Using Single- and Multi-Date UAV and Satellite Imagery to Accurately Monitor Invasive Knotweed Species
Abstract
:1. Introduction
2. Materials and Methods
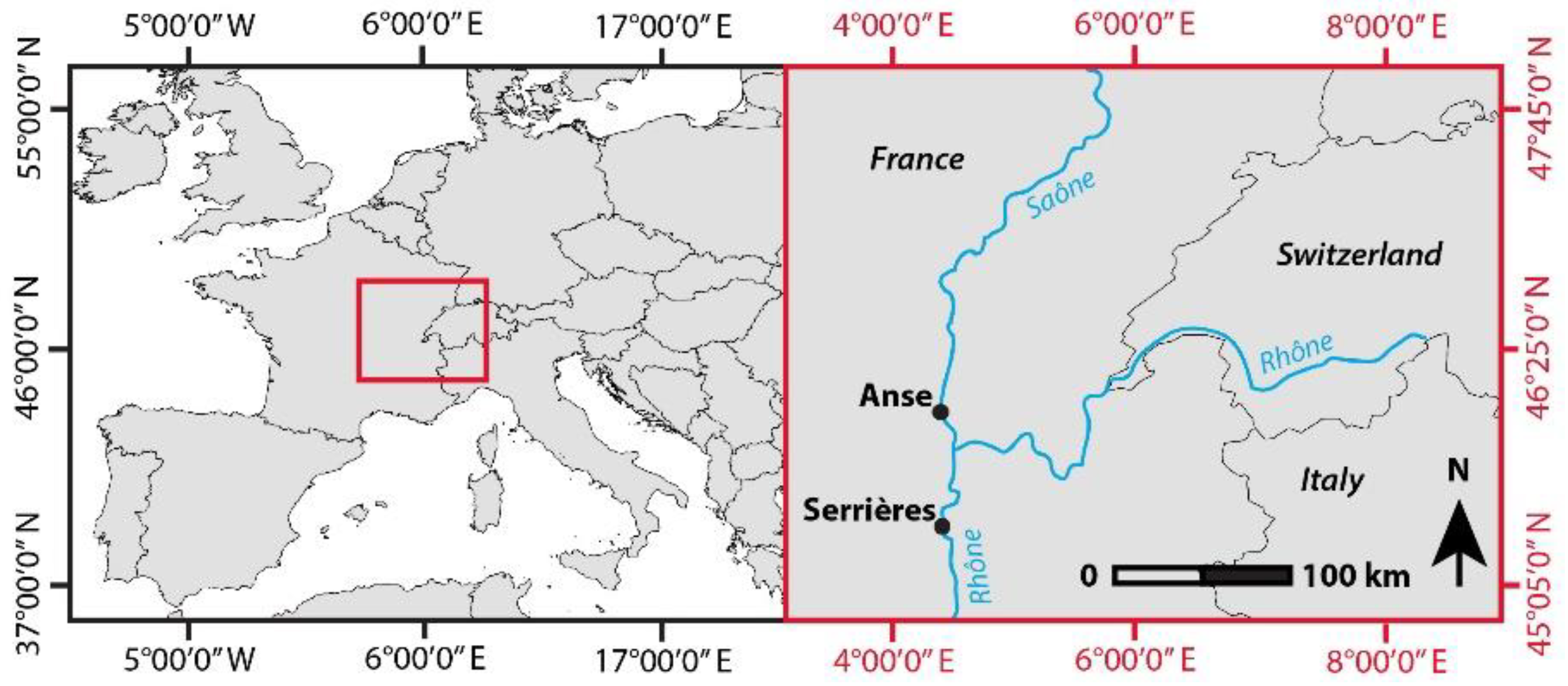
2.1. Study Sites and Image Acquisition
2.2. Image Preprocessing
2.3. Classification Design and Variables
2.4. Classification Procedure
2.5. Validation and Accuracy Assessment
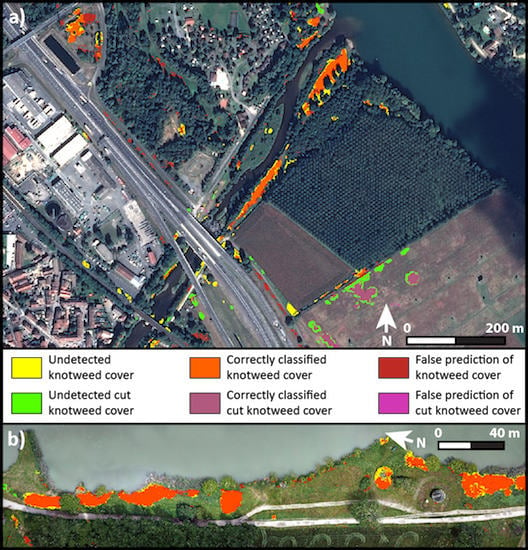
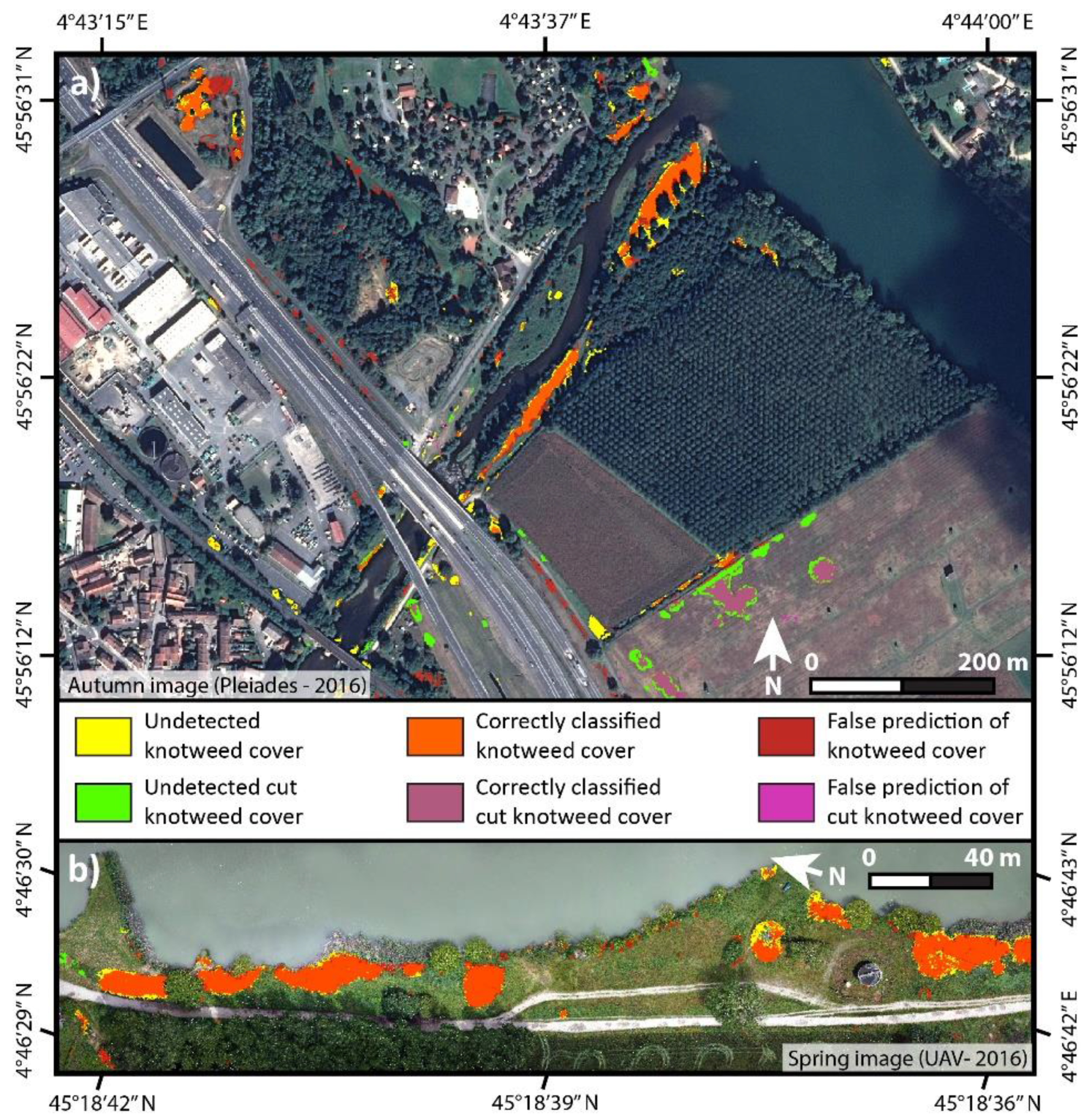
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vilà, M.; Ibáñez, I. Plant invasions in the landscape. Landsc. Ecol. 2011, 26, 461–472. [Google Scholar]
- Holden, M.H.; Nyrop, J.P.; Ellner, S.P. The economic benefit of time-varying surveillance effort for invasive species management. J. Appl. Ecol. 2016, 53, 712–721. [Google Scholar] [CrossRef]
- Byers, J.E.; Reichard, S.; Randall, J.M.; Parker, I.M.; Smith, C.S.; Lonsdale, W.; Atkinson, I.; Seastedt, T.; Williamson, M.; Chornesky, E. Directing research to reduce the impacts of nonindigenous species. Conserv. Biol. 2002, 16, 630–640. [Google Scholar] [CrossRef]
- Huang, C.Y.; Asner, G.P. Applications of remote sensing to alien invasive plant studies. Sensors 2009, 9, 4869–4889. [Google Scholar] [CrossRef] [PubMed]
