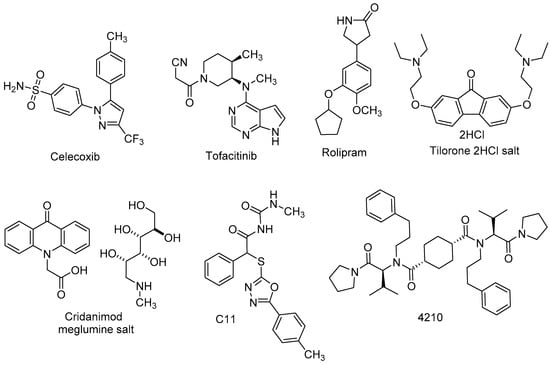
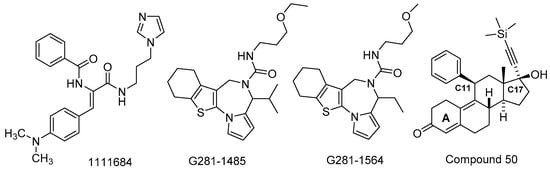
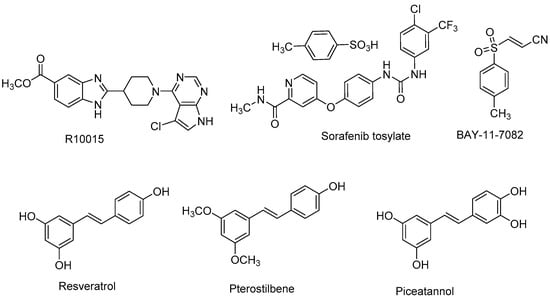
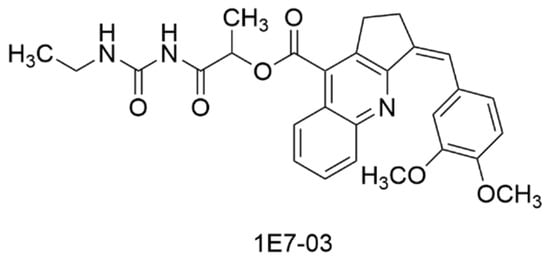
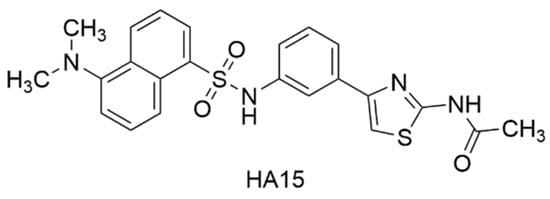
Abstract
Venezuelan, western, and eastern equine encephalitic alphaviruses (VEEV, WEEV, and EEEV, respectively) are arboviruses that are highly pathogenic to equines and cause significant harm to infected humans. Currently, human alphavirus infection and the resulting diseases caused by them are unmitigated due to the absence of approved vaccines or therapeutics for general use. These circumstances, combined with the unpredictability of outbreaks—as exemplified by a 2019 EEE surge in the United States that claimed 19 patient lives—emphasize the risks posed by these viruses, especially for aerosolized VEEV and EEEV which are potential biothreats. Herein, small molecule inhibitors of VEEV, WEEV, and EEEV are reviewed that have been identified or advanced in the last five years since a comprehensive review was last performed. We organize structures according to host- versus virus-targeted mechanisms, highlight cellular and animal data that are milestones in the development pipeline, and provide a perspective on key considerations for the progression of compounds at early and later stages of advancement.
Keywords:
alphavirus; antiviral; small molecule; VEEV; EEEV; WEEV; viral encephalitis; drug discovery 1. Introduction
The positive strand RNA alphaviruses, Eastern (EEEV), Western (WEEV), and Venezuelan Equine Encephalitis (VEEV) viruses are a subset of the Togaviridae family that cause significant disease in humans and equids. Viral transmission predominately occurs subcutaneously through the bite from infected mosquitoes, but other arthropods can also serve as vectors [1]. These “New World” alphaviruses, so termed owing to their historical localization in North, Central, and South America, present in humans with symptoms that range from a combination of fever, headache, muscle aches, and vomiting to a more serious state of infection, resulting in seizure, coma, or death [1,2,3,4]. The clinical manifestation of disease and long-term effects stemming from equine encephalitis virus (EEV) infection are distinguished from that of the “Old World” alphaviruses such as chikungunya (CHIKV), Sindbis (SINV), O’nyong-nyong (ONNV), and Ross River (RRV) viruses that generally cause fever, rash, and arthritogenic effects that can be long-lasting [5,6].
