Nutrigenetic Contributions to Dyslipidemia: A Focus on Physiologically Relevant Pathways of Lipid and Lipoprotein Metabolism
Abstract
:1. Introduction
2. Pathophysiology of Dyslipidemia
3. Genetic Contributions to Dyslipidemia
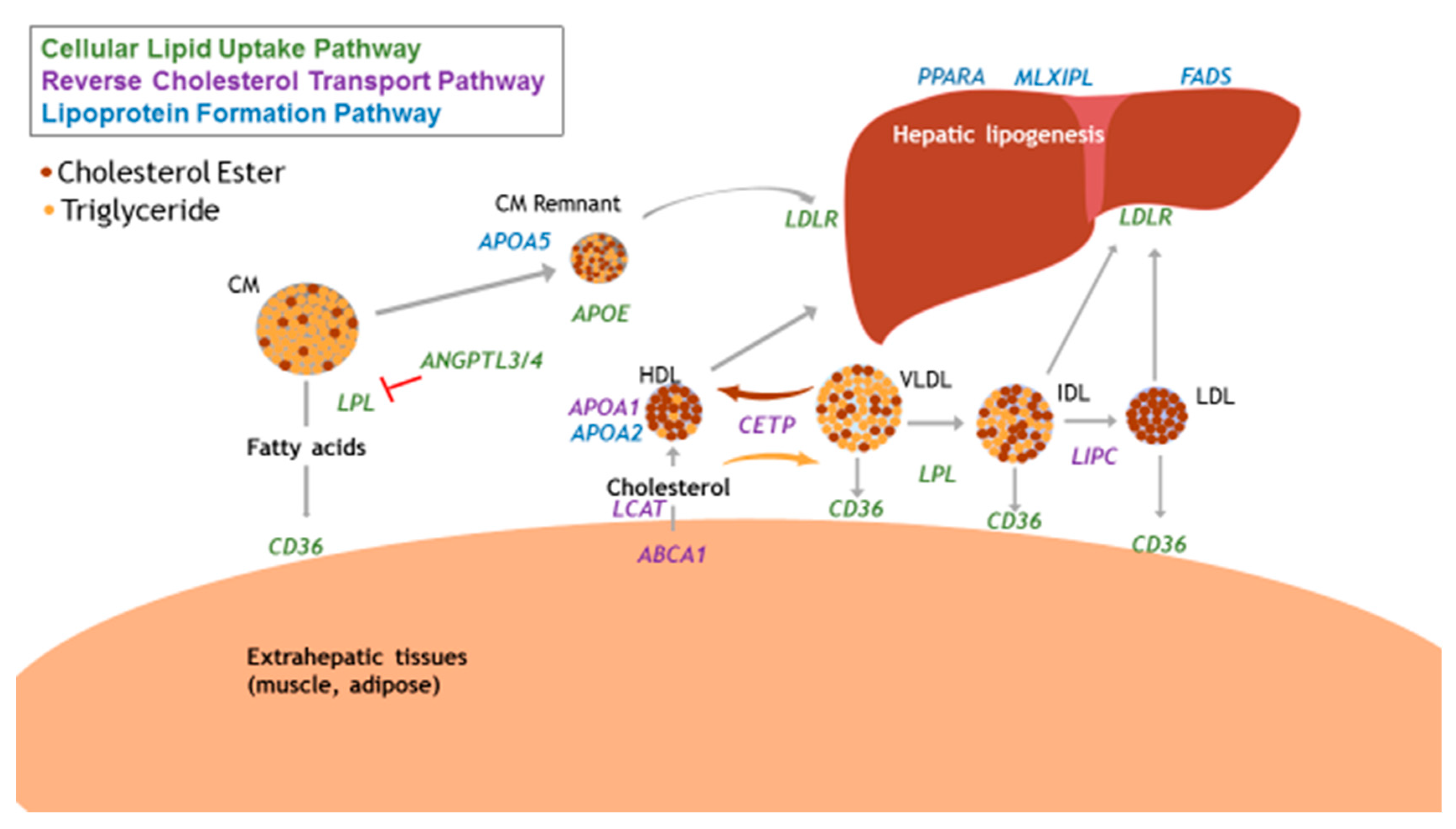
3.1. Focus on Physiological Relevance
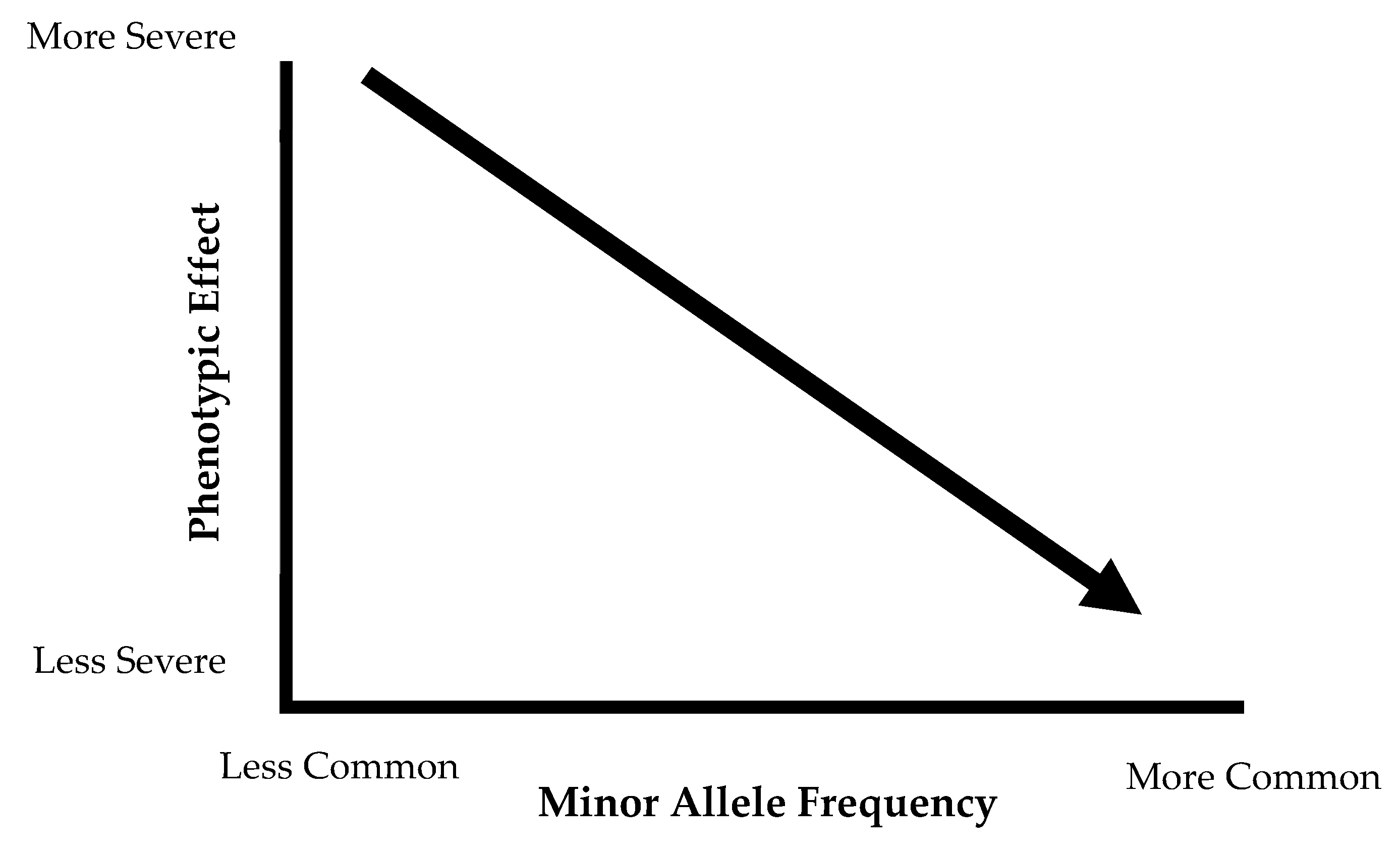
3.2. Differences in Minor Allele Frequency and Special Populations
4. Dietary Contributions to Dyslipidemia
5. Nutrient-Gene Interactions and Dyslipidemia
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Carroll, M.D.; Fryer, C.D.; Nguyen, D.T. High Total and Low High-Density Lipoprotein Cholesterol in Adults: United States, 2015–2016; NCHS Data Brief, No 290; National Center for Health Statistics: Hyattsville, MD, USA, 2017.
- Eckel, R.H.; Jakicic, J.M.; Ard, J.D.; Hubbard, V.S.; de Jesus, J.M.; Lee, I.-M.; Lichtenstein, A.H.; Loria, C.M.; Millen, B.E.; Miller, N.H.; et al. 2013 AHA/ACC Guideline on Lifestyle Management to Reduce Cardiovascular Risk: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2013, 129, S76–S99. [Google Scholar] [CrossRef] [PubMed]
- Bamba, V. Update on screening, etiology, and treatment of dyslipidemia in children. JCEM 2014, 99, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Kussmann, M.; Stover, P.J. Nutrigenomics and Proteomics in Health and Disease: Towards a Systems-Level Understanding of Gene-Diet Interactions, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2017; p. 1119098831. [Google Scholar]
- Tenenbaum, A.; Klempfner, R.; Fisman, E.Z. Hypertriglyceridemia: A too long unfairly neglected major cardiovascular risk factor. Cardiovasc. Diabetol. 2014, 13, 159. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Stone, N.J.; Ballantyne, C.; Bittner, V.; Criqui, M.H.; Ginsberg, H.N.; Goldberg, A.C.; Howard, W.J.; Jacobson, M.S.; Kris-Etherton, P.M.; et al. Triglycerides and cardiovascular disease: A scientific statement from the American Heart Association. Circulation 2011, 123, 2292–2333. [Google Scholar] [CrossRef] [PubMed]
- Wajchenberg, B.L. Subcutaneous and visceral adipose tissue: Their relation to the metabolic syndrome. Endocr. Rev. 2000, 21, 697–738. [Google Scholar] [CrossRef] [PubMed]
- Després, J.-P.; Lemieux, I. Abdominal obesity and metabolic syndrome. Nature 2006, 444, 881. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.G.; Olsson, A.G.; Abt, M.; Ballantyne, C.M.; Barter, P.J.; Brumm, J.; Chaitman, B.R.; Holme, I.M.; Kallend, D.; Leiter, L.A. Effects of dalcetrapib in patients with a recent acute coronary syndrome. NEJM 2012, 367, 2089–2099. [Google Scholar] [CrossRef] [PubMed]
- Rader, D.J.; Hovingh, G.K. HDL and cardiovascular disease. Lancet 2014, 384, 618–625. [Google Scholar] [CrossRef]
- Marais, A.D. Familial hypercholesterolaemia. Clin. Biochem. Rev. 2004, 25, 49. [Google Scholar] [PubMed]
- Oliva, C.P.; Pisciotta, L.; Volti, G.L.; Sambataro, M.P.; Cantafora, A.; Bellocchio, A.; Catapano, A.; Tarugi, P.; Bertolini, S.; Calandra, S. Inherited apolipoprotein AV deficiency in severe hypertriglyceridemia. Arterioscl. Thromb. Vasc. Biol. 2005, 25, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Tolleshaug, H.; Hobgood, K.K.; Brown, M.S.; Goldstein, J.L. The LDL receptor locus in familial hypercholesterolemia: Multiple mutations disrupt transport and processing of a membrane receptor. Cell 1983, 32, 941–951. [Google Scholar] [CrossRef]
- Lusis, A.J.; Fogelman, A.M.; Fonarow, G.C. Genetic basis of atherosclerosis: Part I: New genes and pathways. Circulation 2004, 110, 1868–1873. [Google Scholar] [CrossRef] [PubMed]
- Dron, J.S.; Hegele, R.A. Genetics of lipid and lipoprotein disorders and traits. Curr. Genet. Med. Rep. 2016, 4, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Global Lipids Genetics Consortium; Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274. [Google Scholar] [CrossRef] [Green Version]
- Fielding, C.J.; Fielding, P.E. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 1995, 36, 211–228. [Google Scholar] [PubMed]
- Tall, A.R. An overview of reverse cholesterol transport. Eur. Heart J. 1998, 19 (Suppl. A), A31–A35. [Google Scholar] [CrossRef]