- Bradley, B.A. Remote detection of invasive plants: A review of spectral, textural and phenological approaches. Biol. Invasions 2014, 16, 1411–1425. [Google Scholar] [CrossRef]
- Müllerová, J.; Brůna, J.; Dvořák, P.; Bartaloš, T.; Vítková, M. Does the data resolution/origin matter? Satellite, airborne and UAV imagery to tackle plant invasions. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 903–908. [Google Scholar]
- Dvořák, P.; Müllerová, J.; Bartaloš, T.; Brůna, J. Unmanned aerial vehicles for alien plant species detection and monitoring. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, XL-1/W4, 83–90. [Google Scholar]
- Remondino, F.; Barazzetti, L.; Nex, F.; Scaioni, M.; Sarazzi, D. UAV photogrammetry for mapping and 3d modeling-current status and future perspectives. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2011, 38, C22. [Google Scholar] [CrossRef]
- Alberternst, B.; Böhmer, H. NOBANIS: Invasive Alien Species Fact Sheet—Reynoutria japonica. Online Database of the European Network on Invasive Alien Species. Available online: www.nobanis.org (accessed on 12 January 2018).
- Bailey, J.; Wisskirchen, R. The distribution and origins of Fallopia × bohemica (Polygonaceae) in Europe. Nord. J. Bot. 2006, 24, 173–199. [Google Scholar] [CrossRef]
- Bailey, J.P.; Bímová, K.; Mandák, B. Asexual spread versus sexual reproduction and evolution in Japanese knotweed s.l. Sets the stage for the “battle of the clones”. Biol. Invasions 2009, 11, 1189–1203. [Google Scholar] [CrossRef]
- Buhk, C.; Thielsch, A. Hybridisation boosts the invasion of an alien species complex: Insights into future invasiveness. Perspect. Plant Ecol. Evol. Syst. 2015, 17, 274–283. [Google Scholar] [CrossRef]
- Child, L.; Wade, M. The Japanese Knotweed Manual; Packard Publishing Limited: Chichester, UK, 2000; Volume 1, 123p, ISBN-10 1 85341 127 2. [Google Scholar]
- Bashtanova, U.B.; Beckett, K.P.; Flowers, T.J. Review: Physiological approaches to the improvement of chemical control of Japanese knotweed (Fallopia japonica). Weed Sci. 2009, 57, 584–592. [Google Scholar] [CrossRef]
- McHugh, J.M. A Review of Literature and Field Practices Focused on the Management and Control of Invasive Knotweed. 2006, pp. 1–32. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.512.6014&rep=rep1&type=pdf (accessed on 23 August 2018).
