1. Introduction
Hyperspectral imaging data are vital for understanding the composition of a remote planet’s surface and for gaining deeper insights into its composition. Using reflectance spectroscopy, it is possible to identify minerals by comparing their measured reflectance spectra to laboratory measurements. Knowing the mineralogical composition allows for a more informed study of regions that are of scientific interest. Specific spectral bands are combined to calculate spectral parameters that make up RGB browse products, which give a qualitative overview of surface spectral variability within the image. A high spectral resolution and large bandwidth come at the cost of other desirable qualities, such as spatial coverage. Imaging instruments are therefore designed for specific purposes, as is the case with the Mars Reconnaissance Orbiter (MRO) [
1]. This houses a variety of instruments including the Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) [
2] and the Context Camera (CTX) [
3].
CRISM provides the ability to conduct targeted observations and delivers images with over 400 spectral bands, ranging from visible to near infrared. These are, however, limited in terms of the size of areas that can be imaged and the spatial resolution which can be achieved. Targeted observations are therefore conducted in areas of scientific interest, which were discovered using untargeted observations or other instruments. Thousands of images at full spectral and spatial resolution were captured, covering less than one percent of the Martian surface [
4]. Conversely, CTX is able to capture far larger regions at about three times the spatial resolution, and has been used to record almost the entire surface of Mars. CTX images are exclusively monochromatic and represent the visual part of the spectrum. To determine spectral bands of places that have not been imaged with CRISM requires the extrapolation of knowledge from existing CRISM observations to outer regions.
Automating this process serves as the motivation for this work. To what extent machine learning models are capable of plausibly predicting CRISM spectral reflectance from single-channel CTX images is examined. Given such a model, in combination with the supply of CTX data, it would be possible to obtain estimates of spectral bands across large regions beyond which CRISM observations have been made. Doing this could aid in discovering minerals and substances in regions not yet manually examined, if predictions can be made accurately enough.
For this purpose, an investigation of possible machine learning approaches is presented, with the goal of performing pixel-wise predictions of a subset of CRISM spectral bands from CTX images. How accurate the mapping between CTX and CRISM bands is is determined with common types of neural networks. Their generalization ability when deployed in unseen areas is tested and the produced image quality is judged. One of the hopes is that less desirable features of CRISM images such as noisiness and occasional artifacts are averaged out during training, which in conjunction with higher resolution CTX inputs results in the creation of multispectral images with fewer artifacts.
2. Related Work
In the last two decades, machine learning has become an important tool in the analysis of remotely sensed data on Earth and the planetary sciences in general. Applications on Earth include land cover classification [
5], target detection, unmixing and physical/chemical parameter estimation, employing a wide variety of approaches and model architectures [
6]. On the Moon, machine-learning based systems for the automated detection of small craters is becoming an important tool to investigate the age and composition of lunar surfaces [
7,
8]. On Mars, machine learning is, among other things, used to create geomorphologic maps [
9]. Works regarding CRISM data are mainly focused on the identification and classification of surface mineralogy [
10]. In terms of machine learning, this work broadly falls under the category of image-to-image translation. This corresponds to learning output images based on input images, where the training set consists of coregistered pairs of input and output images. If a generalizing mapping is learned, it can be used to transform new images, for example, from areal photographs to maps or from day to night pictures [
11]. There have been a large number of contributions made in this field, involving tasks such as style transfer, object transfiguration, photo enhancement and semantic image synthesis [
12]. Image colorization, band-to-band translation [
13], and monocular depth estimation are additional problems which are related to our work.
In both image colorization studies and this work, the goal is to predict pixel-wise color information from grayscale values. This task is therefore most similar regarding the structure of input and output samples, meaning the prediction of multiple color channels from a single channel. In colorization, different color spaces such as Lab [
14] are often used, which aims to ensure that the Euclidean distance between colors corresponds to perceived differences ([
15] (p. 68)). This changes the problem to a prediction of a and b color channels from lightness L [
16]. However, color spaces are limited to three channel images and designed around human perception, making them less suited for this task. The problem of colorization has previously been posed as either regression onto a continuous color space or the classification of quantized color values [
17]. Colors in real-world photographs can often be highly ambiguous, which is one of the reasons GANs or conditional generative adversarial networks (CGANs) have more recently been used to obtain sharp and colorful results [
11,
18].
Monocular depth estimation (MDE) is another highly researched topic, with applications involving 3D modeling, robotics and autonomous driving [
19]. MDE aims to predict a depth map from a single, generally RGB color image. This is often phrased as a pixel-wise regression problem [
20]. Alternatively, [
21] the problem can be addressed through ordinal classification [
22]. Model architectures employed for this task are often akin to segmentation models or more generally exhibit an encoder–decoder structure. One of the differences between this work and MDE is the format of input and output samples, since MDE only predicts one channel from three. Furthermore, the range and significance of output values differs, with depth ranging from zero to some areas being “infinitely far away”, where exact values lose importance in many applications. Hyperspectral reflectance values, on the other hand, are more confined and equally important across their range.
Another task that, while not being very similar, provides useful tools is image or semantic segmentation. This corresponds to the classification of each pixel to a semantic class, in order to separate or locate different objects or regions. This amounts to a pixel-wise classification problem, with each semantic label representing one class [
23]. Although this task differs in terms of desired output, models employed within this field can easily be adapted for this work due to their pixel-wise prediction behavior.
The basis of many segmentation architectures is suitable encoding networks that are connected in different ways. An early encoder of this type is VGG [
24], which solely used a series of convolutional layers, pooling operations and non-linearities. It was later found that deep networks are easier to train when utilizing so-called residual blocks with skip connections. Such feature extraction blocks work by adding the given input to their output, thereby only requiring to learn the residual function that differs from identity [
25]. Going further, dense blocks are set up such that each layer takes in all preceding feature maps of the same size, alleviating vanishing gradients and promoting feature propagation and reuse [
26]. Alternatively, EfficientNets [
27] utilize a compound scaling method, in which the network’s depth, width and resolution are uniformly increased via a simple compound coefficient. Another design paradigm, where the widths and depths of well-performing networks can be explained by a quantized linear function, is leveraged to create RegNets [
28].