- Clifford, A.J.; Rincon, G.; Owens, J.E.; Medrano, J.F.; Moshfegh, A.J.; Baer, D.J.; Novotny, J.A. Single nucleotide polymorphisms in CETP, SLC46A1, SLC19A1, CD36, BCMO1, APOA5, and ABCA1 are significant predictors of plasma HDL in healthy adults. Lipids Health Dis. 2013, 12, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Liu, Y.; Li, Y.; Zhang, H.; Yu, M.; Kanu, J.S.; Qiao, Y.; Tang, Y.; Zhen, Q.; Cheng, Y. Association of ATP-binding cassette transporter A1 gene polymorphisms with plasma lipid variability and coronary heart disease risk. Int. J. Clin. Exp. Path 2015, 8, 13441–13449. [Google Scholar]
- Mirmiran, P.; Esfandiar, Z.; Hosseini-Esfahani, F.; Koochakpoor, G.; Daneshpour, M.S.; Sedaghati-Khayat, B.; Azizi, F. Genetic variations of cholesteryl ester transfer protein and diet interactions in relation to lipid profiles and coronary heart disease: A systematic review. Nutr. Metab. 2017, 14, 77. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Niimura, H.; Kuwabara, K.; Takezaki, T.; Morita, E.; Wakai, K.; Hamajima, N.; Nishida, Y.; Turin, T.C.; Suzuki, S.; et al. Gene-Gene combination effect and interactions among ABCA1, APOA1, SR-B1, and CETP polymorphisms for serum high-density lipoprotein-cholesterol in the Japanese population. PLoS ONE 2013, 8, e82046. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Yin, R.-X.; Khounphinith, E.; Zhang, F.-H.; Yang, D.-Z.; Pan, S.-L. Association of the APOA1 rs964184 SNP and serum lipid traits in the Chinese Maonan and Han populations. Lipids Health Dis. 2018, 17, 105. [Google Scholar] [CrossRef] [PubMed]
- Deeb, S.S.; Zambon, A.; Carr, M.C.; Ayyobi, A.F.; Brunzell, J.D. Hepatic lipase and dyslipidemia: Interactions among genetic variants, obesity, gender, and diet. J. Lipid Res. 2003, 44, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Agirbasli, M.; Eren, F.; Agirbasli, D.; White, M.J.; Williams, S.M. Multi-Locus Candidate Gene Analyses of Lipid Levels in a Pediatric Turkish Cohort: Lessons Learned on LPL, CETP, LIPC, ABCA1, and SHBG. OMICS J. Integr. Biol. 2013, 17, 636–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Zhang, D.; Ling, J.; Lu, W.; Zhang, S.; Zhu, Y.; Lai, M. Gender specific effect of LIPC C-514T polymorphism on obesity and relationship with plasma lipid levels in Chinese children. J. Cell. Mol. Med. 2015, 19, 2296–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Ng, S.S.; Bray, G.A.; Ryan, D.H.; Sacks, F.M.; Ning, G.; Qi, L. Dietary Fat Intake Modifies the Effect of a Common Variant in the LIPC Gene on Changes in Serum Lipid Concentrations during a Long-Term Weight-Loss Intervention Trial. J. Nutr. 2015, 145, 1289–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agirbasli, D.; Cirakoglu, B.; Eren, F.; Sumerkan, M.; Aksoy, S.; Aral, C.; Agirbasli, M. Effects of lecithin: Cholesterol acyltransferase genotypes, enzyme levels, and activity on high-density lipoprotein levels. J. Clin. Lipid 2011, 5, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, I.J.; Eckel, R.H.; Abumrad, N.A. Regulation of fatty acid uptake into tissues: Lipoprotein lipase and CD36-mediated pathways. J. Lipid Res. 2009, 50, S86–S90. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, R.L.; Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009, 2, re3. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C.; Reich, D.; Tandon, A.; Akylbekova, E.; Patterson, N.; Waliszewska, A.; Kathiresan, S.; Sarpong, D.; Taylor, H.A.; Wilson, J.G. Genetic Differences between the Determinants of Lipid Profile Phenotypes in African and European Americans: The Jackson Heart Study. PLoS Genet. 2009, 5, e1000342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayyappa, K.A.; Shatwan, I.; Bodhini, D.; Bramwell, L.R.; Ramya, K.; Sudha, V.; Anjana, R.M.; Lovegrove, J.A.; Mohan, V.; Radha, V.; et al. High fat diet modifies the association of lipoprotein lipase gene polymorphism with high density lipoprotein cholesterol in an Asian Indian population. Nutr. Metab. 2017, 14, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairoozy, R.H.; White, J.; Palmen, J.; Kalea, A.Z.; Humphries, S.E. Identification of the functional variant(s) that explain the low-density lipoprotein receptor (LDLR) GWAS SNP rs6511720 association with lower LDL-C and risk of CHD. PLoS ONE 2016, 11, e0167676. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Ihn, H.E.; Medina, M.W.; Krauss, R.M. A common polymorphism in the LDL receptor gene has multiple effects on LDL receptor function. Hum. Mol. Genet. 2013, 22, 1424–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Gusarova, V.; Banfi, S.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Inactivation of ANGPTL3 reduces hepatic VLDL-triglyceride secretion. J. Lipid Res. 2015, 56, 1296–1307. [Google Scholar] [CrossRef] [PubMed]