- Kettunen, M.; Genovesi, P.; Gollasch, S.; Pagad, S.; Starfinger, U.; ten Brink, P.; Shine, C. Technical Support to EU Strategy on Invasive Alien Species (IAS); Assessment of the Impacts of IAS in Europe and the EU (Final Module Report for the European Commission); Institute for European Environmental Policy (IEEP): Brussels, Belgium, 2009; 44p. [Google Scholar]
- Williams, F.; Eschen, R.; Harris, A.; Djeddour, D.; Pratt, C.; Shaw, R.; Varia, S.; Lamontagne-Godwin, J.; Thomas, S.; Murphy, S. The Economic Cost of Invasive Non-Native Species on Great Britain; CABI Proj No. VM10066; CABI: Oxfordshir, UK, 2010; Volume 9, 199p. [Google Scholar]
- Meier, E.S.; Dullinger, S.; Zimmermann, N.E.; Baumgartner, D.; Gattringer, A.; Hülber, K. Space matters when defining effective management for invasive plants. Divers. Distrib. 2014, 20, 1029–1043. [Google Scholar] [CrossRef]
- Fox, J.C.; Buckley, Y.M.; Panetta, F.; Bourgoin, J.; Pullar, D. Surveillance protocols for management of invasive plants: Modelling Chilean needle grass (Nassella neesiana) in Australia. Divers. Distrib. 2009, 15, 577–589. [Google Scholar] [CrossRef]
- Pyšek, P.; Hulme, P.E. Spatio-temporal dynamics of plant invasions: Linking pattern to process. Ecoscience 2005, 12, 302–315. [Google Scholar] [CrossRef]
- Tiébré, M.-S.; Saad, L.; Mahy, G. Landscape dynamics and habitat selection by the alien invasive Fallopia (Polygonaceae) in Belgium. Biodivers. Conserv. 2008, 17, 2357–2370. [Google Scholar] [CrossRef]
- Lass, L.W.; Prather, T.S.; Glenn, N.F.; Weber, K.T.; Mundt, J.T.; Pettingill, J. A review of remote sensing of invasive weeds and example of the early detection of spotted knapweed (Centaurea maculosa) and babysbreath (Gypsophila paniculata) with a hyperspectral sensor. Weed Sci. 2005, 53, 242–251. [Google Scholar] [CrossRef]
- Mulla, D.J. Twenty five years of remote sensing in precision agriculture: Key advances and remaining knowledge gaps. Biosyst. Eng. 2013, 114, 358–371. [Google Scholar] [CrossRef]
- Müllerová, J.; Pergl, J.; Pyšek, P. Remote sensing as a tool for monitoring plant invasions: Testing the effects of data resolution and image classification approach on the detection of a model plant species Heracleum mantegazzianum (giant hogweed). Int. J. Appl. Earth Obs. Geoinf. 2013, 25, 55–65. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Aguiar, F.C.; Silva, J.M.; Ferreira, M.T.; Pereira, J.M. Optimal attributes for the object based detection of giant reed in riparian habitats: A comparative study between airborne high spatial resolution and WorldView-2 imagery. Int. J. Appl. Earth Obs. Geoinf. 2014, 32, 79–91. [Google Scholar] [CrossRef]
- Guo, Y.; Graves, S.; Flory, S.L.; Bohlman, S. Hyperspectral measurement of seasonal variation in the coverage and impacts of an invasive grass in an experimental setting. Remote Sens. 2018, 10, 784. [Google Scholar] [CrossRef]
- Walsh, S.J.; McCleary, A.L.; Mena, C.F.; Shao, Y.; Tuttle, J.P.; González, A.; Atkinson, R. Quickbird and Hyperion data analysis of an invasive plant species in the Galapagos Islands of Ecuador: Implications for control and land use management. Remote Sens. Environ. 2008, 112, 1927–1941. [Google Scholar] [CrossRef]
- Ouyang, Z.T.; Gao, Y.; Xie, X.; Guo, H.Q.; Zhang, T.T.; Zhao, B. Spectral discrimination of the invasive plant Spartina alterniflora at multiple phenological stages in a saltmarsh wetland. PLoS ONE 2013, 8, e67315. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.T.; Zhang, M.Q.; Xie, X.; Shen, Q.; Guo, H.Q.; Zhao, B. A comparison of pixel-based and object-oriented approaches to VHR imagery for mapping saltmarsh plants. Ecol. Inf. 2011, 6, 136–146. [Google Scholar] [CrossRef]