Given these insights and tools, a finalized segmentation architecture is defined by how they are combined. The majority of networks examined in this work exhibit an encoder–decoder structure as a baseline. One of the most persistent modifications to that structure is U-Net’s skip connections. Whether they are implemented as concatenations, additions or other operations, and which of the various possible ways they are connected, this idea is repeatedly used in many segmentation architectures. Going further, LinkNet [
29] changed the way in which encoders and decoders are linked, achieving superior results. Compared to this, Feature Pyramid Networks (FPNs) [
30] leverage that same structure as a feature pyramid with independent predictions at all scales. Another development is the incorporation of self-attention mechanisms at varying scales in the form of attention blocks, as is carried out by MA-Net [
31]. UNet++ [
32] is also presented as a more powerful architecture in which dense convolutional network layers connect the encoder with the decoder, translating the features of the encoder into those of the decoder. In contrast, the Pyramid Scene Parsing Network (PSPNet) [
33] includes a pyramid pooling module that unifies features defined on different spatial scales of the pyramid to achieve a higher accuracy in scene parsing tasks.
In disciplines such as image colorization and monocular depth estimation, a state-of-the-art performance has been achieved by formulating these tasks as pixel-wise classification problems, instead of regressions. This approach involves the discretization of continuous target values into bins, which are then treated as class labels. For more accurate image colorization, ref. [
17] utilized the Lab color space and quantized the two-dimensional target space of a and b color channels into bins with a grid size of 10. After this, methods such as class rebalancing are used to address biases within the distribution of a and b values, which occur in natural images. A CNN is then trained to predict the probability distributions of a and b values for each lightness pixel, using multinomial cross-entropy loss. Ref. [
21] similarly applied this concept to MDE. Target depth values are again discretized into sub-intervals, although since larger depth values are less rich in information, a spacing-increasing discretization with bigger bins towards larger values is utilized. This approach also benefits from ordinal classification [
22], which takes into account the ordering of discrete depth values.
4. Materials
In
Section 4.1, an overview is given of the MRO’s two imaging instruments upon which this work is based, including their technical specifications and scientific objectives. The specific data products used in this work are subsequently named in
Section 4.2 and their properties are discussed. Following this, various steps towards creating a suitable dataset are presented in
Section 4.3. These include the acquisition and choice of data files, coregistration, sample selection, clipping and data normalization. In
Section 4.4, specific data augmentation techniques seen as appropriate are discussed.
4.1. Imaging Instruments of the Mars Reconnaissance Orbiter
The MRO was launched on 12 August 2005 to study the geology and climate of Mars. Its science objectives include the observation of its present climate, search for aqueous activity, mapping and characterization of the Martian surface, and examination of potential future landing sites [
1]. Among its many special-purpose instruments, the CTX and CRISM cameras are the subject of this work and, therefore, are discussed in the following two sections.
Figure 3 shows an exemplary enhanced visible color (EVC) image of CRISM compared to a corresponding cut-out from CTX, in order to illustrate differences in appearance. CTX images, being only single-channel, are visualized as grayscale by mapping the reflectance values of shown scenes to the minimum and maximum brightnesses. Compared to this, EVC shows a composite of only three ( 592
, 533
, and 442
) out of over 400 spectral channels.
4.1.1. CRISM
Images from the CRISM instrument are the primary focus of this work. Its gimbaled optical sensor unit can be operated in multispectral untargeted, or hyperspectral targeted mode. The untargeted mode allows the reconnaissance of large areas due to reduced spatial and spectral resolution in order to discover scientifically interesting locations. Such locations are then mapped with the full spatial resolution of 15
to 19
and full spectral resolution of 362
–3920
at
per channel in targeted mode. CRISM’s mission objectives are to characterize the crustal mineralogy across the entire surface, to map the mineralogy of key areas and to measure spatial and seasonal variations in the atmosphere [
2].
Hyperspectral imaging data, such as those captured by CRISM, are the basis of visible-near infrared reflectance spectroscopy. Each pixel represents a reflectance spectrum with characteristic absorption bands depending on the materials. These spectra are then compared to laboratory measurements to determine the mineralogical composition. The right-hand side of
Figure 4 shows one such spectrum. In order to visually highlight or distinguish minerals within an image or scene, browse products are created. These are synthesized RGB color images, where each channel is assigned to a summary parameter, such as band depth or some other calculated measure of spectral variability, and afterwards stretched to a specific range. The previously mentioned spectrum is taken from a red region of the browse product displayed in the center of
Figure 4, whose red channel is set to the BD1300 parameter, corresponding to iron olivine.
The simplest browse products, such as enhanced visible color (see
Figure 3) or false color, are constructed by setting each RGB channel to one specific reflectance band. These browse products are visualized using a 3-sigma data stretch applied to each band (
crism.jhuapl.edu/msllandingsites/browse/abtBrwPrd1.php, accessed on 24 January 2022). This means that the values of each individual channel
i above
and below
are clipped to that boundary, with
and
being the mean and standard deviation of those values, respectively, and afterwards mapped to the minimum and maximum brightnesses.
4.1.2. CTX
The Context Camera’s primary function is to provide contextual information for other MRO instruments, by making simultaneous observations. It is also used to observe candidate landing sites and conduct investigations into the geologic, geomorphic, and meteorological processes on Mars. Owing to the 5064-pixel wide line array CCD and a 256
large DRAM, the pictures taken are 30
wide and up to 160
long, covering a much larger surface area than CRISM. These images are exclusively monochromatic, filtered through a bandpass ranging from 500
to 700
. The spatial resolution is 6
/
, making it around three times higher than CRISM [
3].
CTX data have been used to create a seamless mosaic of Mars resampled to 5
/
[
36]. This allows the analysis of much larger regions by studying more localized observations made by other instruments and extrapolating these findings to outer areas. The instrument has further collected many stereo pairs, prompting the creation of digital elevation models that aid in quantitative analyses of Martian landforms [
37].
