Anti-Virulence Activity of 3,3′-Diindolylmethane (DIM): A Bioactive Cruciferous Phytochemical with Accelerated Wound Healing Benefits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Growth Curves Profile
2.3. Biofilm Growth in Static Culture
2.4. Biofilm Growth in Dynamic Culture
2.5. Antimicrobial Susceptibility Tests
2.6. Biofilm Visualization
2.7. Qualitative Congo Red Plate Assay of EPS Production
2.8. Qualitative Assay of Swarming Motility
2.9. Quantization of Extracellular Virulence Factor Production by P. aeruginosa PA01
2.10. Bioluminescence Assay for Anti-QS Activity
2.11. RNA Extraction, RNA-Seq, and Transcriptomic Analysis
2.12. Reverse Transcription and Real-Time PCR
2.13. Formulation of DIM-Based Cream
2.14. Full-Thickness P. aeruginosa-Infected Porcine Excision Wound Model
2.15. Quantitative Bacteriology
3. Results
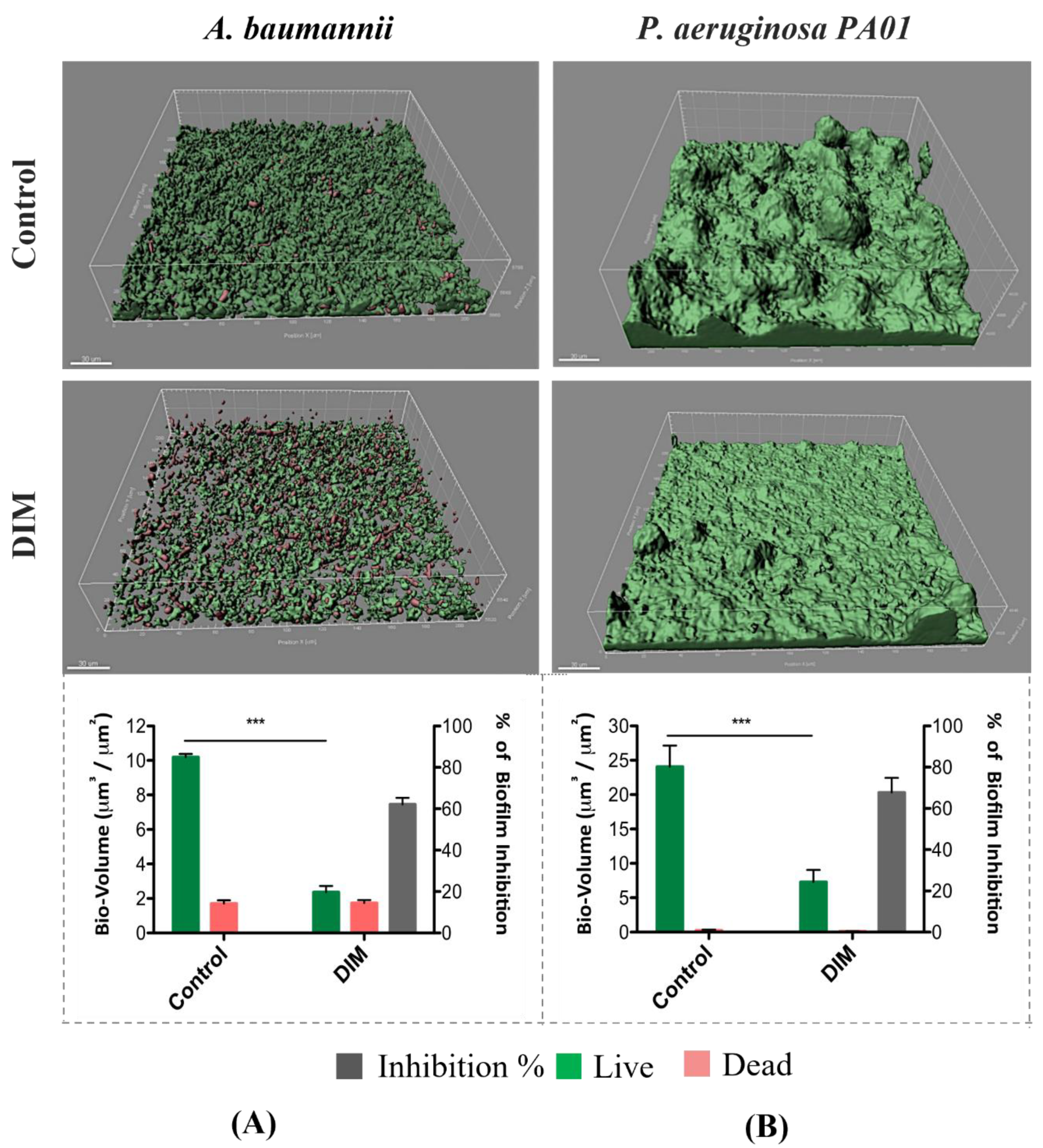
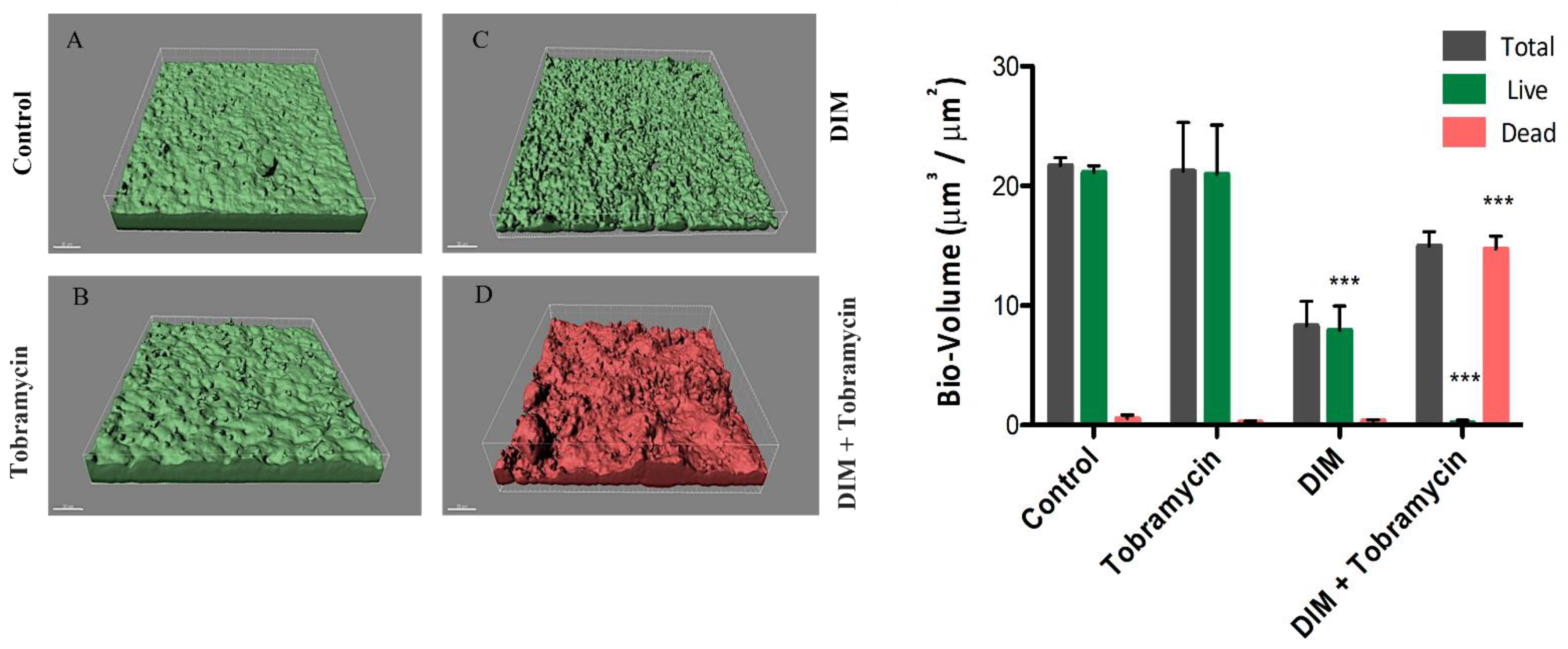
3.1. Assessing Biofilm Formation of Model Bacteria
3.2. Virulence Factors Attenuation
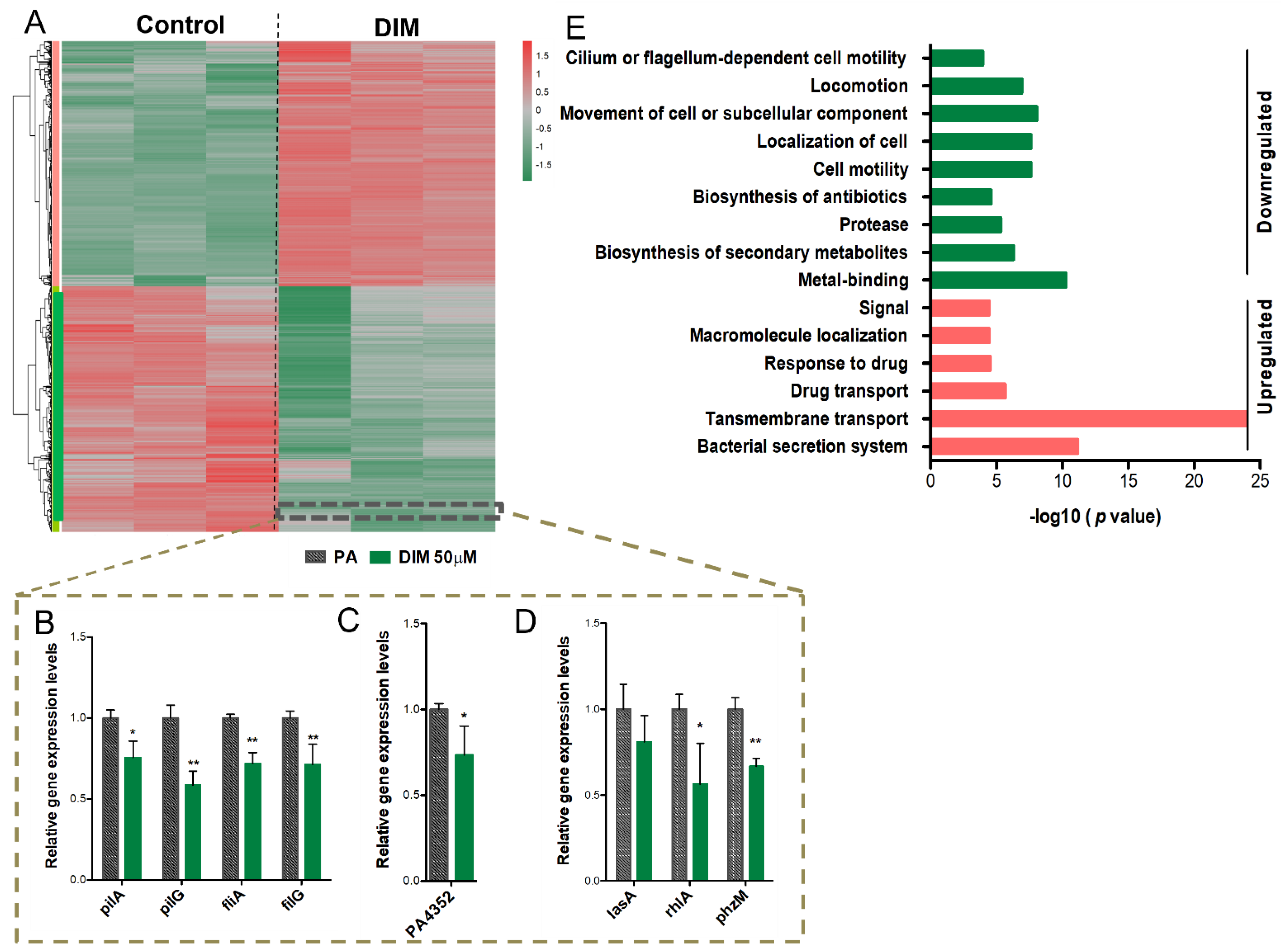
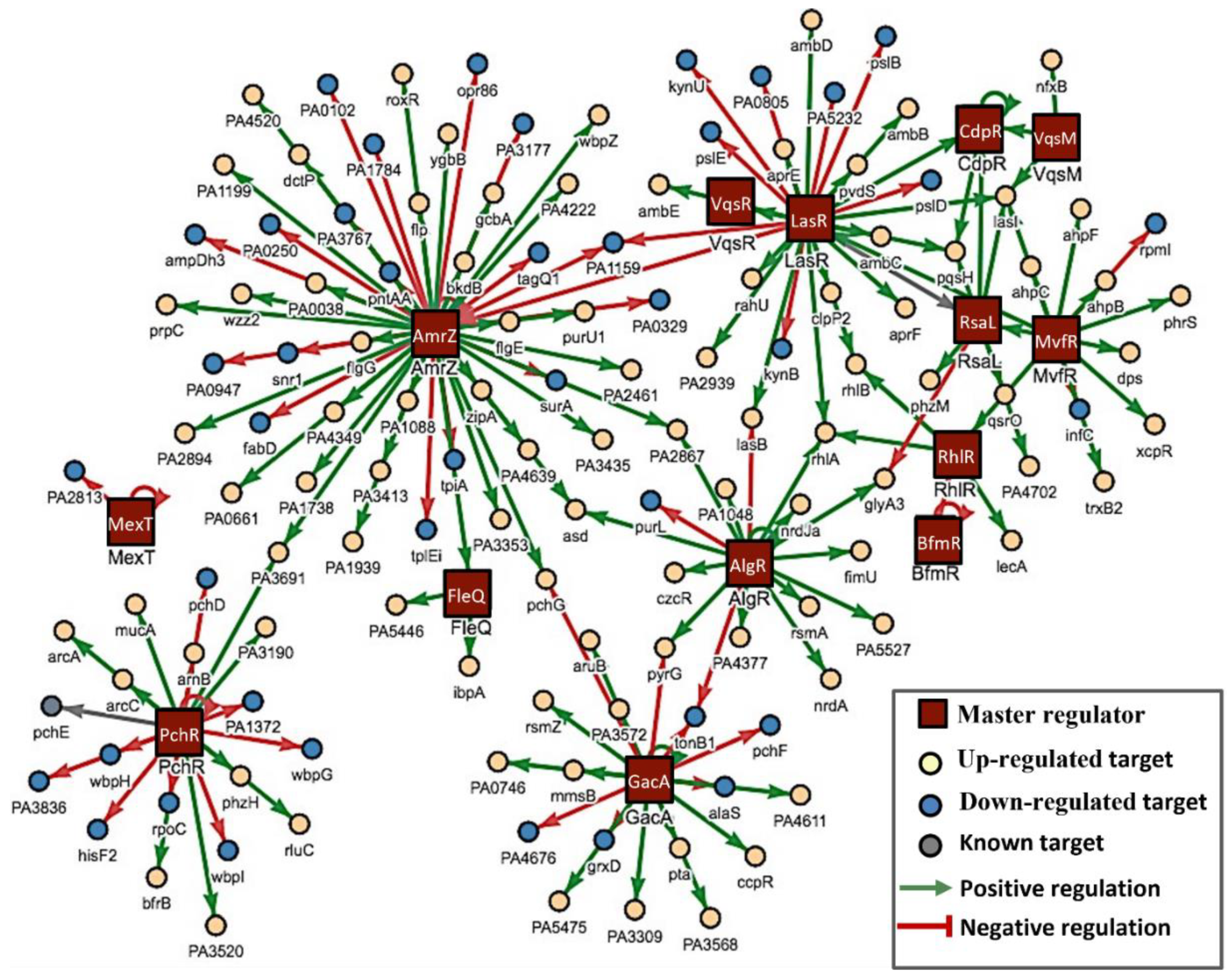
3.3. Mechanisms Underlying the Anti-Pathogenic Activity
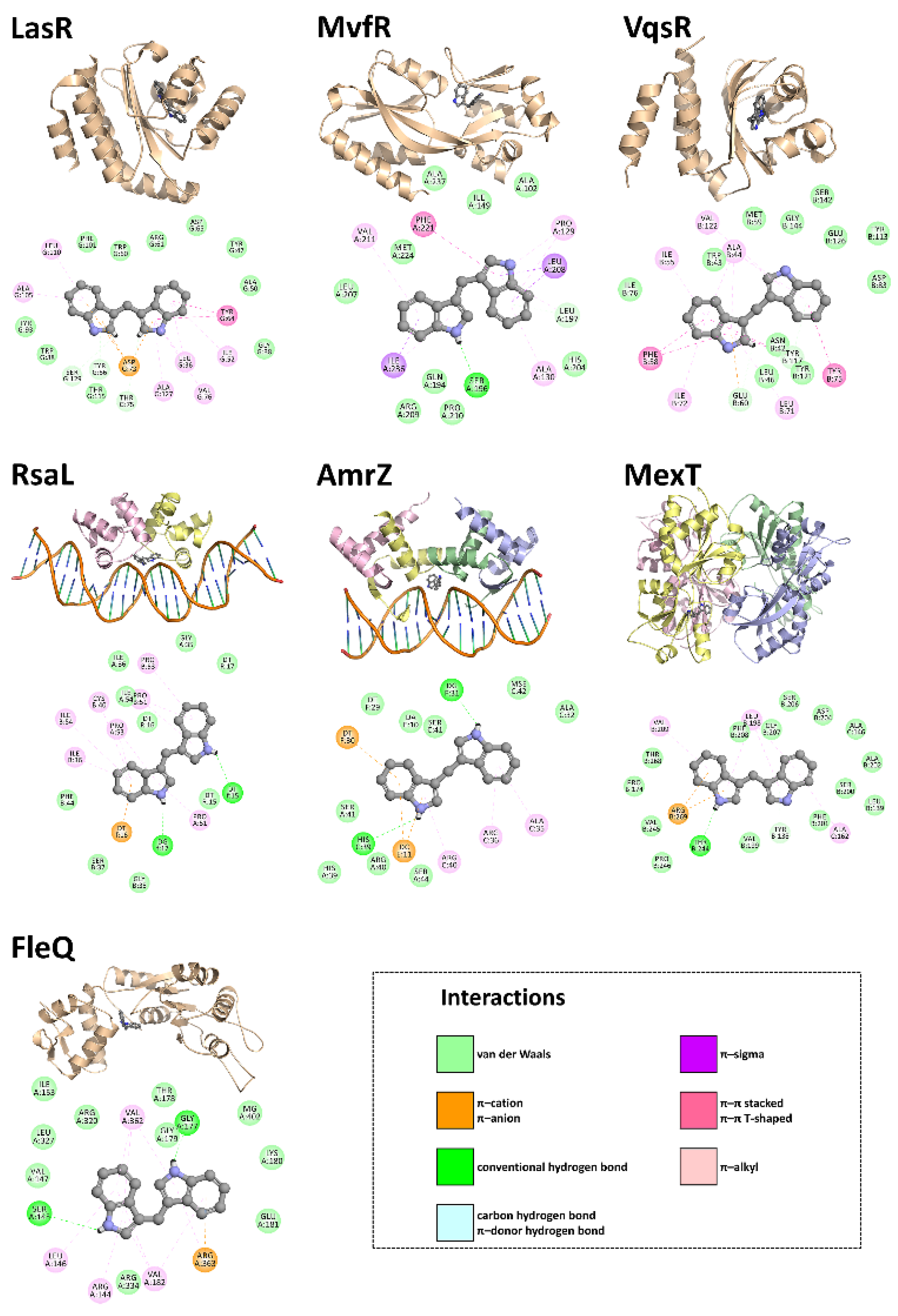
3.4. In Silico Studies
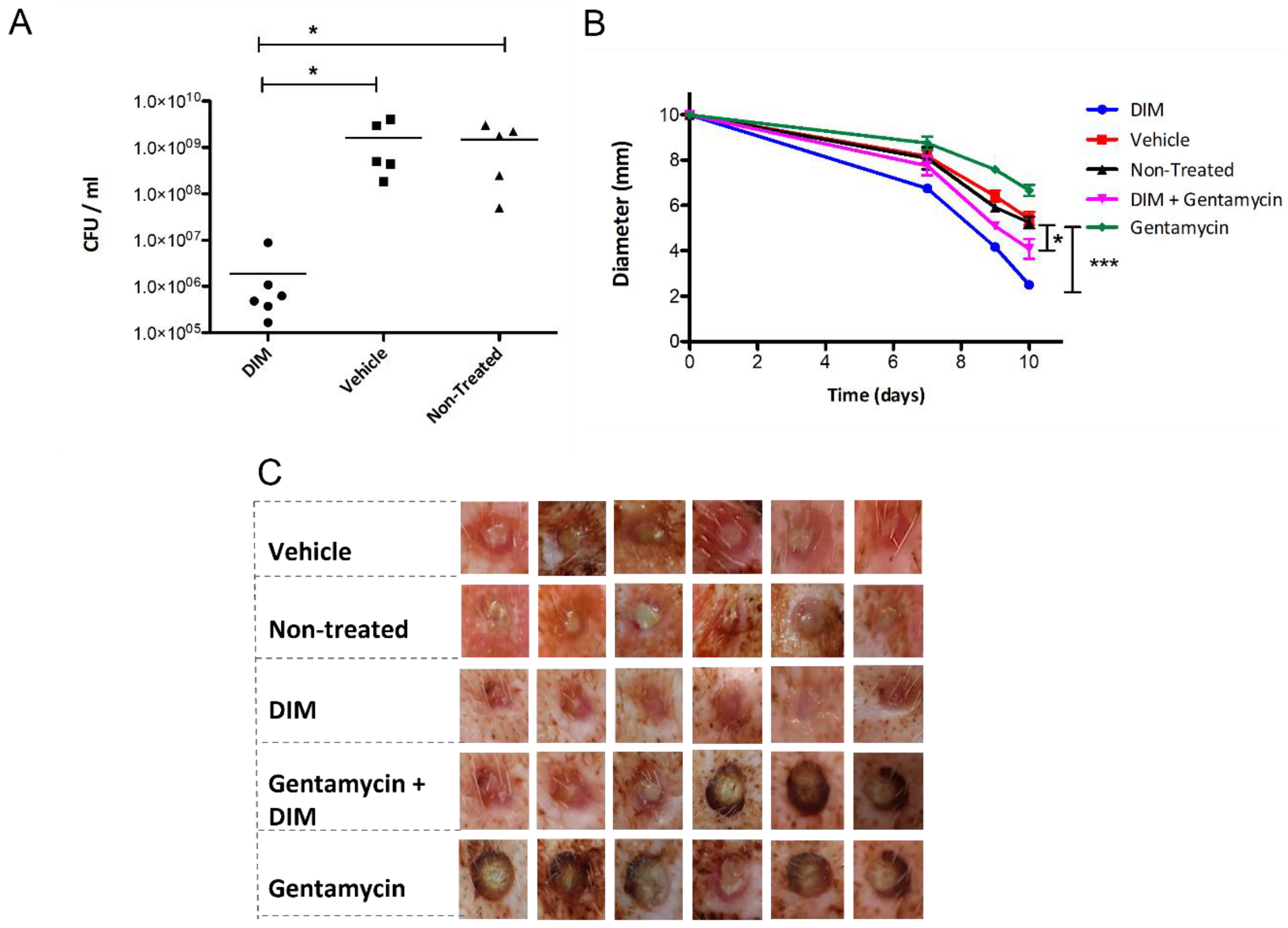
3.5. Dermal Excisional Wound Healing in a Porcine Model Following DIM Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calfee, D.P. Crisis in hospital-acquired, healthcare-associated infections. Annu. Rev. Med. 2012, 63, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Fried, E.D. Hospital-acquired infections. In Patient Safety: A Case-Based Comprehensive Guide; Agrawal, A., Ed.; Springer: New York, NY, USA, 2014; pp. 179–195. ISBN 978-1-4614-7419-7. [Google Scholar]
- Pérez-Rodríguez, F.; Mercanoglu Taban, B. A state-of-art review on multi-drug resistant pathogens in foods of animal origin: Risk factors and mitigation strategies. Front. Microbiol. 2019, 10, 2091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, R.N. Important and emerging β-lactamase-mediated resistances in hospital-based pathogens: The Amp C enzymes. Diagn. Microbiol. Infect. Dis. 1998, 31, 461–466. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef]
- Tello, A.; Austin, B.; Telfer, T.C. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ. Health Perspect. 2012, 120, 1100–1106. [Google Scholar] [CrossRef]