- Hill, D.J.; Tarasoff, C.; Whitworth, G.E.; Baron, J.; Bradshaw, J.L.; Church, J.S. Utility of unmanned aerial vehicles for mapping invasive plant species: A case study on yellow flag iris (Iris pseudacorus L.). Int. J. Remote Sens. 2017, 38, 2083–2105. [Google Scholar] [CrossRef]
- Zaman, B.; Jensen, A.M.; McKee, M. Use of high-resolution multispectral imagery acquired with an autonomous Unmanned Aerial Vehicle to quantify the spread of an invasive wetlands species. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Vancouver, BC, Canada, 24–29 July 2011; pp. 803–806. [Google Scholar]
- Wan, H.W.; Wang, Q.; Jiang, D.; Fu, J.Y.; Yang, Y.P.; Liu, X.M. Monitoring the invasion of Spartina alterniflora using very high resolution unmanned aerial vehicle imagery in Beihai, Guangxi (China). Sci. World J. 2014, 2014, 638296. [Google Scholar] [CrossRef] [PubMed]
- Dorigo, W.; Lucieer, A.; Podobnikar, T.; Carni, A. Mapping invasive Fallopia japonica by combined spectral, spatial, and temporal analysis of digital orthophotos. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 185–195. [Google Scholar] [CrossRef]
- Jones, D.; Pike, S.; Thomas, M.; Murphy, D. Object-based image analysis for detection of Japanese knotweed s.l. Taxa (Polygonaceae) in wales (UK). Remote Sens. 2011, 3, 319–342. [Google Scholar] [CrossRef]
- Müllerová, J.; Brůna, J.; Bartaloš, T.; Dvořák, P.; Vítková, M.; Pyšek, P. Timing is important: Unmanned aircraft vs. satellite imagery in plant invasion monitoring. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar]
- Michez, A.; Piégay, H.; Jonathan, L.; Claessens, H.; Lejeune, P. Mapping of riparian invasive species with supervised classification of unmanned aerial system (UAS) imagery. Int. J. Appl. Earth Obs. Geoinf. 2016, 44, 88–94. [Google Scholar] [CrossRef]
- Casady, G.M.; Hanley, R.S.; Seelan, S.K. Detection of leafy spurge (Euphorbia esula) using multidate high-resolution satellite imagery. Weed Technol. 2005, 19, 462–467. [Google Scholar] [CrossRef]
- Lisein, J.; Pierrot-Deseilligny, M.; Bonnet, S.; Lejeune, P. A photogrammetric workflow for the creation of a forest canopy height model from small unmanned aerial system imagery. Forests 2013, 4, 922–944. [Google Scholar] [CrossRef]
- Whitehead, K.; Hugenholtz, C.H. Remote sensing of the environment with small unmanned aircraft systems (UASs), part 1: A review of progress and challenges. J. Unmanned Veh. Syst. 2014, 2, 69–85. [Google Scholar] [CrossRef]
- Turner, D.; Lucieer, A.; Watson, C. An automated technique for generating georectified mosaics from ultra-high resolution unmanned aerial vehicle (UAV) imagery, based on structure from motion (SfM) point clouds. Remote Sens. 2012, 4, 1392–1410. [Google Scholar] [CrossRef]
- Agisoft Photoscan User Manual: Professional Edition, Version 1.2. Available online: www.agisoft.com/downloads/user-manuals/ (accessed on 14 March 2018).
- Remondino, F.; Spera, M.G.; Nocerino, E.; Menna, F.; Nex, F. State of the art in high density image matching. Photogramm. Rec. 2014, 29, 144–166. [Google Scholar] [CrossRef]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef]
- Benz, U.C.; Hofmann, P.; Willhauck, G.; Lingenfelder, I.; Heynen, M. Multi-resolution, object-oriented fuzzy analysis of remote sensing data for GIS-ready information. ISPRS J. Photogramm. Remote Sens. 2004, 58, 239–258. [Google Scholar] [CrossRef] [Green Version]
- Baatz, M.; Schäpe, A. Multiresolution segmentation: An optimization approach for high quality multi-scale image segmentation. In Angewandte Geographische Informations-Verarbeitung XII; Strobl, J., Blaschke, T., Griesebner, G., Eds.; Herbert Wichmann Verlag: Berlin, Germany, 2000; Volume 58, pp. 12–23. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Trimble eCognition Developer v.8.9.1. Available online: www.ecognition.com (accessed on 1 April 2018).