4.2. Data Products and Properties
All CRISM images were acquired at the most recent, publicly available, calibration level, called Map-projected Targeted Reduced Data Record (MTRDR). Its joined visible/near-infrared plus infrared (VNIR-IR) full spectral image cube holds records in units of corrected I/F as 32-bit real numbers. I/F is the ratio of spectral radiance at the sensor and spectral irradiance of the Sun. The creation pipeline of this product type includes a series of standard spectral corrections and spatial transforms for the purposes of noise reduction and mitigation of data characteristics inherent to CRISM observations (
https://ode.rsl.wustl.edu/mars/pagehelp/Content/Missions_Instruments/Mars%20Reconnaissance%20Orbiter/CRISM/CRISM%20Product%20Primer/CRISM%20Product%20Primer.htm, accessed on 24 January 2022). Out of this product suite, only Full Resolution Targeted (FRT) images were used, ensuring the maximum similarity to corresponding CTX data. Available CRISM FRT products were searched using the Orbital Data Explorer (ODE) REST API (
https://oderest.rsl.wustl.edu/#ODERESTInterface). All relevant files related to FRT observations were downloaded from the Geosciences Node of NASA’s Planetary Data System (PDS) (
https://pds-geosciences.wustl.edu/). Also contained in these products is a text table that lists wavelength information for the spectral image cube. After the removal of spectral channels with suspect radiometry during the creation pipeline, 489 bands with wavelengths ranging from 436.13
to 3896.76
remain.
At the time of writing, there exist a total of 9677 MTRDR CRISM products, with 6479 FRT among them. Older calibration levels such as Targeted Reduced Data Record (TRDR) include even more files, 1,754,260 in total. Having this many samples provides a great opportunity for large-scale deep learning applications, although the amount of data also presents a challenge. Each CRISM IMG file within the dataset measures at around 1 and more. The size of their corresponding and fully processed CTX images varies based on the height between around 200 and 2 or more, even though only a small cut-out is required in the end. Some CTX observations have the fortunate property of being closest in time to multiple CRISM observations, reducing the number of CTX images that need to be processed slightly.
Important to note for later discussions on data augmentation is the spacecraft’s orbit and its impact on images taken. The MRO is placed into a low and near-circular sun-synchronous Mars orbit with a mean local solar time of 3:10 PM to perform its remote sensing investigations [
2]. This means that images captured by its instruments will have a similar angle of incidence, resulting in topological structures always being illuminated on the same side, while casting their shadow on the other [
38]. This consistency is expected to be instrumental for a neural network’s decision-making process and should therefore be conserved.
4.3. Dataset Creation
Training a neural network benefits from a large amount of high-quality data [
39]. The dataset used in this work is composed of multiple CRISM-CTX image pairs, as displayed in
Figure 3, taken from a selection of locations on Mars. This section describes in detail the creation process, starting with location selection and data acquisition. Subsequently, the multitude of preprocessing steps are outlined, which include CTX image creation, coregistration, the extraction of patches and clipping. Finally, three data normalization methods are compared and discussed. These steps serve as basic requirements for creating a dataset that is suitable for machine learning.
4.3.1. Acquisition
Due to the large file sizes of CRISM FRT products and CTX images mentioned in
Section 4.2, this work will focus on six locations and their respective CRISM observations.
Table 1 lists these locations, their associated latitude–longitude bounding box that was searched for observations and the number of MTRDR products that are found within. A complete list of all CRISM images and their corresponding CTX images used along with additional information, such as the image set they are assigned to, can be found in
Table A2,
Table A3,
Table A4,
Table A5,
Table A6 and
Table A7.
These areas are chosen because they are either previous landing sites, candidate landing sites or generally significant landmarks of Mars, and already have a large body of research due to their scientific interest. Bounding boxes were queried using the Orbital Data Explorer’s (ODE) REST API [
40], and all FRT-MTRDRs within them are obtained. Another query was conducted for each CRISM image, retrieving all CTX observations which are fully contained in it in terms of latitude–longitude bounding boxes. Out of these, the one closest in creation time is chosen as its corresponding input. This was done in order to minimize the difference between these images, due to changes over time or seasons. Many but not all CRISM observations have a CTX image taken at the same time, which are marked as ride-along. The exact creation time differences in days between CRISM and CTX pairs are listen in the dt column of
Table A2,
Table A3,
Table A4,
Table A5,
Table A6 and
Table A7. Some CRISM images within the locations overlap, as is illustrated in
Figure 5, which was taken into account for the evaluation process. The test and validation images used to evaluate the generalization capabilities of models were chosen such that they share minimal area with images within the training set. The separated observation on the left side of
Figure 5, for example, would be chosen as a test image. This can be seen in the set affiliation column of
Table A2,
Table A3,
Table A4,
Table A5,
Table A6 and
Table A7 when considering the overlap column that shows the average percentage overlap of each CRISM image with all other CRISM images of that location. Only two images from each location belong to the test and validation sets to maximize the amount of data available for training. A validation set during training can reveal whether the model over- or under-fits. Finally, CRISM images that are deemed too noisy or contain large artifacts are omitted from the training set and marked with a dashed line. One such example along with its corresponding CTX cut-out is displayed in
Figure 6.
4.4. Augmentation
Since the goal of this work is to accurately predict spectral bands, no changes to ground-truth CRISM pixel values such as brightness or gamma are considered. Furthermore, due to the consistency in the illumination direction mentioned in
Section 4.2, vertical and horizontal flipping operations are not included. Such images never appear in the collection of CRISM files, and are therefore not useful training samples. One of the abilities a trained network should illustrate is to detect shaded regions and accurately determine their properties, which flipped samples would disrupt.
The augmentations examined in this work are random croppings or deletions of sections, rotation and zoom. Rotations are limited to a small interval, as to not significantly change the direction of illumination. The degree of rotation is randomly chosen from the interval . The height and width of cropped sections are independently taken from the interval , while the height and width of erased sections are taken from , with d denoting the side length of patches. Translational changes are not needed since the random way in which patches are cut out covers any possible location. Each training patch pair—consisting of CTX input image and CRISM ground truth—is given an equal probability of being left unchanged, rotated, cropped or having a section erased. The aforementioned augmentations are applied to both images equally. Invalid data regions created as a result of cropping, deletion and rotation are masked and not included in the loss calculation.
5. Results
In this section, methods and tools that have been presented and created are applied, and their results are compared. Initially, the task is formulated as a pixel-wise regression problem. This formulation serves to establish a basic training configuration. Afterwards, the problem is reformulated as a pixel-wise classification of binned pixel values in
Section 5.4.3 and compared to the basic configuration. Lastly, in
Section 5.6, the best-performing model is used to assess the capabilities of a machine learning approach more generally and possible areas of application are suggested and demonstrated. For all experiments, the single-input channel from the CTX instrument is used to predict three CRISM channels. In the case of enhanced visible color (EVC) wavelengths 592
, 533
, and 442
are used. In the case of false color (FC) 2503
, 1500
, and 1080
are used (see
Table A1).