- Xiong, W.; Sun, Y.; Ding, X.; Wang, M.; Zeng, Z. Selective pressure of antibiotics on ARGs and bacterial communities in manure-polluted freshwater-sediment microcosms. Front. Microbiol. 2015, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wen, Y.M. The role of bacterial biofilm in persistent infections and control strategies. Int. J. Oral Sci. 2011, 3, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Monroe, D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol. 2007, 5, 2458–2461. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.-C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “House of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, N.A.; Barnard, A.M.L.; Slater, H.; Simpson, N.J.L.; Salmond, G.P.C. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 2001, 25, 365–404. [Google Scholar] [CrossRef] [PubMed]
- Vlamakis, H. The world of biofilms. Virulence 2011, 2, 431–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, S.; Parasramka, M.A.; Sarkar, F.H. Cellular, molecular and biological insight into chemopreventive and therapeutic potential of 3,3′-diindolylmethane (DIM). In Nutraceuticals and Cancer; Sarkar, F.H., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 111–133. ISBN 978-94-007-2630-7. [Google Scholar]
- Thomson, C.A.; Ho, E.; Strom, M.B. Chemopreventive properties of 3,30-diindolylmethane in breast cancer: Evidence from experimental and human studies. Nutr. Rev. 2016, 74, 432–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barba, F.J.; Nikmaram, N.; Roohinejad, S.; Khelfa, A.; Zhu, Z.; Koubaa, M. Bioavailability of glucosinolates and their breakdown products: Impact of processing. Front. Nutr. 2016, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Holst, B.; Williamson, G. A critical review of the bioavailability of glucosinolates and related compounds. Nat. Prod. Rep. 2004, 21, 425–447. [Google Scholar] [CrossRef]
- Wang, S.; Cheng, L.; Liu, Y.; Wang, J.; Jiang, W. Indole-3-carbinol (I3C) and its major derivatives: Their pharmacokinetics and important roles in hepatic protection. Curr. Drug Metab. 2016, 17, 401–409. [Google Scholar] [CrossRef]
- Weng, J.R.; Tsai, C.H.; Kulp, S.K.; Chen, C.S. Indole-3-carbinol as a chemopreventive and anti-cancer agent. Cancer Lett. 2008, 262, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Higdon, J.V.; Delage, B.; Williams, D.E.; Dashwood, R.H. Cruciferous vegetables and human cancer risk: Epidemiologic evidence and mechanistic basis. Pharmacol. Res. 2007, 55, 224–236. [Google Scholar] [CrossRef] [Green Version]