- Legry, V.; Bokor, S.; Cottel, D.; Beghin, L.; Catasta, G.; Nagy, E.; Gonzalez-Gross, M.; Spinneker, A.; Stehle, P.; Molnar, D.; et al. Associations between common genetic polymorphisms in angiopoietin-like proteins 3 and 4 and lipid metabolism and adiposity in European adolescents and adults. JCEM 2009, 94, 5070–5077. [Google Scholar] [CrossRef] [PubMed]
- Paththinige, C.S.; Sirisena, N.D.; Dissanayake, V.H.W. Genetic determinants of inherited susceptibility to hypercholesterolemia—A comprehensive literature review. Lipids Health Dis. 2017, 16, 103. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Bacci, S.; Mlynarski, W.; Gottardo, L.; Soccio, T.; Menzaghi, C.; Iori, E.; Lager, R.A.; Shroff, A.R.; Gervino, E.V.; et al. A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum. Mol. Genet. 2004, 13, 2197–2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, Y.; Kim, Y. Oily Fish Consumption Modifies the Association between CD36 rs6969989 Polymorphism and Lipid Profiles in Korean Women. Prev. Nutr. Food Sci. 2016, 21, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-Arellano, L.E.; Salgado-Bernabe, A.B.; Guzman-Guzman, I.P.; Salgado-Goytia, L.; Munoz-Valle, J.F.; Parra-Rojas, I. CD36 haplotypes are associated with lipid profile in normal-weight subjects. Lipids Health Dis. 2013, 12, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellsworth, J.L.; Erickson, S.K.; Cooper, A.D. Very low and low density lipoprotein synthesis and secretion by the human hepatoma cell line Hep-G2: Effects of free fatty acid. J. Lipid Res. 1986, 27, 858–874. [Google Scholar] [PubMed]
- Hellerstein, M.K.; Schwarz, J.M.; Neese, R.A. Regulation of Hepatic De Novo Lipogenesis in Humans. Ann. Rev. Nutr. 1996, 16, 523–557. [Google Scholar] [CrossRef] [PubMed]
- Kathiresan, S.; Willer, C.J.; Peloso, G.M.; Demissie, S.; Musunuru, K.; Schadt, E.E.; Kaplan, L.; Bennett, D.; Li, Y.; Tanaka, T. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009, 41, 56. [Google Scholar] [CrossRef] [PubMed]
- Raichlen, D.A.; Alexander, G.E. Exercise, APOE genotype, and the evolution of the human lifespan. Trends Neurosci. 2014, 37, 247–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guardiola, M.; Ribalta, J. Update on APOA5 Genetics: Toward a Better Understanding of Its Physiological Impact. Curr. Atheroscler. Rep. 2017, 19, 30. [Google Scholar] [CrossRef] [PubMed]
- Keramat, L.; Sadrzadeh-Yeganeh, H.; Sotoudeh, G.; Zamani, E.; Eshraghian, M.; Mansoori, A.; Koohdani, F. Apolipoprotein A2 -265 T>C polymorphism interacts with dietary fatty acids intake to modulate inflammation in type 2 diabetes mellitus patients. Nutrition 2017, 37, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Zamani, E.; Sadrzadeh-Yeganeh, H.; Sotoudeh, G.; Keramat, L.; Eshraghian, M.; Rafiee, M.; Koohdani, F. The interaction between ApoA2 -265T>C polymorphism and dietary fatty acids intake on oxidative stress in patients with type 2 diabetes mellitus. Eur. J. Nutr. 2017, 56, 1931–1938. [Google Scholar] [CrossRef] [PubMed]
- Bokor, S.; Dumont, J.; Spinneker, A.; Gonzalez-Gross, M.; Nova, E.; Widhalm, K.; Moschonis, G.; Stehle, P.; Amouyel, P.; De Henauw, S.; et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J. Lipid Res. 2010, 51, 2325–2333. [Google Scholar] [CrossRef] [PubMed]