5.1. Model Architecture
A comparison of different model architectures is the first experiment performed. This, however, requires the pre-selection of some parameters and methods for a training process to function. The following list lays out the base configuration used for each model.
Number of training epochs: 360;
Patch size: 256 by 256 ;
Batch size: 16;
Data normalization method: image-wise;
Optimizer: Adam;
Learning rate: ;
Loss function: MSE.
The number of epochs and image-wise normalization are chosen based on prior experiments, while a batch size of 16 guarantees that each models fits into memory. The patch size and learning rate are common default values, whereas MSE and Adam are robust first choices.
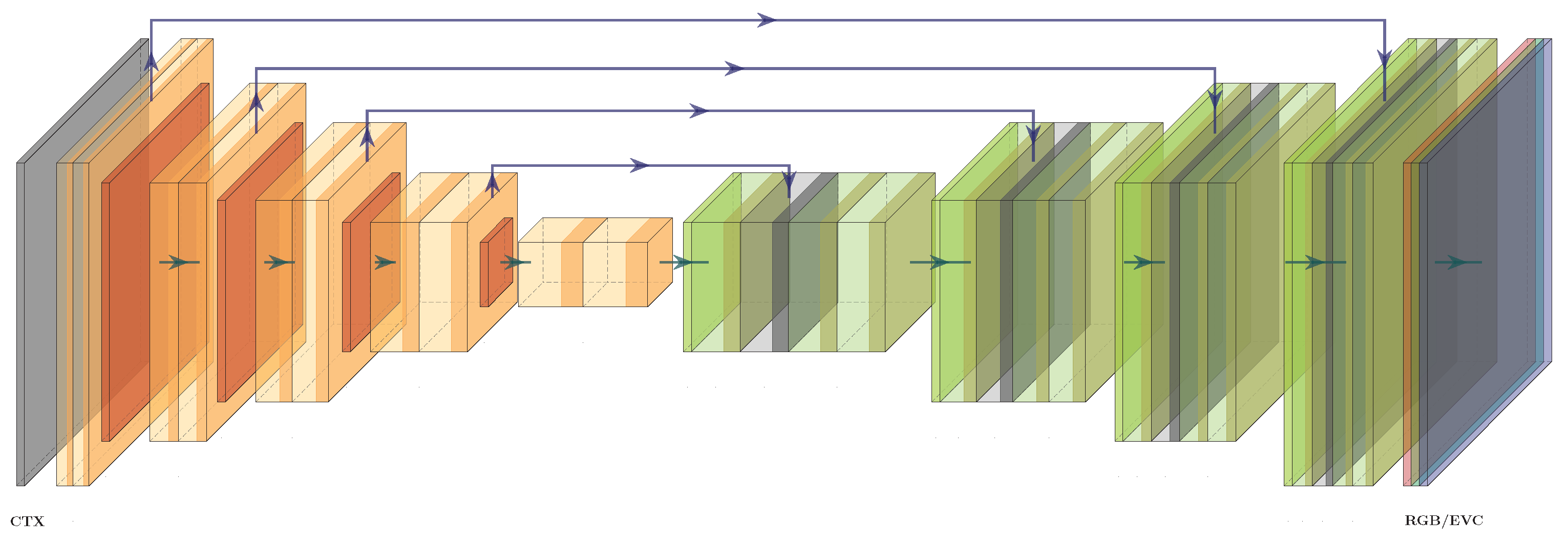
Most of the compared models are taken from an implementation named Segmentation Models PyTorch (
https://github.com/qubvel/segmentation_models.pytorch, accessed on 24 January 2022) (SMP), which provide a selection of architectures intended for image segmentation applications. Each architecture can be initialized with different encoders that are pre-trained on publicly available datasets. In this comparison, every architecture is initialized with the same ResNet18 (see
Section 2) encoder pre-trained on ImageNet [
41]. Two additional U-Nets are compared, a biomedical segmentation model created by [
42] and another one used by [
18] as a CGAN generator for image colorization.
Figure 7 shows six input CTX and ground-truth CRISM patch pairs, one from each of the test images, with their file names in the top left corner. Every subsequent column displays the output of each compared model architecture when given the corresponding CTX patch as input after training. For visualization purposes, CRISM patches are displayed using a channel wise 3-sigma stretch. As mentioned in
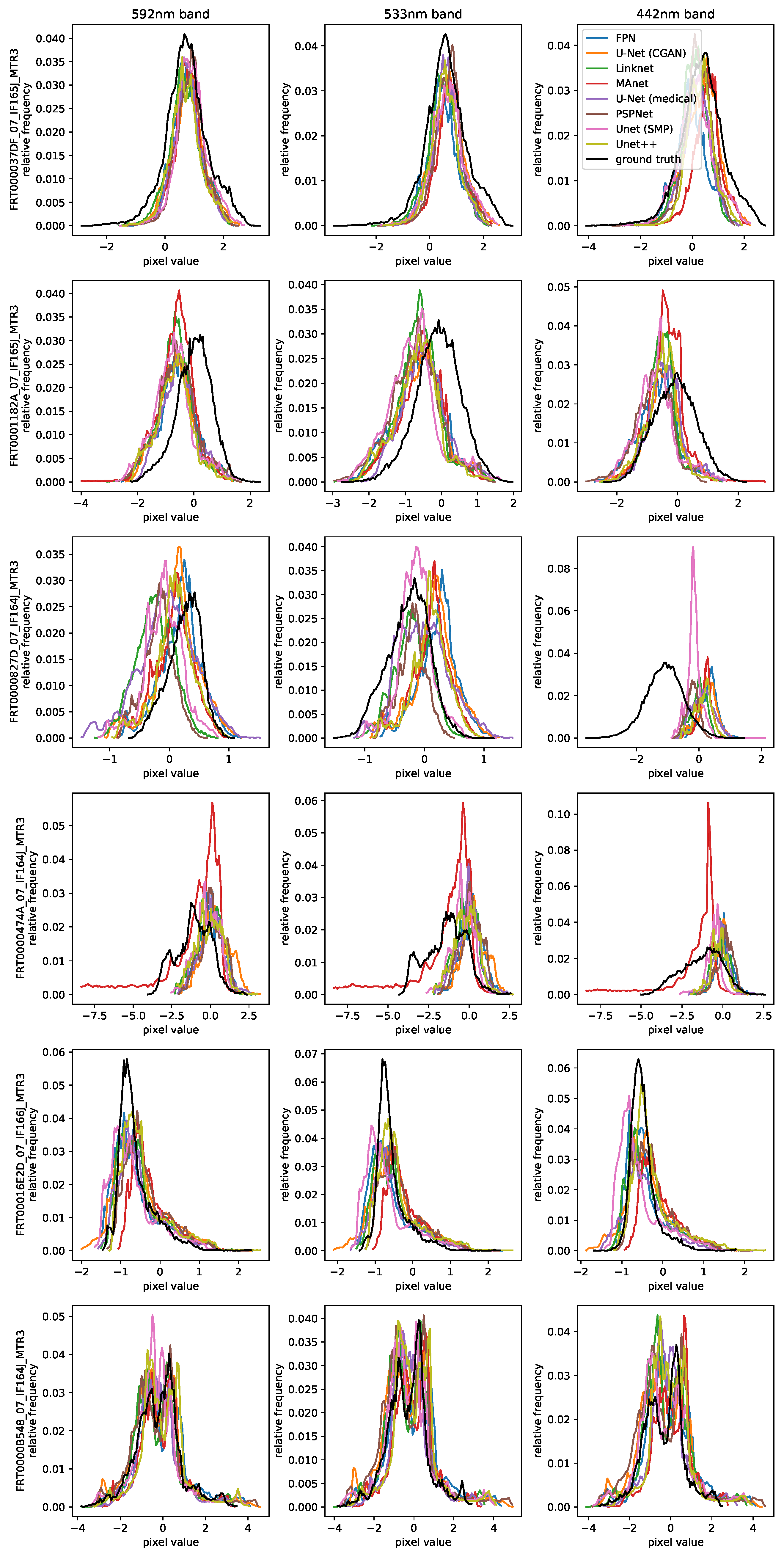
Section 4.3.1, not every CRISM image is taken at the same time as its CTX counterpart, meaning that in these cases the model output and ground-truth CRISM need to be compared with respect to the provided input. For a better assessment of the quality of the predictions, channel-wise histograms are presented in
Figure 8. A comparison of measurable performance metrics is given in
Table 2. They include three metrics—root mean squared error (RMSE), cosine similarity (Cosine), and Pearson correlation coefficient (PCC)—with the average loss of the final epoch over all training samples, the network’s number of trainable parameters, the maximum memory consumption during training, and the average training time per epoch.
The first observation of note is the difficulty faced when relying on regular regression losses when judging the performance of models employed in this task, as a result of artifacts, a lower resolution and the noisiness of CRISM samples compared to CTX. Even a hypothetical ideal output patch that is color accurate, sharp and noise-free would still result in an error compared to the ground truth. Quantifying the quality of unseen validation and test samples when non-global normalization methods are used is especially troubled. This is because values of input–output pixels depicting the same area can be different, depending on from which image or patch they are taken. A network could therefore correctly predict the hue of a given area, but with a wrong magnitude, still resulting in a high loss. A comparison of visual appearance is therefore prioritized.
For one, the test image pair of row three is exemplary of an especially noisy CRISM and CTX observation, as they are occasionally found within the dataset. With such CRISM patches as the ground truth, it is more difficult to judge the accuracy of predictions. Noisy CTX inputs, which can additionally contain large stripes such as artifacts, which can be faintly seen in
