Identification of Hunnivirus in Bovine and Caprine Samples in North America
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Samples
2.3. RNA Extraction
2.4. Metagenomic Sequencing, Assembly, and Analysis
2.5. Real-Time RT-PCR Detection of Ruminant Hunnivirus and 3Dpol Gene Partial Sequencing
2.6. Data Analysis
3. Results
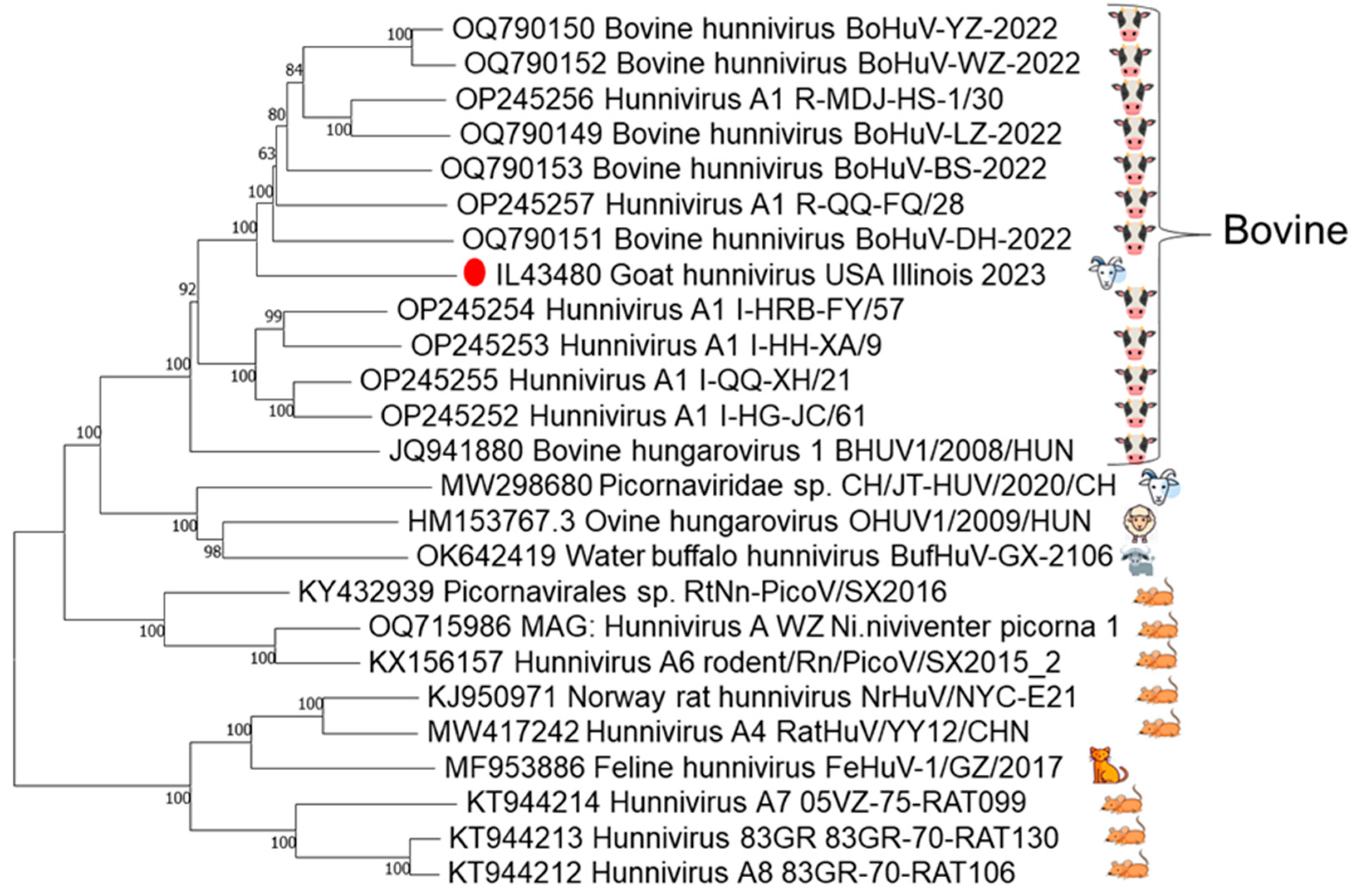
3.1. Identification of Hunnivirus with Metagenomic Sequencing
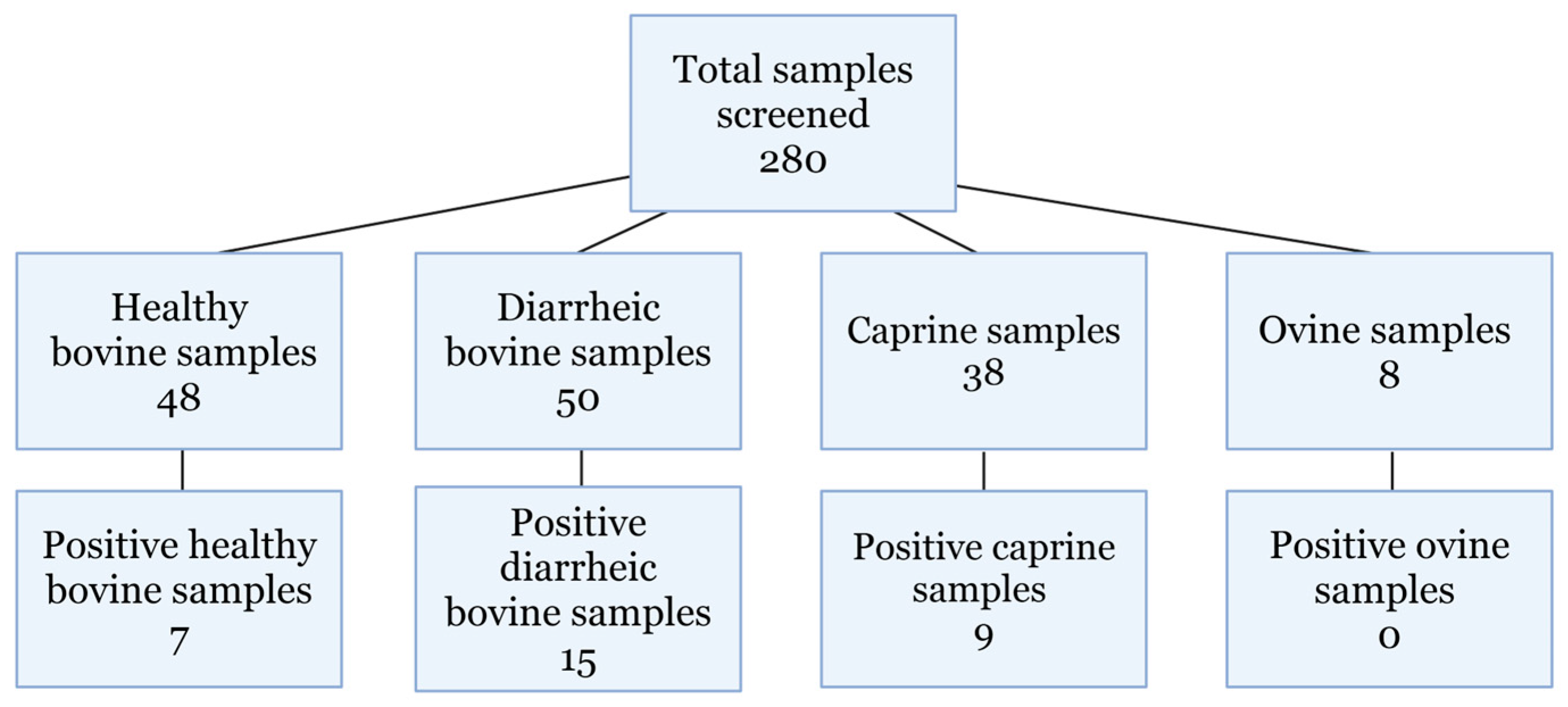
3.2. Real-Time RT-PCR Screening of Archived Samples
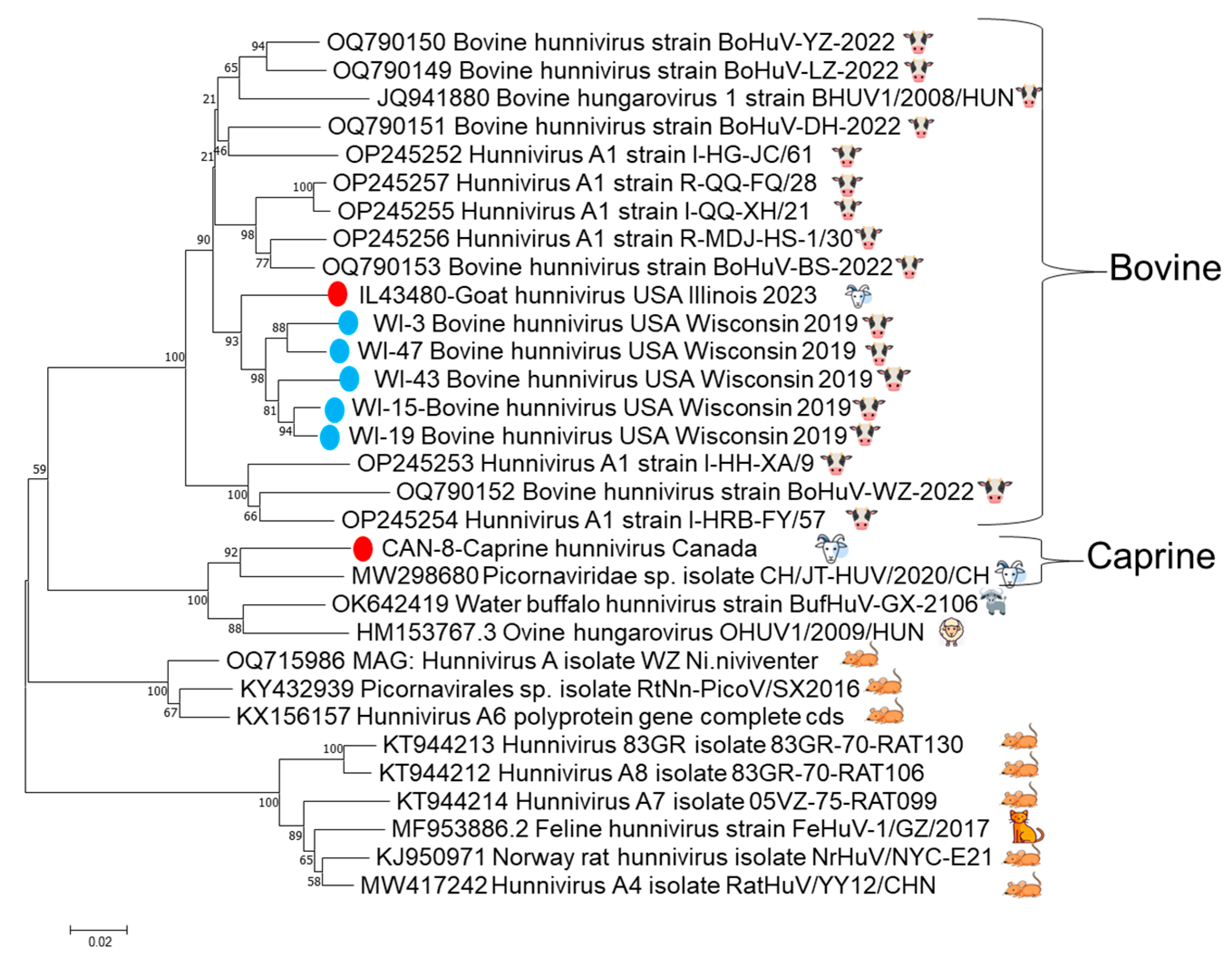