- Bharate, S.B.; Bharate, J.B.; Khan, S.I.; Tekwani, B.L.; Jacob, M.R.; Mudududdla, R.; Yadav, R.R.; Singh, B.; Sharma, P.R.; Maity, S.; et al. Discovery of 3,3′-diindolylmethanes as potent antileishmanial agents. Eur. J. Med. Chem. 2013, 63, 435–443. [Google Scholar] [CrossRef] [Green Version]
- Han, J.C.; Mi, R.S.; Yeo, M.L.; Kim, J.; Kim, J.K.; Sang, G.K.; Park, J.H.Y. 3,3′-diindolylmethane suppresses the inflammatory response to lipopolysaccharide in murine macrophages. J. Nutr. 2008, 138, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Xia, S.; Gao, F.; Zhang, D.; Chen, J.; Zhang, J. 3,3′-diindolylmethane attenuates experimental arthritis and osteoclastogenesis. Biochem. Pharmacol. 2010, 79, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, C.; Christensen, B.B.; Johansen, T.; Nielsen, A.T.; Andersen, J.B.; Givskov, M.; Molin, S. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 1999, 65, 4108–4117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarabhai, S.; Sharma, P.; Capalash, N. Ellagic acid derivatives from Terminalia chebula Retz. Downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Solleti, V.S.; Alhariri, M.; Halwani, M.; Omri, A. Antimicrobial properties of liposomal azithromycin for Pseudomonas infections in cystic fibrosis patients. J. Antimicrob. Chemother. 2015, 70, 784–796. [Google Scholar] [CrossRef] [Green Version]
- Jakobsen, T.H.; van Gennip, M.; Phipps, R.K.; Shanmugham, M.S.; Christensen, L.D.; Alhede, M.; Skindersoe, M.E.; Rasmussen, T.B.; Friedrich, K.; Uthe, F.; et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012, 56, 2314–2325. [Google Scholar] [CrossRef] [Green Version]
- Guimarães, P.P.G.; de Menezes, A.C.; Teixeira, K.I.R.; Denadai, Â.M.L.; Fills, R.A.; Cortés, M.E.; Sinisterra, R.D. Enhanced efficacy against bacterial biofilms via host: Guest cyclodextrin-doxycycline inclusion complexes. J. Incl. Phenom. Macrocycl. Chem. 2021, 99, 197–207. [Google Scholar] [CrossRef]
- Pamp, S.J.; Sternberg, C.; Tolker-Nielsen, T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytom. Part A 2009, 75, 90–103. [Google Scholar] [CrossRef]
- Taccetti, G.; Francalanci, M.; Pizzamiglio, G.; Messore, B.; Carnovale, V.; Cimino, G.; Cipolli, M. Cystic fibrosis: Recent insights into inhaled antibiotic treatment and future perspectives. Antibiotics 2021, 10, 338. [Google Scholar] [CrossRef]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Van Delden, C.; Iglewski, B.H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 1998, 4, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Frederiksen, R.F.; Paspaliari, D.K.; Larsen, T.; Storgaard, B.G.; Larsen, M.H.; Ingmer, H.; Palcic, M.M.; Leisner, J.J. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology 2013, 159, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, P.; Vallet-Gely, I.; Soscia, C.; Genin, S.; Filloux, A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 2005, 151, 985–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, R.D.; Roddam, L.F.; Bettiol, S.; Sanderson, K.; Reid, D.W. Biosignificance of bacterial cyanogenesis in the CF lung. J. Cyst. Fibros. 2010, 9, 158–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilaplana, L.; Marco, M.P. Phenazines as potential biomarkers of Pseudomonas aeruginosa infections: Synthesis regulation, pathogenesis and analytical methods for their detection. Anal. Bioanal. Chem. 2020, 412, 5897–5912. [Google Scholar] [CrossRef]
- Minandri, F.; Imperi, F.; Frangipani, E.; Bonchi, C.; Visaggio, D.; Facchini, M.; Pasquali, P.; Bragonzi, A.; Visca, P. Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect. Immun. 2016, 84, 2324–2335. [Google Scholar] [CrossRef] [Green Version]
- Boes, N.; Schreiber, K.; Härtig, E.; Jaensch, L.; Schobert, M. The Pseudomonas aeruginosa universal stress protein PA4352 is essential for surviving anaerobic energy stress. J. Bacteriol. 2006, 188, 6529–6538. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Shao, X.; Xie, Y.; Wang, T.; Zhang, Y.; Wang, X.; Deng, X. An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Schellhammer, I.; Rarey, M. TrixX: Structure-based molecule indexing for large-scale virtual screening in sublinear time. J. Comput.-Aided Mol. Des. 2007, 21, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Henzler, A.M.; Urbaczek, S.; Hilbig, M.; Rarey, M. An integrated approach to knowledge-driven structure-based virtual screening. J. Comput.-Aided Mol. Des. 2014, 28, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Flachsenberg, F.; Meyder, A.; Sommer, K.; Penner, P.; Rarey, M. A consistent scheme for gradient-based optimization of protein–ligand poses. J. Chem. Inf. Model. 2020, 60, 6502–6522. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J.; Muraglia, E.; Bazzo, R.; Carfì, A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. [Google Scholar] [CrossRef] [Green Version]