- Mathias, R.A.; Sergeant, S.; Ruczinski, I.; Torgerson, D.G.; Hugenschmidt, C.E.; Kubala, M.; Vaidya, D.; Suktitipat, B.; Ziegler, J.T.; Ivester, P.; et al. The impact of FADS genetic variants on ω6 polyunsaturated fatty acid metabolism in African Americans. BMC Genet. 2011, 12, 50–50. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.M.; Johnston, H.; Clarke, S.; Roke, K.; Nielsen, D.; Badawi, A.; El-Sohemy, A.; Ma, D.W.; Mutch, D.M. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol. Genet. Metab. 2011, 103, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Sone, Y.; Kido, T.; Ainuki, T.; Sonoda, M.; Ichi, I.; Kodama, S.; Sone, H.; Kondo, K.; Morita, Y.; Egawa, S.; et al. Genetic variants of the fatty acid desaturase gene cluster are associated with plasma LDL cholesterol levels in Japanese males. J. Nutr. Sci. Vitaminol. 2013, 59, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Vidal, I.; Voruganti, V.S.; Hannon, B.A.; Andrade, F.C.D.; Aradillas-Garcia, C.; Nakamura, M.T.; Teran-Garcia, M. Serum Lipid Concentrations and FADS Genetic Variants in Young Mexican College Students: The UP-AMIGOS Cohort Study. Lifestyle Genom. 2018, 1–9. [Google Scholar] [CrossRef]
- Yilmaz-Aydogan, H.; Kurnaz, O.; Kucukhuseyin, O.; Akadam-Teker, B.; Kurt, O.; Eronat, A.P.; Tekeli, A.; Bugra, Z.; Ozturk, O. Different effects of PPARA, PPARG and ApoE SNPs on serum lipids in patients with coronary heart disease based on the presence of diabetes. Gene 2013, 523, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Alsaleh, A.; Frost, G.S.; Griffin, B.A.; Lovegrove, J.A.; Jebb, S.A.; Sanders, T.A.; O’Dell, S.D. PPARgamma2 gene Pro12Ala and PPARalpha gene Leu162Val single nucleotide polymorphisms interact with dietary intake of fat in determination of plasma lipid concentrations. J. Nutrigenet. Nutrigenom. 2011, 4, 354–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AlSaleh, A.; Sanders, T.A.; O’Dell, S.D. Effect of interaction between PPARG, PPARA and ADIPOQ gene variants and dietary fatty acids on plasma lipid profile and adiponectin concentration in a large intervention study. Proc. Nutr. Soc. 2012, 71, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Aung, L.-H.-H.; Yin, R.-X.; Wu, J.-Z.; Wu, D.-F.; Wang, W.; Li, H. Association between the MLX interacting protein-like, BUD13 homolog and zinc finger protein 259 gene polymorphisms and serum lipid levels. Sci. Rep. 2014, 4, 5565. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.D.; Burchard, E.G.; De la Vega, F.M. Genomics for the world. Nature 2011, 475, 163–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar-Salinas, C.A.; Gomez-Perez, F.J.; Rull, J.; Villalpando, S.; Barquera, S.; Rojas, R. Prevalence of dyslipidemias in the Mexican National Health and Nutrition Survey 2006. Salud Publica Mexico 2010, 52 (Suppl. 1), S44–S53. [Google Scholar] [CrossRef]
- The Genomes Project Consortium; Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; et al. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posadas-Sanchez, R.; Ocampo-Arcos, W.A.; Lopez-Uribe, A.R.; Posadas-Romero, C.; Villarreal-Molina, T.; Leon, E.A.; Perez-Hernandez, N.; Rodriguez-Perez, J.M.; Cardoso-Saldana, G.; Medina-Urrutia, A.; et al. Hepatic lipase (LIPC) C-514T gene polymorphism is associated with cardiometabolic parameters and cardiovascular risk factors but not with fatty liver in Mexican population. Exp. Mol. Pathol. 2015, 98, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Weissglas-Volkov, D.; Aguilar-Salinas, C.A.; Nikkola, E.; Deere, K.A.; Cruz-Bautista, I.; Arellano-Campos, O.; Muñoz-Hernandez, L.L.; Gomez-Munguia, L.; Ordoñez-Sánchez, M.L.; Reddy, P.M.V.L.; et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J. Med. Genet. 2013, 50, 298–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dainis, A.M.; Ashley, E.A. Cardiovascular Precision Medicine in the Genomics Era. JACC Basic Transl. Sci. 2018, 3, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Despres, J.P.; Fullerton, H.J.; Howard, V.J.; et al. Heart disease and stroke statistics—2015 update: A report from the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, I. Alcohol and Dyslipidemia. In Alcohol, Nutrition, and Health Consequences; Watson, R.R., Preedy, V.R., Zibadi, S., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 329–339. [Google Scholar]
- Feinman, R.D.; Volek, J.S. Carbohydrate restriction as the default treatment for type 2 diabetes and metabolic syndrome. Scand. Cardiovasc. J. 2008, 42, 256–263. [Google Scholar] [CrossRef] [PubMed]
- US Department of Health and Human Services. 2015–2020 Dietary Guidelines for Americans; USDA: Washington, DC, USA, 2015. [Google Scholar]