- Laliberte, A.S.; Herrick, J.E.; Rango, A.; Winters, C. Acquisition, orthorectification, and object-based classification of unmanned aerial vehicle (UAV) imagery for rangeland monitoring. Photogramm. Eng. Remote Sens. 2010, 76, 661–672. [Google Scholar] [CrossRef]
- Congalton, R.G.; Green, K. Assessing the Accuracy of Remotely Sensed Data: Principles and Practices; CRC Press: Boca Raton, FL, USA, 2009; 210p, ISBN 978-1-4200-5512-2. [Google Scholar]
- Foody, G.M. Status of land cover classification accuracy assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Stehman, S.V.; Czaplewski, R.L. Design and analysis for thematic map accuracy assessment: Fundamental principles. Remote Sens. Environ. 1998, 64, 331–344. [Google Scholar] [CrossRef]
- Pontius Jr, R.G.; Millones, M. Death to kappa: Birth of quantity disagreement and allocation disagreement for accuracy assessment. Int. J. Remote Sens. 2011, 32, 4407–4429. [Google Scholar] [CrossRef]
- Foody, G.M. Harshness in image classification accuracy assessment. Int. J. Remote Sens. 2008, 29, 3137–3158. [Google Scholar] [CrossRef] [Green Version]
- Theiler, J. Sensitivity of anomalous change detection to small misregistration errors. In Proceedings of the Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery XIV, Orlando, FL, USA, 2 May 2008; Volume 6966, p. 6966OX. [Google Scholar]
- ESRI Arcgis 10.3. Available online: www.arcgis.com (accessed on 22 November 2017).
- Velnajovski, T.; Đurić, N.; Kanjir, U.; Oštir, K. Sub-object examination aimed at improving detection and distinction of objects with similar attribute characteristics. In Proceedings of the 4th GEOBIA Conference, Rio de Janeiro, Brazil, 7–9 May 2008; pp. 543–550. [Google Scholar]
- Laba, M.; Downs, R.; Smith, S.; Welsh, S.; Neider, C.; White, S.; Richmond, M.; Philpot, W.; Baveye, P. Mapping invasive wetland plants in the Hudson river national estuarine research reserve using QuickBird satellite imagery. Remote Sens. Environ. 2008, 112, 286–300. [Google Scholar] [CrossRef]
- Smith, J.H.; Stehman, S.V.; Wickham, J.D.; Yang, L. Effects of landscape characteristics on land-cover class accuracy. Remote Sens. Environ. 2003, 84, 342–349. [Google Scholar] [CrossRef]
- Dommanget, F.; Cavaillé, P.; Evette, A.; Martin, F.M. Asian knotweeds—An example of a raising theat. In Introduced Tree Species in European Forests: Opportunities and Challenges; Krumm, F., Vítková, L., Eds.; European Forest Institute: Freiburg, Germany, 2016; Volume 1, pp. 202–211. ISBN 978-952-5980-32-5. [Google Scholar]
- Hantson, W.; Kooistra, L.; Slim, P.A. Mapping invasive woody species in coastal dunes in the Netherlands: A remote sensing approach using LiDAR and high-resolution aerial photographs. Appl. Veg. Sci. 2012, 15, 536–547. [Google Scholar] [CrossRef]
- Hartfield, K.A.; Landau, K.I.; Van Leeuwen, W.J. Fusion of high resolution aerial multispectral and LiDAR data: Land cover in the context of urban mosquito habitat. Remote Sens. 2011, 3, 2364–2383. [Google Scholar] [CrossRef]
- Gaulton, R.; Malthus, T.J. LiDAR mapping of canopy gaps in continuous cover forests: A comparison of canopy height model and point cloud based techniques. Int. J. Remote Sens. 2010, 31, 1193–1211. [Google Scholar] [CrossRef]
- Aasen, H.; Burkart, A.; Bolten, A.; Bareth, G. Generating 3d hyperspectral information with lightweight UAV snapshot cameras for vegetation monitoring: From camera calibration to quality assurance. ISPRS J. Photogramm. Remote Sens. 2015, 108, 245–259. [Google Scholar] [CrossRef]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; João Sousa, J. Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef]