- Zender, M.; Witzgall, F.; Kiefer, A.; Kirsch, B.; Maurer, C.K.; Kany, A.M.; Xu, N.; Schmelz, S.; Börger, C.; Blankenfeldt, W.; et al. Flexible fragment growing boosts potency of quorum-sensing inhibitors against Pseudomonas aeruginosa virulence. ChemMedChem 2020, 15, 188–194. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Wang, K.; Su, T.; Wang, F.; Gu, L.; Xu, S. Crystal structure of the N-terminal domain of VqsR from Pseudomonas aeruginosa at 2.1 Å resolution. Acta Crystallogr. Sect. Struct. Biol. Commun. 2017, 73, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Volkamer, A.; Griewel, A.; Grombacher, T.; Rarey, M. Analyzing the topology of active sites: On the prediction of pockets and subpockets. J. Chem. Inf. Model. 2010, 50, 2041–2052. [Google Scholar] [CrossRef]
- Kang, H.; Gan, J.; Zhao, J.; Kong, W.; Zhang, J.; Zhu, M.; Li, F.; Song, Y.; Qin, J.; Liang, H. Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing. Nucleic Acids Res. 2017, 45, 699–710. [Google Scholar] [CrossRef] [Green Version]
- Pryor, E.E.; Waligora, E.A.; Xu, B.; Dellos-Nolan, S.; Wozniak, D.J.; Hollis, T. The transcription factor AmrZ utilizes multiple DNA binding modes to recognize activator and repressor sequences of Pseudomonas aeruginosa virulence genes. PLoS Pathog. 2012, 8, e1002648. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Kim, S.H.; Ahn, J.; Jo, I.; Lee, Z.W.; Choi, S.H.; Ha, N.C. Crystal structure of the regulatory domain of mext, a transcriptional activator of the MexEF-OprN efflux pump in Pseudomonas aeruginosa. Mol. Cells 2019, 42, 1–8. [Google Scholar] [CrossRef]
- Banerjee, P.; Chanchal; Jain, D. Sensor I regulated ATPase activity of FleQ is essential for motility to biofilm transition in Pseudomonas aeruginosa. ACS Chem. Biol. 2019, 14, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Kirketerp-Møller, K.; Jensen, P.Ø.; Madsen, K.G.; Phipps, R.; Krogfelt, K.; Høiby, N.; Givskov, M. Why chronic wounds will not heal: A novel hypothesis. Wound Repair Regen. 2008, 16, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sperandio, V. Indole signaling at the host-microbiota-pathogen interface. MBio 2019, 10, e01031-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melander, R.J.; Minvielle, M.J.; Melander, C. Controlling bacterial behavior with indole-containing natural products and derivatives. Tetrahedron 2014, 70, 6363–6372. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Yamasaki, R.; Song, S.; Wood, T.K. Interkingdom signal indole inhibits Pseudomonas aeruginosa persister cell waking. J. Appl. Microbiol. 