- Bach-Faig, A.; Berry, E.M.; Lairon, D.; Reguant, J.; Trichopoulou, A.; Dernini, S.; Medina, F.X.; Battino, M.; Belahsen, R.; Miranda, G.; et al. Mediterranean diet pyramid today: Science and cultural updates. Public Health Nutr. 2011, 14, 2274–2284. [Google Scholar] [CrossRef] [PubMed]
- Willett, W.C.; Sacks, F.; Trichopoulou, A.; Drescher, G.; Ferro-Luzzi, A.; Helsing, E.; Trichopoulos, D. Mediterranean diet pyramid: A cultural model for healthy eating. Am. J. Clin. Nutr. 1995, 61, 1402S–1406S. [Google Scholar] [CrossRef] [PubMed]
- De Lorgeril, M.; Salen, P.; Martin, J.-L.; Monjaud, I.; Delaye, J.; Mamelle, N. Mediterranean diet, traditional risk factors, and the rate of cardiovascular complications after myocardial infarction: Final report of the Lyon Diet Heart Study. Circulation 1999, 99, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.-I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J. Primary prevention of cardiovascular disease with a Mediterranean diet. NEJM 2013, 368, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Hannon, B.A.; Thompson, S.V.; An, R.; Teran-Garcia, M. Clinical outcomes of dietary replacement of saturated fatty acids with unsaturated fat sources in adults with overweight and obesity: A systematic review and meta-analysis of randomized control trials. Ann. Nutr. Metab. 2017, 71, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.; Martin, N.; Abdelhamid, A.; Davey Smith, G. Reduction in saturated fat intake for cardiovascular disease. Cochrane Database Syst. Rev. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kris-Etherton, P.M. Monounsaturated fatty acids and risk of cardiovascular disease. Circulation 1999, 100, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Lichtenstein, A.H.; Wu, J.H.Y.; Appel, L.J.; Creager, M.A.; Kris-Etherton, P.M.; Miller, M.; Rimm, E.B.; Rudel, L.L.; Robinson, J.G.; et al. Dietary Fats and Cardiovascular Disease: A Presidential Advisory From the American Heart Association. Circulation 2017. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.; Bryan, J.; Hodgson, J.; Murphy, K. Definition of the Mediterranean diet: A literature review. Nutrients 2015, 7, 9139–9153. [Google Scholar] [CrossRef] [PubMed]
- Kastorini, C.M.; Milionis, H.J.; Esposito, K.; Giugliano, D.; Goudevenos, J.A.; Panagiotakos, D.B. The effect of Mediterranean diet on metabolic syndrome and its components: A meta-analysis of 50 studies and 534,906 individuals. J. Am. Coll. Cardiol. 2011, 57, 1299–1313. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, A.S.; Martin, N.; Bridges, C.; Brainard, J.S.; Wang, X.; Brown, T.J.; Hanson, S.; Jimoh, O.F.; Ajabnoor, S.M.; Deane, K.H.O.; et al. Polyunsaturated fatty acids for the primary and secondary prevention of cardiovascular disease. Cochrane Databse Syst. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Erkkilä, A.; de Mello, V.D.F.; Risérus, U.; Laaksonen, D.E. Dietary fatty acids and cardiovascular disease: An epidemiological approach. Prog. Lipid Res. 2008, 47, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Siscovick, D.S.; Barringer, T.A.; Fretts, A.M.; Wu, J.H.Y.; Lichtenstein, A.H.; Costello, R.B.; Kris-Etherton, P.M.; Jacobson, T.A.; Engler, M.B.; Alger, H.M.; et al. Omega-3 polyunsaturated fatty acid (fish oil) supplementation and the prevention of clinical cardiovascular disease. Sci. Advis. Am. Heart Assoc. 2017, 135, e867–e884. [Google Scholar] [CrossRef]
- Lamarche, B.; Couture, P. Dietary fatty acids, dietary patterns, and lipoprotein metabolism. Curr. Opin. Lipidol. 2015, 26, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.L.; West, K.L. Mechanisms by which dietary fatty acids modulate plasma lipids. J. Nutr. 2005, 135, 2075–2078. [Google Scholar] [CrossRef] [PubMed]
- Teran-Garcia, M.; Rufo, C.; Nakamura, M.T.; Osborne, T.F.; Clarke, S.D. NF-Y involvement in the polyunsaturated fat inhibition of fatty acid synthase gene transcription. Biochem. Piophys. Res. Commun. 2002, 290, 1295–1299. [Google Scholar] [CrossRef] [PubMed]