3.3. The 3Dpol Partial Genome Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zell, R.; Delwart, E.; Gorbalenya, A.E.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV Virus Taxonomy Profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef] [PubMed]
- Ehrenfeld, E.; Domingo, E.; Roos, R.P. The Picornaviruses; ASM Press: Washitington, DC, USA, 2010. [Google Scholar] [CrossRef]
- De Castro, E.; Hulo, C.; Masson, P.; Auchincloss, A.; Bridge, A.; Mercier, P. Le ViralZone 2024 Provides Higher-Resolution Images and Advanced Virus-Specific Resources. Nucleic Acids Res. 2024, 52, D817–D821. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, X.; Dong, Q.; Qiao, C.; Luo, Y.; Liu, Y.; Zou, Y.; Liu, H.; Wu, C.; Su, J.; et al. Isolation and Phylogenetic Analysis of a Hunnivirus Strain in Water Buffaloes From China. Front. Vet. Sci. 2022, 9, 851743. [Google Scholar] [CrossRef] [PubMed]
- Kerkvliet, J.; Edukulla, R.; Rodriguez, M. Novel Roles of the Picornaviral 3D Polymerase in Viral Pathogenesis. Adv. Virol. 2010, 2010, 368068. [Google Scholar] [CrossRef] [PubMed]
- Genus: Hunnivirus|ICTV. Available online: https://ictv.global/report/chapter/picornaviridae/picornaviridae/hunnivirus (accessed on 17 June 2025).