2019, 127, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Wood, T.K.; Lee, J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015, 23, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Campana, R.; Favi, G.; Baffone, W.; Lucarini, S. Marine alkaloid 2,2-bis(6-bromo-3-indolyl) ethylamine and its synthetic derivatives inhibit microbial biofilms formation and disaggregate developed biofilms. Microorganisms 2019, 7, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, B.; Hu, Y.; Zhao, Y. New insights into gut-bacteria-derived indole and its derivatives in intestinal and liver diseases. Front. Pharmacol. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Busbee, P.B.; Menzel, L.; Alrafas, H.R.; Dopkins, N.; Becker, W.; Miranda, K.; Tang, C.; Chatterjee, S.; Singh, U.P.; Nagarkatti, M.; et al. Indole-3-carbinol prevents colitis and associated microbial dysbiosis in an IL-22–dependent manner. JCI Insight 2020, 5, e127551. [Google Scholar] [CrossRef] [Green Version]
- Arora, P.K.; Sharma, A.; Bae, H. Microbial degradation of indole and its derivatives. J. Chem. 2015, 2015, 129159. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, M.; Yang, L.; Pamp, S.J.; Tolker-Nielsen, T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2010, 59, 253–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mah, T.F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef]
- Alanis, A.J. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 2005, 36, 697–705. [Google Scholar] [CrossRef]
- Pantosti, A.; Moro, M.L. Antibiotic use: The crystal ball for predicting antibiotic resistance. Clin. Infect. Dis. 2005, 40, 1298–1300. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Gajbhiye, R.; Mandal, M.; Pal, C.; Meyyapan, A.; Mukherjee, J.; Jaisankar, P. Synthesis and antibacterial evaluation of 3,3′-diindolylmethane derivatives. Med. Chem. Res. 2014, 23, 1371–1377. [Google Scholar] [CrossRef]
- Merritt, J.H.; Brothers, K.M.; Kuchma, S.L.; O’Toole, G.A. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 2007, 189, 8154–8164. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Barken, K.B.; Pamp, S.J.; Yang, L.; Gjermansen, M.; Bertrand, J.J.; Klausen, M.; Givskov, M.; Whitchurch, C.B.; Engel, J.N.; Tolker-Nielsen, T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008, 10, 2331–2343. [Google Scholar] [CrossRef]