- Brousseau, M.E.; Ordovas, J.M.; Osada, J.; Fasulo, J.; Robins, S.J.; Nicolosi, R.J.; Schaefer, E.J. Dietary monounsaturated and polyunsaturated fatty acids are comparable in their effects on hepatic apolipoprotein mRNA abundance and liver lipid concentrations when substituted for saturated fatty acids in cynomolgus monkeys. J. Nutr. 1995, 125, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Milagro, F.I.; Allayee, H.; Chmurzynska, A.; Choi, M.S.; Curi, R.; De Caterina, R.; Ferguson, L.R.; Goni, L.; Kang, J.X.; et al. Guide for current nutrigenetic, nutrigenomic, and nutriepigenetic approaches for precision nutrition involving the prevention and management of chronic diseases associated with Oobesity. Lifestyle Genom. 2017, 10, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. BMJ 2018, 361. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; El-Sohemy, A.; Cahill, L.; Ferguson, L.R.; French, T.-A.C.; Tai, E.S.; Milner, J.; Koh, W.-P.; Xie, L.; Zucker, M.; et al. Nutrigenetics and nutrigenomics: Viewpoints on the current status and applications in nutrition research and practice. J. Nutrigenet. Nutrigenom. 2011, 4, 69–89. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, C.; Sparks, D.L. Hepatic lipase, high density lipoproteins, and hypertriglyceridemia. Am. J. Pathol. 2011, 178, 1429–1433. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M.; Corella, D.; Demissie, S.; Cupples, L.A.; Couture, P.; Coltell, O.; Wilson, P.W.F.; Schaefer, E.J.; Tucker, K.L. Dietary fat intake determines the effect of a common polymorphism in the hepatic lipase gene promoter on high-density lipoprotein metabolism: Evidence of a strong dose effect in this gene-nutrient interaction in the Framingham study. Circulation 2002, 106, 2315–2321. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Moreno, C.; Ordovás, J.M.; Smith, C.E.; Baraza, J.C.; Lee, Y.-C.; Garaulet, M. APOA5 gene variation interacts with dietary fat intake to modulate obesity and circulating triglycerides in a Mediterranean population. J. Nutr. 2011, 141, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Merched, A.J.; Chan, L. Nutrigenetics and nutrigenomics of atherosclerosis. Curr. Atheroscler. Rep. 2013, 15, 328–328. [Google Scholar] [CrossRef] [PubMed]
- Tai, E.S.; Corella, D.; Demissie, S.; Cupples, L.A.; Coltell, O.; Schaefer, E.J.; Tucker, K.L.; Ordovas, J.M. Polyunsaturated fatty acids interact with the PPARA-L162V polymorphism to affect plasma triglyceride and apolipoprotein C-III concentrations in the Framingham Heart Study. J. Nutr. 2005, 135, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Q.; Corella, D.; Demissie, S.; Cupples, L.A.; Adiconis, X.; Zhu, Y.; Parnell, L.D.; Tucker, K.L.; Ordovas, J.M. Dietary intake of n-6 fatty acids modulates effect of apolipoprotein A5 gene on plasma fasting triglycerides, remnant lipoprotein concentrations, and lipoprotein particle size: The Framingham Heart Study. Circulation 2006, 113, 2062–2070. [Google Scholar] [CrossRef] [PubMed]
- Garaulet, M.; Smith, C.E.; Hernandez-Gonzalez, T.; Lee, Y.C.; Ordovas, J.M. PPARgamma Pro12Ala interacts with fat intake for obesity and weight loss in a behavioural treatment based on the Mediterranean diet. Mol. Nutr. Food Res. 2011, 55, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Qi, Q.; Zhang, C.; Smith, S.R.; Hu, F.B.; Sacks, F.M.; Bray, G.A.; Qi, L. FTO Genotype and 2-Year Change in Body Composition and Fat Distribution in Response to Weight-Loss Diets: The POUNDS LOST Trial. Diabetes 2012, 61, 3005–3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Qi, Q.; Bray, G.A.; Hu, F.B.; Sacks, F.M.; Qi, L. APOA5 genotype modulates 2-y changes in lipid profile in response to weight-loss diet intervention: The Pounds Lost Trial. Am. J. Clin. Nutr. 2012, 96, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Corella, D.; Peloso, G.; Arnett, D.K.; Demissie, S.; Cupples, L.A.; Tucker, K.; Lai, C.Q.; Parnell, L.D.; Coltell, O.; Lee, Y.C.; et al. APOA2, dietary fat, and body mass index: Replication of a gene-diet interaction in 3 independent populations. Arch. Intern. Med. 2009, 169, 1897–1906. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Tucker, K.L.; Smith, C.E.; Lee, Y.C.; Huang, T.; Richardson, K.; Parnell, L.D.; Lai, C.Q.; Young, K.L.; Justice, A.E.; et al. Lipoprotein lipase variants interact with polyunsaturated fatty acids for obesity traits in women: Replication in two populations. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 1323–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robitaille, J.; Brouillette, C.; Lemieux, S.; Perusse, L.; Gaudet, D.; Vohl, M.C. Plasma concentrations of apolipoprotein B are modulated by a gene–diet interaction effect between the LFABP T94A polymorphism and dietary fat intake in French-Canadian men. Mol. Genet. Metab. 