Figure 6, further result in widely varying predictions of lower quality. Otherwise, immediately noticeable when examining the fourth row of patches is that the MAnet introduces large artifacts with extreme values, which is reflected in its high test loss. Since the remaining test losses are very similar and not correlated with the image quality or accuracy of outputs, test loss appears to be an insufficient performance metric in this case, due to the aforementioned challenges. Time and memory requirements vary significantly depending on the model architecture. Among the most time-consuming and memory-intensive models are the medical U-Net with its two-layered blocks at full resolution, the CGAN U-Net with its large size, and Unet++ with its complex skip connections. In general, all remaining SMP models exhibit a lower memory footprint and training time because of their shared ResNet18 encoder that uses a 3 × 3 pooling operation at the beginning of the model.
PSPNet, like some other segmentation networks, includes an upsampling operation at the end, making it unable to create sharp images. Other networks suffer from occasional striping artifacts, inaccurate colors, or desaturation, such as U-Net (medical), U-Net (CGAN) and FPN, respectively. Both Unet and Unet++ produce some of the most visually appealing images, although the Unet++ requires close to twice the memory and time due to its complexity. The regular Unet is therefore chosen. However, sometimes it produces artifacts at corners, as can be seen in the third row. Since the problem occurs at the edge, it can be speculated that the standard zero-padding procedure causes this issue. Switching to a reflection-based padding removes these artifacts, as can be seen when comparing the bottom left corner of the third row of the Unet column of
Figure 7 with the third row of the resnet18 column of
Figure 9.
5.2. Encoders
With Unet being chosen, different encoders can be compared in the same way. Out of the encoders provided by SMP, a selection of prominent architectures with a similar number of parameters were taken. Scaled-up versions of these models exist, but the smallest configuration was used for this comparison. Each of them was pre-trained on ImageNet. In the same manner as before, the results can be seen in
Figure 9 and
Table 3.
There is far less difference in appearance between encoders compared to models. A particularly low train loss of the VGG11 encoder stands out, although this is neither noticeably reflected in the appearance of the predicted patches nor in the metrics. One problem that emerged during training is that some models showed a diverging behavior. GerNet, DenseNet, and both RegNets show spikes in validation loss that far exceed the usual values. These models sometimes produce artifacts with extreme values in their outputs, which happen to not be present in
Figure 9. Three exemplary artifacts from each of the models are shown in
Figure 10. Such divergences could potentially be solved with regularization techniques such as weight decay by lowering the learning rate or they could stem from some incompatibility between the model architecture and this problem formulation. Despite these issues, the patches predicted by these models that do not suffer from artifacts are still comparable to any of the other outputs. The remaining encoders, ResNet, EfficientNet and VGG, produce similar images, but EfficientNet was chosen for further tests, as it is also among the lowest in parameter count, giving it more room for expansion.
