, R.; Shetti, U.; Sanjeev, G.; Venkataraman, A. Photo-bio-synthesis of irregular shaped functionalized gold nanoparticles using edible mushroom Pleurotus florida and its anticancer evaluation. J. Photochem. Photobiol. B 2013, 125, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Vamanu, E. Antioxidant properties and chemical compositions of various extracts of the edible commercial mushroom, Pleurotus ostreatus, in Romanian markets. Rev. Chim. 2013, 64, 49–54. [Google Scholar]
- Krithiga, N.; Rajalakshmi, A.; Jayachitra, A. Green synthesis of silver nanoparticles using leaf extracts of Clitoriaternatea and Solanum nigrum and study of its antibacterial effect against common nosocomial pathogens. J. Nanosci. 2015, 2015, 928204. [Google Scholar] [CrossRef]
- Ficai, M.; Nedelcu, I.A.; Ficai, D.; Vasile, B.S.; Ficai, A.; Andronescu, E. The Influence of ultrasonication on the formation of COLL/HA composite materials. Mater. Plast. 2011, 48, 23–26. [Google Scholar]
- Vamanu, E.; Pelinescu, D.; Sarbu, I. Comparative fingerprinting of the human microbiota in diabetes and cardiovascular disease. J. Med. Food 2016, 19, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Markus, J.; Mathiyalagan, R.; Kim, Y.J.; Abbai, R.; Singh, P.; Ahn, S.; Perez, Z.E.J.; Hurh, J.; Yang, D.C. Intracellular synthesis of gold nanoparticles with antioxidant activity by probiotic Lactobacillus kimchicus DCY51T isolated from Korean kimchi. Enzym. Microb. Technol. 2016, 95, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Tułodziecka, A.; Szydłowska-Czerniak, A. Development of a novel gold nanoparticle-based method to determine antioxidant capacity of Brassica oil seeds, white flakes and meal. Food Chem. 2016, 208, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Takebayashi, J.; Chen, J.; Tai, A. A method for evaluation of antioxidant activity based on inhibition of free radical-induced erythrocyte hemolysis. Methods Mol. Biol. 2010, 594, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Ho, E.; Galougahi, K.K.; Liu, C.C.; Bhindi, R.; Figtree, G.A. Biological markers of oxidative stress: Applications to cardiovascular research and practice. Redox Biol. 2013, 1, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Ketabi, A.; Dieleman, L.A.; Ganzle, M.G. Influence of isomalto-oligosaccharides on intestinal microbiota in rats. J. Appl. Microbiol. 2011, 110, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Turcu, I.; Zarafu, I.; Popa, M.; Chifiriuc, M.C.; Bleotu, C.; Culita, D.; Ghica, C.; Ionita, P. Lipoic acid gold nanoparticles functionalized with organic compounds as bioactive materials. Nanomaterials 2017, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Sârbu, I.; Pelinescu, D.; Stoica, I.; Măruţescu, L.; Vassu, T. Phenotypic profiles of virulence in different Candida species isolated from vulvovaginal infections. Roum. Arch. Microbiol. Immunol. 2013, 72, 225–233. [Google Scholar] [PubMed]
- Mulvaney, P. Surface plasmon spectroscopy of nanosized metal particles. Langmuir 1996, 12, 788–800. [Google Scholar] [CrossRef]
- Palanisamy, S.; Rajasekar, P.; Vijayaprasath, G.; Manikandan, R.; Prabhu, N.M. A green route to synthesis silver nanoparticles using Sargassum polycystum and its antioxidant and cytotoxic effects: An in vitro analysis. Mater. Lett. 2017, 189, 196–200. [Google Scholar] [CrossRef]
- Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Homogentisic_acid#section=Tissue-Locations (accessed on 4 May 2018).
- Jyoti, K.; Baunthiyal, M.; Singh, A. Characterization of silver nanoparticles synthesized using Urtica dioica Linn. leaves and their synergistic effects with antibiotics. J. Radiat. Res. Appl. Sci. 2016, 9, 217–227. [Google Scholar] [CrossRef]
- Pawar, O.; Deshpande, N.; Dagade, S.; Waghmode, S.; Joshi, P.N. Green synthesis of silver nanoparticles from purple acid phosphatase apoenzyme isolated from a new source Limonia acidissima. J. Exp. Nanosci. 2015, 11, 28–37. [Google Scholar] [CrossRef]
- Ravichandran, V.; Vasanthi, S.; Shalini, S.; Shah, S.A.A.; Harish, R. Green synthesis of silver nanoparticles using Atrocarpus altilis leaf extract and the study of their antimicrobial and antioxidant activity. Mater. Lett. 2016, 180, 264–267. [Google Scholar] [CrossRef]
- Mahendran, G.; Ranjitha Kumari, B.D. Biological activities of silver nanoparticles from Nothapodytes nimmoniana (Graham) Mabb. fruit extracts. Food Sci. Hum. Wellness 2016, 5, 207–218. [Google Scholar] [CrossRef]