2004, 82, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.E.; Arnett, D.K.; Corella, D.; Tsai, M.Y.; Lai, C.Q.; Parnell, L.D.; Lee, Y.C.; Ordovas, J.M. Perilipin polymorphism interacts with saturated fat and carbohydrates to modulate insulin resistance. Nutr. Metab. Cardiovasc. Dis. 2012, 22, 449–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdullah, M.M.; Jones, P.J.; Eck, P.K. Nutrigenetics of cholesterol metabolism: Observational and dietary intervention studies in the postgenomic era. Nutr. Rev. 2015, 73, 523–543. [Google Scholar] [CrossRef] [PubMed]
- Nuno, N.B.; Heuberger, R. Nutrigenetic associations with cardiovascular disease. Rev. Cardiovasc. Med. 2014, 15, 217–225. [Google Scholar] [PubMed]
- de Goede, J.; Geleijnse, J.M.; Ding, E.L.; Soedamah-Muthu, S.S. Effect of cheese consumption on blood lipids: A systematic review and meta-analysis of randomized controlled trials. Nutr. Rev. 2015, 73, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Lordan, R.; Tsoupras, A.; Mitra, B.; Zabetakis, I. Dairy fats and cardiovascular disease: Do we really need to be concerned? Foods 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Otto, M.C.; Lemaitre, R.N.; Song, X.; King, I.B.; Siscovick, D.S.; Mozaffarian, D. Serial measures of circulating biomarkers of dairy fat and total and cause-specific mortality in older adults: The Cardiovascular Health Study. Am. J. Clin. Nutr. 2018, 108, 476–484. [Google Scholar] [CrossRef]
- De Toro-Martín, J.; Arsenault, B.; Després, J.-P.; Vohl, M.-C. Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef] [PubMed]
- Ohlhorst, S.D.; Russell, R.; Bier, D.; Klurfeld, D.M.; Li, Z.; Mein, J.R.; Milner, J.; Ross, A.C.; Stover, P.; Konopka, E. Nutrition research to affect food and a healthy lifespan. Adv. Nutr. 2013, 4, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudkowska, I.; Dewailly, E.; Hegele, R.A.; Boiteau, V.; Dube-Linteau, A.; Abdous, B.; Giguere, Y.; Chateau-Degat, M.L.; Vohl, M.C. Gene-diet interactions on plasma lipid levels in the Inuit population. Br. J. Nutr. 2013, 109, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Jacobo-Albavera, L.; Posadas-Romero, C.; Vargas-Alarcón, G.; Romero-Hidalgo, S.; Posadas-Sánchez, R.; González-Salazar, M.D.C.; Carnevale, A.; Canizales-Quinteros, S.; Medina-Urrutia, A.; Antúnez-Argüelles, E.; et al. Dietary fat and carbohydrate modulate the effect of the ATP-binding cassette A1 (ABCA1) R230C variant on metabolic risk parameters in premenopausal women from the Genetics of Atherosclerotic Disease (GEA) Study. Nutr. Metab. 2015, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Nettleton, J.A.; Steffen, L.M.; Ballantyne, C.M.; Boerwinkle, E.; Folsom, A.R. Associations between HDL-cholesterol and polymorphisms in hepatic lipase and lipoprotein lipase genes are modified by dietary fat intake in African American and White adults. Atherosclerosis 2007, 194, e131–e140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Feskens, E.J.; Dolle, M.E.; Imholz, S.; Verschuren, W.M.; Muller, M.; Boer, J.M. Dietary n-3 and n-6 polyunsaturated fatty acid intake interacts with FADS1 genetic variation to affect total and HDL-cholesterol concentrations in the Doetinchem Cohort Study. Am. J. Clin. Nutr. 2010, 92, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Dumont, J.; Huybrechts, I.; Spinneker, A.; Gottrand, F.; Grammatikaki, E.; Bevilacqua, N.; Vyncke, K.; Widhalm, K.; Kafatos, A.; Molnar, D.; et al. FADS1 genetic variability interacts with dietary alpha-linolenic acid intake to affect serum non-HDL-cholesterol concentrations in European adolescents. J. Nutr. 2011, 141, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Azorin, C.; Sorli, J.V.; Estruch, R.; Asensio, E.M.; Coltell, O.; Gonzalez, J.I.; Martinez-Gonzalez, M.A.; Ros, E.; Salas-Salvado, J.; Fito, M.; et al. Amino acid change in the carbohydrate response element binding protein is associated with lower triglycerides and myocardial infarction incidence depending on level of adherence to the Mediterranean diet in the PREDIMED trial. Circ. Cardiovasc. Genet. 2014, 7, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Volcik, K.A.; Nettleton, J.A.; Ballantyne, C.M.; Boerwinkle, E. Peroxisome proliferator–activated receptor α genetic variation interacts with n–6 and long-chain n–3 fatty acid intake to affect total cholesterol and LDL-cholesterol concentrations in the Atherosclerosis Risk in Communities Study. Am. J. Clin. Nutr. 2008, 87, 1926–1931. [Google Scholar] [CrossRef] [PubMed]