5.3. Evaluation of Full Images
Figure 11 depicts an entire input CTX cut-out alongside its ground-truth CRISM image in the top row, together with the current network’s prediction in the mid-left image, generated via the aforementioned method. The quantitative results are presented in
Table 4.
The Gaussian averaging strategy performs best in providing visually seamless results when comparing the lower two images. Even though the left image requires
as many patches to be processed, the right image still appears slightly more or just as seamless, showing the importance of weighting pixels towards the edge less. By examining the accuracy of this full image prediction, it can be said that hue and saturation are consistently plausible for images from the Eberswalde crater and are comparable with the ground truth. Out of the six locations that are used for training, each of which exhibits a different kind of color scheme, the model is able to predict an accurate set of colors for this unseen image. Only two spots at the top edge are colored overly blue. The separation between a more brown half at the top and a mostly turquoise region at the bottom as seen in the ground truth is also lost in the prediction. This can be attributed to insufficient context being provided to the model because of too-small patches, resulting in an inability to capture larger trends. Another less likely cause is the time difference of 16 days between the CTX and CRISM observations, which can mean that the reflectance properties during the CTX’s recording are different from what is seen in its corresponding CRISM image. On average, the time difference between each image pair is 29 days (cf.
Table A2,
Table A3,
Table A4,
Table A5,
Table A6 and
Table A7).
5.4. Ablation Study
After selecting the Unet with an EfficientNet Encoder, we conducted a set of ablation studies to test how incremental changes to the normalization method, the loss function, and adding additional input data affect the metrics. The results are summarized in
Table 5 and discussed in the following sections.
5.4.1. Normalization Methods
Using a Unet with an EfficientNet encoder, the three data normalization methods in
Appendix A.2 are compared. With the network being fixed, the batch size is increased to 32, while all other parameters remain. Since the magnitude of pixel values is vastly different between comparands, a direct comparison of losses is not possible. Lower values result in lower losses and vice versa. The only remaining metric that is different across normalization methods is the training time, where a patch-wise normalization necessitates on the fly calculations that can not be performed prior, increasing the training time slightly. The mean and standard deviation of each CRISM and CTX channel across the entire dataset that are used for global normalization can be seen in
Table A1. Predictions of six patches are shown in
Figure 12.
Un-normalized data are confirmed to be unsuitable for this application. The network creates unfitting colors, as shown in the first row, line artifacts at the bottom of patches and miscolored spots in the last row. This is reflected in the first row of
Table 5, which shows the second highest RMSE and the lowest PCC and COSS. Globally transformed data result in more stable and accurate outputs. Obvious miscolorizations and artifacts in the first and last rows are remedied with this method. However, the issue of pixel value distributions of CRISM bands being different orders of magnitude and often constrained to small intervals still proves challenging for the model. The metrics in the second row of
Table 5 coincide with these issues, as this method results in the highest RMSE and second lowest PCC. Both globally and non-normalized data seem to be particularly problematic in combination with noisy input and output pairs, as shown in the third row, where predictions appear especially blurry and spotted. Image-wise and patch-wise normalizations produce the most consistent results in comparison, with both being quite similar. Patch-wise outputs are sometimes more color-accurate, such as when predicting the blue area in the bottom left quantile of the second row patch. It also appears sharper in some instances, as in the bottom left of the fourth row. These two methods are listed in the third and fourth rows in
Table 5, and score much better than the first two methods discussed. Major differences, however, emerge if an evaluation of entire CTX cut-outs is attempted, which are shown in
Figure 13.
Without normalization, not much colorization or processing is applied to the input image and some patches produce widely spanning artifacts. Global normalization yields much more plausibly colored results, but some patches stand out again as being miscolored. Both these results indicate an unstable training process and a non-robust model, which drastically changes its output if the input shows a slightly different region. With patch-wise normalization, each individual output appears plausible, but it is not directly possible to reassemble the image. With each patch occupying a similar range of values, discontinuities are strongly apparent, as would be the case when putting together ground-truth patches. In this predicted reassembly, some patches appear desaturated and gray, while others exhibit a broad range of colors that are not fitting with those given areas. Compared to this, an image-wise normalization allows the creation of a coherent overall image, since ground-truth patches now form a continuum across each image. The more favorable distribution of pixel values compared to global normalization also improved the robustness of the network, hence why this method was chosen for further comparisons.
5.4.2. Digital Elevation Model
Elevation data can improve network performance if used in training as an additional cue. In cases where certain minerals correlate with the characteristic topology, pixel-wise elevation data of sufficient resolution can be helpful to a network in identifying them. At a smaller scale, positive or negative gradients along the axis of illumination determine whether a region is shaded or illuminated. If given enough context, then the detection of hills and valleys can also factor into a prediction. For this test, the HRSC and MOLA blended digital elevation model (DEM) by [
44] with a resolution of 200
/
is used, which is far lower than the CTX resolution. This can therefore only benefit the prediction of larger landmarks and regions. Each CTX-CRISM image pair’s bounding box is used to cut out that region from the global DEM and the resulting image is added to the dataset. The elevation values are given in meters and range within the thousands, making them unusable without normalization. For this, an image-wise normalization analogously to CTX is used. The input is now made up of CTX patches that are channel-wise concatenated with their corresponding DEM patches. In accordance with this, the number of input channels of the network is changed to two.
To examine the impact of including DEMs in the input at a larger scale, a comparison of full CTX predictions was conducted, specifically of Mawrth Vallis’ validation pair depicted in
Figure 14. The top row shows the input CTX and un-normalized DEM cut-out on the left, and ground-truth CRISM on the right. This pair was chosen because the DEM exhibits a clear horizontal gradation, sectioning the ground truth into two distinct halves. This information seems to have benefited the model that utilized it during training, as seen in the bottom right image, which is closer to ground truth in terms of color accuracy. In contrast, the models trained without that information in the bottom left predict colors less accurately and create an overall less coherent image.
5.4.3. Classification
The goal of this section is to determine whether reformulating the problem as a classification task achieves better results (see
Section 3.2). With this formulation, the model is tasked with assigning pixel-wise probabilities to discrete spectral values in each spectral band. Ground-truth CRISM reflectance values are normalized on an image-wise basis and afterwards assigned to
equidistant bins within the interval
. The various additional steps such as discretization, the prediction of more channels and output reconstruction make this approach more computationally expensive than regression. Having shown the overall best performance in the previous section, a U-Net with an EfficientNet-B0 encoder is used again to predict EVC spectral bands. To obtain the desired output, the final convolutional layer’s number of output channels is changed to
, with
being the number of spectral bands predicted. Its output tensor of shape
is subsequently reshaped to
in order to simplify the loss calculation and image reconstruction.