- Galloway, W.R.J.D.; Hodgkinson, J.T.; Bowden, S.; Welch, M.; Spring, D.R. Applications of small molecule activators and inhibitors of quorum sensing in gram-negative bacteria. Trends Microbiol. 2012, 20, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, S.H.; Byun, Y.; Park, H.D. 6-gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci. Rep. 2015, 5, 8656. [Google Scholar] [CrossRef]
- Taga, M.E.; Bassler, B.L. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 2003, 100, 14549–14554. [Google Scholar] [CrossRef] [Green Version]
- Adonizio, A.; Kong, K.F.; Mathee, K. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by south Florida plant extracts. Antimicrob. Agents Chemother. 2008, 52, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Duan, K.; Surette, M.G. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 2007, 189, 4827–4836. [Google Scholar] [CrossRef] [Green Version]
- Winson, M.K.; Swift, S.; Fish, L.; Throup, J.P.; Jørgensen, F.; Chhabra, S.R.; Bycroft, B.W.; Williams, P.; Stewart, G.S.A.B. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998, 163, 185–192. [Google Scholar] [CrossRef]
- Attinger, C.; Wolcott, R. Clinically addressing biofilm in chronic wounds. Adv. Wound Care 2012, 1, 127–132. [Google Scholar] [CrossRef] [Green Version]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.D.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef]
- Liu, J.; Li, T.; Zhang, H.; Zhao, W.; Qu, L.; Chen, S.; Wu, S. Materials today bio (L-lactic-acid) nanoyarns for constructing advanced nanotextile tissue scaffolds. Mater. Today Bio 2022, 14, 100243. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.F.; Gui, D.; Wong, M.; Döring, A.; Rogach, A.L.; He, T.; Wong, W.T. A self-indicating cellulose-based gel with tunable performance for bioactive agent delivery. J. Drug Deliv. Sci. Technol. 2021, 63, 102428. [Google Scholar] [CrossRef]
- Norouzi, M.; Nazari, B.; Miller, D.W. Injectable hydrogel-based drug delivery systems for local cancer therapy. Drug Discov. Today 2016, 21, 1835–1849. [Google Scholar] [CrossRef] [PubMed]
- Henderson, P.W.; Singh, S.P.; Krijgh, D.D.; Yamamoto, M.; Rafii, D.C.; Sung, J.J.; Rafii, S.; Rabbany, S.Y.; Spector, J.A. Stromal-derived factor-1 delivered via hydrogel drug-delivery vehicle accelerates wound healing in vivo. Wound Repair Regen. 2011, 19, 420–425. [Google Scholar] [CrossRef]
- Ren, Y.; Zhao, X.; Liang, X.; Ma, P.X.; Guo, B. Injectable hydrogel based on quaternized chitosan, gelatin and dopamine as localized drug delivery system to treat Parkinson’s disease. Int. J. Biol. Macromol. 2017, 105, 1079–1087. [Google Scholar] [CrossRef]