| Phenotype | Disorder | Gene Affected | Prevalence |
|---|---|---|---|
| High LDL | Hyperlipoproteinemia Type 2A | LDLR | 0.2% |
| Autosomal Dominant Hypercholesterolemia | PCSK9, APOE | 0.5% | |
| Low HDL | Tangier Disease | ABCA1 | <100 cases reported worldwide |
| Familial LCAT deficiency | LCAT | 70 reported cases | |
| High TG | Familial Chylomicronemia | LPL, APOC2 | <0.0001 |
| Severe Hypertriglyceridemia | APOA5, LMF1 | <0.5% |
| Gene | Locus | Protein Function | Previous Nutrient-Gene Interaction with Blood Lipids | SNP | Function of Variant | Risk Allele | MAF Global |
|---|---|---|---|---|---|---|---|
| Reverse Cholesterol Transport Pathway | |||||||
| CETP | 16q13 | Facilitates the exchange of cholesterol esters for TG between lipoproteins in circulation | Total fat and TG [98]; total fat and TG [108] | rs5882 | Missense variant | G | 0.37 |
| ABCA1 | 9q31.1 | HDL-C bound protein that transports intracellular cholesterol onto HDL-C | Total fat and HDL [109] | rs9282541 | Missense variant | T | 0.01 |
| SFA and TG [108] | rs2230806 | Missense variant | T | 0.32 | |||
| LIPC | 15q21.3 | Hepatic triglyceride lipase, also involved in lipoprotein uptake | SFA and HDL, TG [98]; total fat and HDL [110] | rs1800588 | Intron variant in promotor region, associated with lowered LIPC activity | T | 0.29 |
| APOA1 | 11q23.3 | Predominant apolipoprotein on HDL; activator of LCAT | SFA, total fat, and TC [108] | rs670 | Upstream intronic variant | T | 0.18 |
| Total fat and HDL [108] | rs5070 | Intron variant | G | 0.44 | |||
| Cellular Lipid Uptake Pathway | |||||||
| APOE | 19q13.32 | Present on TG-rich lipoproteins (chylomicrons, VLDL) | Total fat, SFA, and HDL [98] | rs405509 | Upstream variant in promoter region | T | 0.47 |
| CD36 | 7q21.11 | Scavenger receptor, binds to oxidized LDL and LCFA. | Oily fish (n-3 PUFA) and HDL [40] | rs6969989 | Intron variant | G | 0.33 |
| LPL | 8p21.3 | Hydrolyzes TG to allow fatty acids from lipoproteins into circulation | Total fat and HDL [33,110] | rs328 | Nonsense variant | G | 0.10 |
| Lipid/Lipoprotein Formation Pathway | |||||||
| APOA5 | 11q23.3 | Present on HDL particles, stimulates LPL, major determinant of plasma TG concentrations | Total fat and TC, LDL, HDL [96]; | rs964184 | 3’ untranslated region (UTR) variant | G | 0.22 |
| Total fat and TG [90] | rs662799 | Upstream variant in promoter region | G | 0.16 | |||
| FADS Complex | 11q12-13.1 | Desaturation of long-chain fatty acids | n-3, n-6 PUFAs and HDL [111]; alpha-linolenic acid and non-HDL cholesterol [112] | rs174546 | 3’ UTR variant | T | 0.28 |
| MLXIPL | 7q11.23 | Activates carbohydrate-responsive element binding protein and promotes hepatic TG synthesis | Mediterranean diet and TG [113] | rs3812316 | Missense variant | G | 0.11 |
| PPARA | 22q13.31 | Nuclear receptor in liver, ligand for PUFAs | n-3 PUFA and TC, LDL | rs6008259 | Non-coding transcript variant | A | 0.32 |
| n-6 PUFA and TC, LDL [114] | rs3892755 | Non-coding transcript variant | A | 0.09 | |||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hannon, B.A.; Khan, N.A.; Teran-Garcia, M. Nutrigenetic Contributions to Dyslipidemia: A Focus on Physiologically Relevant Pathways of Lipid and Lipoprotein Metabolism. Nutrients 2018, 10, 1404. https://doi.org/10.3390/nu10101404
Hannon BA, Khan NA, Teran-Garcia M. Nutrigenetic Contributions to Dyslipidemia: A Focus on Physiologically Relevant Pathways of Lipid and Lipoprotein Metabolism. Nutrients. 2018; 10(10):1404. https://doi.org/10.3390/nu10101404
Chicago/Turabian StyleHannon, Bridget A., Naiman A. Khan, and Margarita Teran-Garcia. 2018. "Nutrigenetic Contributions to Dyslipidemia: A Focus on Physiologically Relevant Pathways of Lipid and Lipoprotein Metabolism" Nutrients 10, no. 10: 1404. https://doi.org/10.3390/nu10101404