In humans, the incidence of natural VEEV infection is the highest of the three EEVs, resulting in hundreds of thousands of cases each year [7,8]; however, this is accompanied by the lowest case mortality rate of the three viruses at <1% [9]. Importantly, VEE symptoms are similar to that of other viral diseases such as Dengue fever, and in areas where both viruses circulate, it is estimated that ~10% of the Dengue fever cases are misdiagnosed and are actually caused by VEEV [8,9]. Compared to VEEV, WEEV infection occurs less frequently in humans but has a higher case mortality rate of 3–15% [10,11]. Natural enzootic EEEV infection in humans is relatively rare; however, there is a higher documented mortality rate of 30–50% [4]. In fact, the typical average of 11 EEE cases per year was dwarfed by the occurrence of 38 human cases in the United States during 2019 that resulted in the death of 19 patients [2,12,13]. Survivors of equine encephalitic infections frequently contend with persistent neurological complications [1,2,14,15], and these infections in the elderly and young children are associated with poorer outcomes [4,16]. As noted, infection can occur naturally through environmental vectors; however, for VEEV and EEEV, an intentional release of an aerosolized virus has been studied as a potential biowarfare agent [17,18,19].
Inactivated vaccines are available to protect equids, and either an inactivated or live, attenuated VEE TC83 vaccine has been provided for select laboratory and military personnel who risked exposure to these viruses; however, these vaccines are not suitable for use by the general public due to questions of efficacy and safety [20,21]. As such, there are currently no FDA-approved vaccines or therapeutics in hand for any alphavirus infection, leaving patients only with supportive care options [22] and creating a void of opportunity for these viruses to find advantage. Research efforts have focused on the development of vaccines [23,24,25], monoclonal antibodies [26,27,28], and small molecules as possible therapeutic or prophylactic measures. Having mechanistically divergent approaches available to address VEEV, WEEV, and EEEV is an attractive, and likely required, strategy to derive an antiviral approach that is independent of viral strain and infection stage, avoids the emergence of resistance, and is safe and effective for a patient population that is heterogeneous with respect to age, sex, and underlying medical conditions. Small molecules play an important role in the development of antiviral agents—as drugs themselves—or as chemical probes [29,30] that serve as a molecular lens through which novel aspects of virology are discovered, leading to intervention opportunities. Several published reviews describe small molecules that affect alphaviruses [17,31,32,33], including a comprehensive 2017 account that was contributed by Ching and co-workers [34].
This review focuses on small molecules that, since that 2017 report [34], have been newly discovered as VEEV, WEEV, or EEEV inhibitors or were previously reported, but have advanced to a new development milestone such as structure–activity relationship exploration, broader antiviral spectrum, mechanism of action studies, or have demonstrated efficacy in animals. Compound structures and associated data are generally organized by host-targeting or direct acting antivirals, and the mechanism underlying the compound activity is described when possible. When assessing the development landscape, it should be recognized that there is variability in assay conditions; therefore, comparing data across tabulated compounds should be done with caution due to differences in assay duration and endpoint, assay type and readout, viral strain, multiplicity of infection (MOI), and cell line, to name only a few with respect to in vitro assessments. For in vivo efficacy, several of the factors outlined for cell assays, along with the mouse strain and age, route and dose of viral challenge, timepoint in initiating treatment and its duration, compound route of administration and dose, formulation, and a myriad of compound related factors (e.g., solubility, stability, protein binding, tissue distribution, pharmacokinetics, etc.) play a role in the outcome of the evaluation. As such, the compilation of structures and data herein may be useful in selecting validated compound controls and aligning parameters for future experiments in this area.
2. Alphavirus Structure and Life Cycle
A single, positive-sense RNA strand contains the alphavirus genome. Within two open reading frames are encoded six structural proteins and four non-structural proteins (nsPs). A capsid protein (CP), envelope proteins (E1, E2, E3), a viroporin 6K protein, and a virulence factor known as the transframe (TF) protein [35] represent the structural proteins needed for cell entry, host defense mitigation, virulence, encapsidation, and budding events [36,37]. The non-structural proteins, nsP1, nsP2, nsP3, and nsP4, are involved in capping of RNA molecules, helicase and protease activity, avoiding host immune responses, processing of the viral polyproteins, and viral replication [38,39]. The viral RNA and capsid proteins form a nucleocapsid core that is surrounded by a protective lipid bilayer, or envelope, that is derived from the host plasma membrane during the budding and egress of newly formed virions [40]. The envelope displays glycoproteins essential to host cell receptor or attachment factor recognition, attachment, and entry (E2 glycoproteins) or post-entry membrane fusion (E1 glycoproteins) [41,42,43]. Generally, alphaviruses utilize clathrin-mediated endocytosis to infect host cells [44], though caveolae-mediated endocytosis [45] of Mayaro virus (MAYV), micropinocytosis [46] of CHIKV in human muscle cells, and pH-dependent pore formation with SINV [47] has also been described [48]. Once inside the host cell, the virus-containing endosome matures and undergoes a neutral to acidic pH shift, thereby leading to viral envelope/endosomal membrane fusion and release of the viral nucleocapsid into the cytosol [42,47]. Alphaviruses appropriate host cell proteins to translate the viral polyprotein p1234 which is further cleaved into individual nsPs. The nsPs have individual roles in the viral life cycle; however, an important function of these proteins involves their formation of a replication complex that facilitates negative strand RNA synthesis. Genomic and subgenomic RNA is subsequently synthesized, producing structural proteins that aid in virion assembly and release.