- Reuter, G.; Pankovics, P.; Knowles, N.J.; Boros, Á. Two Closely Related Novel Picornaviruses in Cattle and Sheep in Hungary from 2008 to 2009, Proposed as Members of a New Genus in the Family Picornaviridae. J. Virol. 2012, 86, 13295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, Q.; Wu, F.; Ou, Z.; Li, Y.; You, F.; Chen, Q. Epidemiology, Genetic Characterization, and Evolution of Hunnivirus Carried by Rattus Norvegicus and Rattus Tanezumi: The First Epidemiological Evidence from Southern China. Pathogens 2021, 10, 661. [Google Scholar] [CrossRef] [PubMed]
- Firth, C.; Bhat, M.; Firth, M.A.; Williams, S.H.; Frye, M.J.; Simmonds, P.; Conte, J.M.; Ng, J.; Garcia, J.; Bhuva, N.P.; et al. Detection of Zoonotic Pathogens and Characterization of Novel Viruses Carried by Commensal Rattus Norvegicus in New York City. mBio 2014, 5, e01933-14. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.Y.; Tang, F.; Zhang, M.Q.; Zhang, J.T.; Zhang, Y.F.; Si, G.Q.; Fu, B.K.; Wang, G.; Li, S.; Zhang, L.; et al. Virome Characterization of Field-Collected Rodents in Suburban City. NPJ Biofilms Microbiomes 2025, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Huang, M.; Chen, X.; Sun, Y.; Huang, J.; Hu, R.; Li, S. Identification and Genome Characterization of a Novel Feline Picornavirus Proposed in the Hunnivirus Genus. Infect. Genet. Evol. 2019, 71, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Karayel-Hacioglu, I.; Duran-Yelken, S.; Alkan, F. Molecular Detection of Picornaviruses in Diarrheic Small Ruminants at a Glance: Enterovirus, Hunnivirus, and Kobuvirus in Türkiye. Kafkas Univ. Vet. Fak. Derg. 2022, 28, 499–506. [Google Scholar] [CrossRef]
- Wang, L.; Stuber, T.; Camp, P.; Robbe-Austerman, S.; Zhang, Y. Whole-Genome Sequencing of Porcine Epidemic Diarrhea Virus by Illumina MiSeq Platform. In Animal Coronaviruses; Springer: New York, NY, USA, 2016; pp. 201–208. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Taxon Details|ICTV. Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202402012&taxon_name=Hunnivirus (accessed on 23 July 2025).