H and
W, in this case, are the height and width of the patches used.
Number of Bins
The first and most fundamental parameter to determine is the number of bins
. A small number of bins results in fewer possible colors and larger quantization errors, while more bins requires additional computational effort and diminishes the advantage of discretization. The network is trained with four different bin counts and a constant batch size of 10. The resulting predictions of test patches are shown in
Figure 15, when outputs are reconstructed using a weighted sum, as described in
Section 3.2. The according metrics are listed in
Table 6.
Judging the quality and accuracy of predictions in
Figure 15, bins counts of 20 or 40 produce some of the better images, for example, in the second and third rows of
and in the fifth row of
. Ten bins are not sufficient to produce detailed results and 80 bins may be too many as more artifacts arise, such as the turquoise spot in the last row. This classification approach in general has shown to carry the risk of producing artifacts like this, which were not present in the regression approach. Examining test and train loss in
Table 6 shows how more bins leads to higher losses, which is expected since the same prediction with more classes results in a higher cross-entropy loss. As a result of this, a judgement based on the cross-entropy loss is not possible in this comparison. Another noticeable trend is the increased demand of memory and time with more bins, because the final convolutional layer is required to output more channels. Even though the resulting increase in the number of parameters is very small, these weights are used to convolve many more full-resolution feature channels, which takes longer and requires more memory. Based on the quality of outputs and ease of computation, 20 bins were chosen for further comparisons.
An advantage of this approach is that a measure of confidence is provided for each prediction. If the predicted probability density displays a single sharp peak, it can be said to be confident. A broad peak or multiple maxima conversely indicate uncertainty and are a hint for faulty predictions. Both cases are illustrated in
Figure 16, where the bottom right prediction from
Figure 15 is shown again on the left without a 3-sigma stretch. Two pixels are marked, (1) taken from a region within an artifact and (2) being an accurate prediction. Their predicted probability densities of each channel across bins are displayed to the right. Pixel (1) exhibits broader peaks in each channel and two maxima within the 592
channel, with the one at bin zero being clearly faulty and distorting the results no matter the used reconstruction method. The peaks of pixel (2), conversely, are sharper, which corresponds to more confident, and in this case accurate, predictions.
Loss Modification
Additionally, we examined three modifications or additions to the problem formulation, which were used to address the shortcomings of the classification approach or offer improvements in general. Some of these are found within the literature, as mentioned in
Section 2, and are adapted to this task. An overview of six test patches and the baseline model’s predictions with 20 bins is presented in
Figure 17, which is compared to the model’s output if any of the modifications are enabled.
Regular cross-entropy loss does not take into account the distance of a wrong prediction from the ground truth. Only when the bell curve of predicted probabilities, as seen in the right plot in
Figure 16, enters the correct bin does the loss measurably decrease with lower distance. One way to increase the loss of predictions that stray further from the ground truth is to introduce a regression loss component, where the absolute difference between the predicted value at the highest probability and ground truth value is additionally back-propagated. This is equivalent to reconstructing the network’s output using the argmax method and calculating the MAE between it and the ground-truth image. Due to the difference in magnitude between both types of losses, MAE is weighted by 0.9 and cross-entropy is weighted by 0.1 to arrive at similar scales, after which both are summed and back-propagated. In the last column of
Figure 17, it is shown that there is not much difference between the model that utilizes this addition compared to the baseline model, except the superior colorization of the fifth patch. Examining larger areas, however, reveals some improvements, as is shown in
Figure 18. This figure depicts the validation image of the Jezero crater and both model’s predictions. The model with a MAE loss component produces a more colorful image, especially in the yellow regions. Further, features such as the brown streak going from the center left to the top right and the crater’s wind streak are predicted more accurately.
Spacing-Increasing Discretization
Whether this technique reduces the quantization loss in practice can be tested by evaluating the MAE between ground-truth CRISM samples and predicted samples reconstructed via the argmax method. Doing this for one pass of the training set and comparing the model with and without SID reveals that the MAE drops from 0.32 to 0.3 if SID is used, a decrease of 6%. This shows that utilizing a SID slightly reduces the difference between predicted discrete and ground-truth continuous values.
5.5. False Color Prediction
Up to this point, only channels that make up enhanced visible color (EVC) browse products were predicted, which lie within the visible spectrum. This section examines the prediction of false color (FC) channels, situated in the infrared part of the spectrum. The three exact bands that constitute this browse product are 2503 nm, 1500 nm and 1080
, which correspond to red, green and blue color channels, respectively.
Figure 19 shows six EVC test patches and predictions made by a model that is only trained on them. Two additional columns on the right show the same patches in FC, one being the ground truth and the other being predictions made by the same type of model trained only on FC channels. The patch size in this and following comparisons is increased to 384 by 384 pixels, meaning that different regions within each of the six test images are shown. This figure shows that learning a mapping between CTX and FC bands provides additional challenges compared to EVC. The general trend of blue, gray and pink shades in specific areas such as dunes in the third patch or the general appearance of the last patch is predicted adequately. Some areas, however, such as the green region of the second patch that indicates the presence of clay or potentially carbonates [
45], is lost in the prediction. This is likely due to an imbalance in the dataset, because such green sections are almost exclusively found within the Jezero crater subset. These deviations from the norm are more localized and rare, which makes the mapping between CTX and CRISM’s infrared reflectance harder to learn. CRISM channels within the visible spectrum in comparison tend to exhibit wide spanning and clearer tendencies towards certain colors, such as the dark blue dunes or general beige terrain in
Figure 3. The network may also be too small or unsuited to learn the features necessary for making a nuanced prediction such as this possible. A larger and potentially more balanced dataset, as well as a more powerful machine learning model, would presumably be better able to accurately predict such features.