- Lateef, A.; Azeez, M.A.; Asafa, T.B.; Yekeen, T.A.; Akinboro, A.; Oladipo, I.C.; Azeez, L.; Ajibade, S.E.; Ojo, S.A.; Gueguim-Kana, E.B.; et al. Biogenic synthesis of silver nanoparticles using a pod extract of Cola nitida: Antibacterial and antioxidant activities and application as a paint additive. J. Taibah Univ. Sci. 2016, 10, 551–562. [Google Scholar] [CrossRef]
- Shaker, M.A.; Shaaban, M.I. Synthesis of silver nanoparticles with antimicrobial and anti-adherence activities against multidrug-resistant isolates from Acinetobacter baumannii. J. Taibah Univ. Med. Sci. 2017, 12, 291–297. [Google Scholar] [CrossRef]
- Phanjom, P.; Ahmed, G. Biosynthesis of silver nanoparticles by Aspergillus oryzae (MTCC No. 1846) and its characterizations. Nanosci. Nanotechnol. 2015, 5, 14–21. [Google Scholar] [CrossRef]
- Lavakumar, V.; Masilamani, K.; Ravichandiran, V.; Venkateshan, N.; Saigopal, D.V.R.; Ashok Kumar, C.K.; Sowmya, C. Promising upshot of silver nanoparticles primed from Gracilaria crassa against bacterial pathogens. Chem. Cent. J. 2015, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Patil Shriniwas, P.; Kumbhar Subhash, T. Antioxidant, antibacterial and cytotoxic potential of silver nanoparticles synthesized using terpenes rich extract of Lantana camara L. leaves. Biochem. Biophys. Rep. 2017, 10, 76–81. [Google Scholar] [CrossRef]
- Chen, L.Q.; Fang, L.; Ling, J.; Ding, C.Z.; Kang, B.; Huang, C.Z. Nanotoxicity of silver nanoparticles to red blood cells: Size dependent adsorption, uptake, and hemolytic activity. Chem. Res. Toxicol. 2015, 28, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; McDonald, J.A.K.; Petrof, E.O.; Allen-Vercoe, E.; Walker, V.K. Nanosilver-mediated change in human intestinal microbiota. J. Nanomed. Nanotechnol. 2014, 5, 5. [Google Scholar] [CrossRef]
- Van den Brule, S.; Ambroise, J.; Lecloux, H.; Levard, C.; Soulas, R.; De Temmerman, P.J.; Palmai-Pallag, M.; Marbaix, E.; Lison, D. Dietary silver nanoparticles can disturb the gut microbiota in mice. Part. Fibre Toxicol. 2016, 13, 38. [Google Scholar] [CrossRef] [PubMed]
- Xiu, Z.M.; Zhang, Q.B.; Puppala, H.L.; Colvin, V.L.; Alvarez, P.J. Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 2012, 12, 4271. [Google Scholar] [CrossRef] [PubMed]
- Fröhlichand, E.E.; Fröhlich, E. Cytotoxicity of nanoparticles contained in food on intestinal cells and the gut microbiota. Int. J. Mol. Sci. 2016, 17, 509. [Google Scholar] [CrossRef] [PubMed]
- Mocan, T. Hemolysis as Expression of nanoparticles-induced cytotoxicity in red blood cells. Biotechnol. Mol. Biol. Nanomed. 2013, 1, 7–12. [Google Scholar]
- Villalobos, M.C.; Serradilla, M.J.; Martín, A.; Ordiales, E.; Ruiz-Moyano, S.; de Guía Córdoba, M. Antioxidant and antimicrobial activity of natural phenolic extract from defatted soybean flour by-product for stone fruit postharvest application. J. Sci. Food Agric. 2016, 96, 2116–2124. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Kwon, D.N.; Kim, J.H. Enhanced antibacterial and anti-biofilm activities of silver nanoparticles against Gram-negative and Gram-positive bacteria. Nanoscale. Res. Lett. 2014, 9, 373. [Google Scholar] [CrossRef] [PubMed]
| Samples | Staphylococcus aureus | Escherichia coli | Candida albicans |
|---|---|---|---|
| AgNO3 (1 mM) | 250 µM | 62.5 µM | 7.8 µM |
| Extract (10% v/v) | 2.5% | 2.5% | 1.25% |
| AgNPs (200 mg/mL) | 1.56 mg/mL | 0.39 mg/mL | ˂0.098 mg/mL |
| Samples | Staphylococcus aureus | Escherichia coli | Candida albicans |
|---|---|---|---|
| AgNO3 | 250 µM | 62.5 µM | 7.8 µM |
| Extract | 0.312% | 0.156% | ˂0.0045% |
| AgNPs | 1.562 mg/mL | 0.39 mg/mL | 50 mg/mL |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vamanu, E.; Ene, M.; Biță, B.; Ionescu, C.; Crăciun, L.; Sârbu, I. In Vitro Human Microbiota Response to Exposure to Silver Nanoparticles Biosynthesized with Mushroom Extract. Nutrients 2018, 10, 607. https://doi.org/10.3390/nu10050607
Vamanu E, Ene M, Biță B, Ionescu C, Crăciun L, Sârbu I. In Vitro Human Microbiota Response to Exposure to Silver Nanoparticles Biosynthesized with Mushroom Extract. Nutrients. 2018; 10(5):607. https://doi.org/10.3390/nu10050607
Chicago/Turabian StyleVamanu, Emanuel, Mihaela Ene, Bogdan Biță, Cristina Ionescu, Liviu Crăciun, and Ionela Sârbu. 2018. "In Vitro Human Microbiota Response to Exposure to Silver Nanoparticles Biosynthesized with Mushroom Extract" Nutrients 10, no. 5: 607. https://doi.org/10.3390/nu10050607