| PCR Type | Primer Name | Oligonucleotide Sequence (5′ > 3′) | Amplicon |
|---|---|---|---|
| Real-time RT-PCR | Rum-HunniV-For-1 | CAGGTGAATGAACGACTGTC | 189 nt |
| Rum-HunniV-For-2 | CCTGTGAATGAACGGCTGTC | ||
| Rum-HunniV-Rev-1 | CTTCCATCATAAGCATCAGTTC | ||
| Rum-HunniV-Rev-2 | CTTCCATCATGAGCATCAGTTC | ||
| Rum-HunniV-Probe | FAM/TGCACGAGG/ZEN/CAGTCTTTGGAAC/IABkFQ | ||
| Conventional PCR | HunniV-Pan-For | GAGGCAGTCTTTGGRACWGACAA | 803 nt |
| HunniV-Pan-Rev-2 | CCACTCTTTGAAGCTGGDGTAAT |
| Animal ID | Species | Animal Location | Hunnivirus Ct | Salmonella spp. Ct | Bovine Kobuvirus Ct | Age in Weeks * |
|---|---|---|---|---|---|---|
| 1 | Bovine | IL | 31.80 | ND | ND | Unknown |
| 2 | Bovine | WI | 0.00 | 36.80 | 20.58 | 1 |
| 3 | Bovine | WI | 0.00 | ND | ND | Unknown |
| 4 | Bovine | WI | 28.56 | 0.00 | 19.48 | 1 |
| 5 | Bovine | WI | 0.00 | 0.00 | 32.14 | 16 |
| 6 | Bovine | OH | 0.00 | 0.00 | 0.00 | 16 |
| 7 | Bovine | WI | 0.00 | 0.00 | 0.00 | 52 * |
| 8 | Bovine | WI | 0.00 | 39.00 | 0.00 | <1 |
| 9 | Bovine | WI | 0.00 | 0.00 | 0.00 | 104 * |
| 10 | Bovine | WI | 0.00 | 0.00 | 0.00 | 104 * |
| 11 | Bovine | OH | 35.73 | 0.00 | 0.00 | 5 |
| 12 | Bovine | WI | 30.10 | 0.00 | 28.00 | 32 * |
| 13 | Bovine | WI | 0.00 | 0.00 | 19.93 | 3 |
| 14 | Bovine | WI | 0.00 | 0.00 | 22.37 | 2 |
| 15 | Bovine | WI | 29.76 | 0.00 | 20.38 | 1 |
| 16 | Bovine | WI | 38.24 | 0.00 | 23.10 | 1 |
| 17 | Bovine | WI | 37.35 | 0.00 | 22.00 | 1 |
| 18 | Bovine | WI | 38.65 | 0.00 | 18.90 | 1 |
| 19 | Bovine | WI | 25.36 | 0.00 | 24.55 | 1 |
| 20 | Bovine | NM | 0.00 | 33.40 | 25.29 | 1 |
| 21 | Bovine | NM | 0.00 | 34.00 | 18.22 | 1 |
| 22 | Bovine | NM | 0.00 | 31.90 | 19.28 | 1 |
| 23 | Bovine | NM | 0.00 | 0.00 | 18.68 | 1 |
| 24 | Bovine | NM | 0.00 | 36.90 | 19.29 | 1 |
| 25 | Bovine | NM | 32.29 | 37.10 | 19.67 | 1 |
| 26 | Bovine | WI | 38.01 | 0.00 | 20.06 | Unknown |
| 27 | Bovine | WI | 0.00 | 0.00 | 21.46 | 1 |
| 28 | Bovine | WI | 0.00 | 26.50 | 0.00 | <1 |
| 29 | Bovine | OH | 33.09 | 0.00 | 36.10 | 10 |
| 30 | Bovine | WI | 0.00 | 0.00 | 24.98 | 1 |
| 31 | Bovine | WI | 0.00 | 0.00 | 29.23 | Unknown |
| 32 | Bovine | WI | 0.00 | 0.00 | 39.12 | Unknown |
| 33 | Bovine | WI | 0.00 | 29.10 | 0.00 | 208 * |
| 34 | Bovine | WI | 0.00 | 39.00 | 0.00 | 208 * |
| 35 | Bovine | WI | 0.00 | 0.00 | 0.00 | Unknown |
| 36 | Bovine | WI | 0.00 | 0.00 | 38.46 | Uunknown |
| 37 | Bovine | WI | 0.00 | 0.00 | 0.00 | Unknown |
| 38 | Bovine | WI | 0.00 | 0.00 | 0.00 | Unknown |
| 39 | Bovine | WI | 0.00 | 0.00 | 39.34 | 468 * |
| 40 | Bovine | WI | 0.00 | 0.00 | 0.00 | Unknown |
| 41 | Bovine | WI | 0.00 | 24.00 | 37.77 | 156 * |
| 42 | Bovine | WI | 0.00 | 0.00 | 39.75 | Unknown |
| 43 | Bovine | WI | 29.77 | 0.00 | 24.01 | 3 |
| 44 | Bovine | WI | 0.00 | 0.00 | 0.00 | Unknown |
| 45 | Bovine | WI | 0.00 | 30.30 | 0.00 | Unknown |
| 46 | Bovine | OH | 0.00 | 0.00 | 22.15 | 2 |
| 47 | Bovine | WI | 27.18 | 0.00 | 0.00 | Unknown |
| 48 | Bovine | WI | 0.00 | 0.00 | 0.00 | Unknown |
| 49 | Bovine | MI | 30.41 | 0.00 | 20.10 | 1 |
| 50 | Bovine | WI | 0.00 | 0.00 | 16.13 | 1 |
| 51 | Caprine | IL | 0.00 | ND | ND | 4 |