5.6. Assessment of Capabilities and Applications
Concluding this chapter, a general assessment of the machine learning model’s capabilities is provided. Images from the training set and unseen images were evaluated to determine what factors are required for high-quality predictions and generalization abilities. Quantitative results are presented in
Table 7. The final two examples show the restoration of a faulty CRISM observation and the evaluation of entire CTX images as possible areas of application. For this evaluation, the training configuration from
Section 5.5 with rotational augmentation from
Section 4.4 was used. It was trained for 500 epochs with a reduced learning rate of
to obtain the following results.
Examining the predictions made by the model on CTX images that it was trained on, it can be seen that the model’s capacity is sufficient to memorize most of the training data. As long as the input and ground truth training data are of high quality, meaning minimally noisy and devoid of artifacts, reconstructions of these images are consistently accurate. The top two images in
Figure 20 show the evaluation of two training images, which are very close to the ground truth and exhibit a promising continuation beyond the regions where CRISM data are available. Therefore, no over-fitting is observed in these cases, and predictions are more accurate the nearer they are to training locations. Inaccuracies arise when lower-quality data are used for training, and when the model is tasked to predict outputs that are infrequent within the dataset and only constitute a minor deviation from the norm. This second aspect is illustrated in the third image of
Figure 20, which shows an overall very accurate reconstruction of an image from the training set, with plausible predictions beyond borders. However, the pink hue towards the top left is not captured in the prediction, as it is a less frequent deviation within the dataset. This same problem is observed more clearly in the prediction of false color bands.
The evaluation of unseen CTX images that have not been trained on and that share little to no area with the training set revealed the generalization capabilities to be adequate in most cases.
Figure 20 shows one such example from the validation set, where the predicted color scheme is plausible throughout and the image quality is high. This second characteristic is naturally reliant on high-quality inputs, while the first characteristic requires enough similar data to be trained on. Because there exists at least one training image pair in
Figure 20 that exhibits the same type of terrain and mineralogy, a plausible prediction can be made.
In
Figure 21, we present two examples from the test set. Although variations in color are obvious, the resulting metrics presented in
Table 7 show similar values to those from the training set. The proposed approach is thus able to generalize the spectral characteristics learned from the training set on unseen data.
One possible area of application that can be shown using the present dataset is the restoration of a faulty CRISM observation, as is presented in
Figure 20. It can be seen how the CRISM image contains bright-blue horizontal striping artifacts that span the entire image. Since the input CTX image is faultless and the model is trained on data from that location, a prediction can be made. While it is difficult to verify the accuracy of this prediction, some aspects such as the central beige dunes and the blue-tinted bottom left region appear reasonable.
Finally, the evaluation of entire CTX images can now be attempted, instead of the much smaller cut-outs that have been examined and trained on thus far. The CTX image is normalized as a whole, which changes the pixel values compared to the image-wise normalized cut-outs that are used for training. A global normalization in comparison would preserve the same values. A successful prediction of a full CTX image is shown in
Figure 22. Depending on the surface character of CTX, outputs are colored in accordance with images provided during training, as can be seen when comparing with first and the last but one row in
Figure 20. These results show that plausible EVC images of large areas can be created within regions where high-quality training data are provided.
6. Discussion
Within the context of this work, it was found that common segmentation networks are capable of learning a generalizing mapping between CTX and CRISM reflectance within the visible spectrum. The capacity of models is sufficient to memorize training samples, while predictions beyond areas with CRISM data continue accurately. Unseen areas can be evaluated plausibly if high-quality training data of similar mineralogy were provided. Extending this task to the prediction of infrared channels yielded inferior results, due to the localized spectral variability. Upscaled networks and more data of under-represented features are likely required to obtain superior infrared predictions.
Regarding data normalization, it was observed that scaling each image and channel individually results in the most accurate and robust predictions, which also form a continuum across each image. In contrast, the normalization of each individual patch does not allow for a direct reassembly of an image, while patches normalized via global statistics remain constrained to unfavorable intervals and exhibit biases in certain spectral bands. With an image-wise normalization, however, it is not possible to apply an inverse transformation to obtain original reflectance values in areas where none are already available. A consequences of this is the inability to calculate the spectral parameters of most browse products in the usual way. For this reason, another normalization method or method of predicting spectral reflectance that preserves the ability to regain original values needs to be developed.
When comparing the machine learning methods examined in this work, it was found that posing the problem as a regression task in which square patches are predicted performs best overall. Reformulating the problem as a classification of binned spectral reflectance did not improve the quality of outputs, while increasing the demand on time and memory. Some functionality can be gained from these approaches, such as providing a measure of confidence when predicting probability densities across bins. Further research into classification methods is required if these approaches are to be used.
7. Conclusions
This work investigated the potential of machine learning approaches to predict pixel-wise CRISM spectral reflectance from CTX images. In pursuit of this, a dataset of 67 CRISM-CTX image pairs from six locations was created, and the methodologies of data acquisition, co-registration, extraction of patches, clipping, normalization and possible data augmentation methods were detailed. Various common machine learning approaches used in similar fields of study were presented and adjusted to this task. A pixel-wise regression approach that utilizes segmentation networks was the most extensively examined configuration. Different popular model architectures, encoders, and data normalization methods were compared to obtain a best-performing baseline configuration. Upon the baseline, various modifications and additions to the training process were tested. Performance was mainly judged on the basis of visual appearance, which includes color accuracy, consistency, sharpness and potential artifacts. This method was prioritized due to the difficulty of relying on losses when evaluating lower-quality and individually normalized image pairs. Metrics of quality, such as average training or test losses, and measures of computational effort, such as training duration and GPU memory requirements, were also considered. The beneficial and detrimental aspects of different configurations and techniques were discussed and hypotheses for successes and failures were provided. On the basis of these results, a classification approach was tested, in which continuous spectral reflectance is binned and pixel-wise probability densities across bins are predicted. Number of bins, methods of output reconstruction and other modifications or additions to the training process were tested and compared. Finally, the evaluation of entire CTX observations was performed, which constitutes one of the possible areas of application which were discussed. An extended dataset is likely to help with many of the shortcomings experienced in this work. A robust and automated pipeline for the acquisition, re-projection and coregistration of CRISM and CTX images can be used to create a larger dataset that is either focused on a specific location or is representative of the entire Martian surface. With more samples that contain rare deviations from the norm, predictions could be made more accurate. Alternatively, methods that emphasize these deviations during training may be explored. Finally, the prediction of all spectral bands that targeted CRISM observations provided can be attempted. Utilizing the similarity between and continuity along spectral bands can aid in this regard.