| Site Name | Latitude | Longitude | Season | Image Acquisition Date | Area of the Pleiades Study Site | Area of the UAV Study Site | |
|---|---|---|---|---|---|---|---|
| Pleiades | UAV | ||||||
| Anse | 45.936 | 4.722 | Spring | 19 April 2016 | 26 May 2016 | 213 ha | 4.8 ha |
| Summer | 18 July 2016 | Crashed | |||||
| Fall | 3 October 2016 | 22 September 2016 | |||||
| Serrières | 45.319 | 4.763 | Spring | 6 April 2016 | 25 May 2016 | 263 ha | 7.1 ha |
| Summer | 18 July 2016 | Crashed | |||||
| Fall | 29 September 2016 | 5 October 2016 | |||||
| Classification Name | Image Being Classified | Data Used to Derive “Additional Variable” | Type of “Additional Variable” | |
|---|---|---|---|---|
| Pleiades imagery | ||||
| Summer-alone | Summer | - | ||
| Summer-spring | Summer | + | Spring | MBTBR |
| Summer-fall | Summer | + | Fall | MBTBR |
| Summer-all-dates | Summer | + | Spring + Fall | MBTBR |
| Fall-alone | Fall | - | ||
| Fall-spring | Fall | + | Spring | MBTBR |
| Fall-summer | Fall | + | Summer | MBTBR |
| Fall-all-dates | Fall | + | Spring + Summer | MBTBR |
| UAV imagery | ||||
| Spring-alone | Spring | - | ||
| Spring-phenology | Spring | + | Fall | MBTBR |
| Spring-CHM | Spring | + | Spring CHM | CHM |
| Spring-biCHM | Spring | + | Spring CHM + Fall CHM | CHM |
| Spring-all-dates | Spring | + | Spring CHM + Fall + Fall CHM | Both |
| Fall-alone | Fall | - | ||
| Fall-phenology | Fall | + | Spring | MBTBR |
| Fall-CHM | Fall | + | Fall CHM | CHM |
| Fall-biCHM | Fall | + | Fall CHM + Spring CHM | CHM |
| Fall-all-dates | Fall | + | Fall CHM + Spring + Spring CHM | Both |
| Image Type | Site | Classification Name | Crisp Boundary Results | Buffer Boundary Results | ||
|---|---|---|---|---|---|---|
| PA (%) | UA (%) | 2-pixels PA (%) | 10-pixels PA (%) | |||
| Satellite (Pléiades) | Anse | Summer-alone | 59 | 28 | 75 | 88 |
| Summer-sping | 55 | 28 | 71 | 86 | ||
| Summer-fall | 58 | 31 | 74 | 87 | ||
| Summer-all-dates | 56 | 35 | 72 | 87 | ||
| Fall-alone | 50 | 25 | 64 | 81 | ||
| Fall-spring | 50 | 25 | 64 | 81 | ||
| Fall-summer | 61 | 34 | 77 | 90 | ||
| Fall-all-dates | 58 | 33 | 74 | 88 | ||
| UAV | Anse | Spring-alone | 49 | 56 | 62 | 84 |
| Spring-phenology | 57 | 47 | 70 | 84 | ||
| Spring-CHM | 68 | 48 | 81 | 89 | ||
| Spring-biCHM | 72 | 53 | 84 | 95 | ||
| Spring-all-dates | 69 | 50 | 82 | 93 | ||
| Fall-alone | 46 | 34 | 69 | 92 | ||
| Fall-phenology | 50 | 42 | 68 | 88 | ||
| Fall-CHM | 68 | 37 | 80 | 93 | ||
| Fall-biCHM | 49 | 21 | 79 | 99 | ||
| Fall-all-dates | 69 | 48 | 81 | 94 | ||
| UAV | Serrières | Spring-alone | 82 | 48 | 91 | 98 |
| Spring-phenology | 81 | 51 | 90 | 98 | ||
| Spring-CHM | 84 | 72 | 92 | 99 | ||
| Spring-biCHM | 83 | 80 | 91 | 98 | ||
| Spring-all-dates | 86 | 78 | 93 | 99 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, F.-M.; Müllerová, J.; Borgniet, L.; Dommanget, F.; Breton, V.; Evette, A. Using Single- and Multi-Date UAV and Satellite Imagery to Accurately Monitor Invasive Knotweed Species. Remote Sens. 2018, 10, 1662. https://doi.org/10.3390/rs10101662
Martin F-M, Müllerová J, Borgniet L, Dommanget F, Breton V, Evette A. Using Single- and Multi-Date UAV and Satellite Imagery to Accurately Monitor Invasive Knotweed Species. Remote Sensing. 2018; 10(10):1662. https://doi.org/10.3390/rs10101662
Chicago/Turabian StyleMartin, François-Marie, Jana Müllerová, Laurent Borgniet, Fanny Dommanget, Vincent Breton, and André Evette. 2018. "Using Single- and Multi-Date UAV and Satellite Imagery to Accurately Monitor Invasive Knotweed Species" Remote Sensing 10, no. 10: 1662. https://doi.org/10.3390/rs10101662