| 52 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 53 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 54 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 55 | Caprine | CAN | 36.31 | ND | ND | 8 * |
| 56 | Caprine | CAN | 23.85 | ND | ND | 8 * |
| 57 | Caprine | CAN | 33.37 | ND | ND | 8 * |
| 58 | Caprine | CAN | 0.00 | ND | ND | 52 * |
| 59 | Caprine | CAN | 0.00 | ND | ND | 52 * |
| 60 | Caprine | CAN | 0.00 | ND | ND | 52 * |
| 61 | Caprine | CAN | 36.37 | ND | ND | 52 * |
| 62 | Caprine | CAN | 0.00 | ND | ND | 52 * |
| 63 | Caprine | CAN | 35.99 | ND | ND | 52 * |
| 64 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 65 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 66 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 67 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 68 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 69 | Caprine | CAN | 37.63 | ND | ND | 104 * |
| 70 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 71 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 72 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 73 | Caprine | CAN | 0.00 | ND | ND | 104 * |
| 74 | Caprine | WI | 0.00 | ND | ND | 52 * |
| 75 | Caprine | WI | 0.00 | ND | ND | 104 * |
| 76 | Caprine | WI | 0.00 | ND | ND | 104 * |
| 77 | Caprine | WI | 39.35 | ND | ND | 156 * |
| 78 | Caprine | WI | 38.46 | ND | ND | Unknown |
| 79 | Caprine | WI | 0.00 | ND | ND | Unknown |
| 80 | Caprine | WI | 0.00 | ND | ND | 104 * |
| 81 | Caprine | WI | 0.00 | ND | ND | 260 * |
| 82 | Caprine | WI | 0.00 | ND | ND | 520 * |
| 83 | Caprine | WI | 0.00 | ND | ND | 260 * |
| 84 | Caprine | WI | 0.00 | ND | ND | 156 * |
| 85 | Caprine | WI | 35.26 | ND | ND | 104 * |
| 86 | Caprine | WI | 0.00 | ND | ND | 104 * |
| 87 | Caprine | WI | 0.00 | ND | ND | Unknown |
| 88 | Caprine | WI | 0.00 | ND | ND | Unknown |
| 89 | Ovine | IL | 0.00 | ND | ND | Unknown |
| 90 | Ovine | WI | 0.00 | ND | ND | 208 * |
| 91 | Ovine | WI | 0.00 | ND | ND | Unknown |
| 92 | Ovine | CAN | 0.00 | ND | ND | 104 * |
| 93 | Ovine | CAN | 0.00 | ND | ND | 104 * |
| 94 | Ovine | CAN | 0.00 | ND | ND | 104 * |
| 95 | Ovine | CAN | 0.00 | ND | ND | 104 * |
| 96 | Ovine | CAN | 0.00 | ND | ND | 104 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Storms, S.; Lim, A.; Savard, C.; Olivera, Y.R.; Kayastha, S.; Wang, L. Identification of Hunnivirus in Bovine and Caprine Samples in North America. Viruses 2025, 17, 1491. https://doi.org/10.3390/v17111491
Storms S, Lim A, Savard C, Olivera YR, Kayastha S, Wang L. Identification of Hunnivirus in Bovine and Caprine Samples in North America. Viruses. 2025; 17(11):1491. https://doi.org/10.3390/v17111491
Chicago/Turabian StyleStorms, Suzanna, Ailam Lim, Christian Savard, Yaindrys Rodriguez Olivera, Sandipty Kayastha, and Leyi Wang. 2025. "Identification of Hunnivirus in Bovine and Caprine Samples in North America" Viruses 17, no. 11: 1491. https://doi.org/10.3390/v17111491
APA StyleStorms, S., Lim, A., Savard, C., Olivera, Y. R., Kayastha, S., & Wang, L. (2025). Identification of Hunnivirus in Bovine and Caprine Samples in North America. Viruses, 17(11), 1491. https://doi.org/10.3390/v17111491