- Hamilton, M.J.; Karp, J.M.; Zhang, S.; Ermann, J.; Succi, M.D.; Zhou, A.; Hamilton, M.J.; Cao, B.; Korzenik, J.R.; Glickman, J.N.; et al. An inflammation-targeting hydrogel for local drug delivery in inflammatory bowel disease. Sci. Transl. Med. 2015, 7, 1–11. [Google Scholar]
- Yan, J.; Yao, Y.; Yan, S.; Gao, R.; Lu, W.; He, W. Chiral protein supraparticles for tumor suppression and synergistic immunotherapy: An enabling strategy for bioactive supramolecular chirality construction. Nano Lett. 2020, 20, 5844–5852. [Google Scholar] [CrossRef]
- Xiao, J. Phytochemicals in medicine and food. Phytochem. Rev. 2015, 14, 317–320. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.L.; Xu, Y.; Lu, L.; Li, Y.; Han, Z.; Zhang, J.; Shao, C.L.; Wang, C.Y.; Qian, P.Y. Low-toxicity diindol-3-ylmethanes as potent antifouling compounds. Mar. Biotechnol. 2015, 17, 624–632. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, J.; Park, S.; Lee, J. The anticancer agent 3,3′-diindolylmethane inhibits multispecies biofilm formation by acne-causing bacteria and candida albicans. Microbiol. Spectr. 2022, 10, e02056-21. [Google Scholar] [CrossRef]
- Mikkelsen, H.; Duck, Z.; Lilley, K.S.; Welch, M. Interrelationships between colonies, biofilms, and planktonic cells of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2411–2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golberg, K.; Markus, V.; Kagan, B.-e.; Barzanizan, S.; Yaniv, K.; Teralı, K.; Kramarsky-Winter, E.; Marks, R.S.; Kushmaro, A. Anti-Virulence Activity of 3,3′-Diindolylmethane (DIM): A Bioactive Cruciferous Phytochemical with Accelerated Wound Healing Benefits. Pharmaceutics 2022, 14, 967. https://doi.org/10.3390/pharmaceutics14050967
Golberg K, Markus V, Kagan B-e, Barzanizan S, Yaniv K, Teralı K, Kramarsky-Winter E, Marks RS, Kushmaro A. Anti-Virulence Activity of 3,3′-Diindolylmethane (DIM): A Bioactive Cruciferous Phytochemical with Accelerated Wound Healing Benefits. Pharmaceutics. 2022; 14(5):967. https://doi.org/10.3390/pharmaceutics14050967
Chicago/Turabian StyleGolberg, Karina, Victor Markus, Bat-el Kagan, Sigalit Barzanizan, Karin Yaniv, Kerem Teralı, Esti Kramarsky-Winter, Robert S. Marks, and Ariel Kushmaro. 2022. "Anti-Virulence Activity of 3,3′-Diindolylmethane (DIM): A Bioactive Cruciferous Phytochemical with Accelerated Wound Healing Benefits" Pharmaceutics 14, no. 5: 967. https://doi.org/10.3390/pharmaceutics14050967