As with other viruses, small molecules have been discovered or designed to intervene in key events in the EEV life cycle. Generally, these can be categorized into two groups: those compounds that target essential, virus-recruited host proteins or compounds that directly engage viral proteins or processes. A compound that operates by a host-targeted mechanism has the potential benefit of working against a broader spectrum of viruses that all require the same host protein; however, this can lead to potential host related toxicity if the biological target is essential for host cell homeostasis and lacks protein or pathway redundancy. Compounds that target viral proteins can potentially avoid host cell toxicity as their mechanisms of action are independent of critical host proteins; however, viruses can evolve resistance mechanisms to overcome viral protein or pathway blockage, thereby reducing their utility. Of course, not all compounds suffer or are immune from these consequences in either category, and notably, as genomic and biochemical studies continue to elucidate how some small molecules operate, the lines between these groupings can blur. Nonetheless, the small molecules discussed in this review are presented with this classification in mind to aid the reader in navigating the small molecule EEV landscape.
4. Direct Targeting of EEV Non-Structural Proteins (nsPs) with Small Molecules
4.1. Non-Structural Protein 1 as a Target
As alphaviral mRNA is transcribed, the non-structural protein 1 (nsP1) facilitates the biosynthesis of a 7-methylguanosine nucleotide (m7-G), tethered by a triphosphate moiety (ppp, or TP), to the 5′ end of the mRNA strand. This 5′ cap prevents cellular enzyme-driven RNA degradation and aids in host immune response evasion, thereby blunting barriers to viral protein synthesis [123,124,125]. The 5′ cap is generated through a series of chemical reactions. This includes nsP2-mediated cleavage of the 5′ end terminal gamma-phosphate by RNA triphophatase (RTPase). The nsP1 mediates the N-methylation of GTP via a methyltransferase [MTase reaction] to generate m7-GTP. This is followed by guanylylation of nsP1 to form an nsP1-m7-GMP adduct [GT reaction] between viral nsP1 and m7-GTP with concomitant extrusion of inorganic pyrophosphate [123,126]. Transfer of the m7-GMP moiety to the 5′-diphosphate appendage of the RNA affords the capped strand, m7G(5′)ppp(5′)RNA. Given the importance of this process to viral replication, understanding of alphavirus-specific capping mechanisms and identification of inhibitors has been explored to design new antiviral agents [127,128]. For instance, Ferreira-Ramos and co-workers [124] developed a high-throughput ELISA assay based on the detection of the VEEV nsP1-m7-GMP adduct to identify new inhibitors of the GT reaction. A screen of 1220 Prestwick library compounds in the ELISA assay afforded hits that were further evaluated in a Western blot (WB) assay that quantified guanylylated VEEV nsP1. Pyrimethamine, a folic acid antagonist commonly used for chemoprophylaxis of malaria or to treat toxoplasmosis infection, emerged as an inhibitor of the VEEV GT reaction from the WB assay with an IC50 = 2.7 μM (Figure 7, Table 2). A set of seven structurally related 2,4-diaminopyrimidines were less potent by a factor of at least 6-fold with several analogs not showing inhibition of the GT reaction by WB analysis (>200 μM). The hydrated tartrate salt of ketanserin, an antihypertensive serotonin receptor antagonist featuring a quinazoline-2,4-dione core, was also identified by the WB assay as a GT reaction inhibitor, albeit with weaker potency than observed for pyrimethamine (IC50 = 14.6 μM). The pyridopyrimidin-4-one, pirenpirone, was also identified as a hit compound (IC50 = 39.6 μM) and, like ketanserin, is a known serotonin receptor antagonist that contains a 4-fluorobenzoylpiperidine moiety linked by two methylene units to a core nitrogen atom. Of the various commercial fragments and analogs of these hits that were assessed, only altanserin—a 2-thioxo derivative of ketanserin—showed comparable GT reaction inhibition (IC50 = 9.3 μM). Several compounds were also assessed separately for inhibition of the nsP1-mediated MTase reaction. At a concentration of 50 μM, pyrimethamine and ketanserin showed significant inhibition (74–79%) of the VEEV nsP1 MTase reaction while pirenperone showed less inhibition (45%). All three of these compounds demonstrated little to no inhibition of a human methyltransferase at that same concentration. Several known GT reaction inhibitors were used as assay controls, including triazolopyrimidinones MADTP-393 and MADTP-314 [129]. Several MADTP series compounds lost their inhibitory effect on VEEV nsP1 with the introduction of a D34S mutation; however, pyrimethamine and pirenperone both retained the ability to inhibit GTase activity with this variant, thereby suggesting that these compounds inhibit or bind VEEV nsP1 differentially compared to that of the MADTP series.
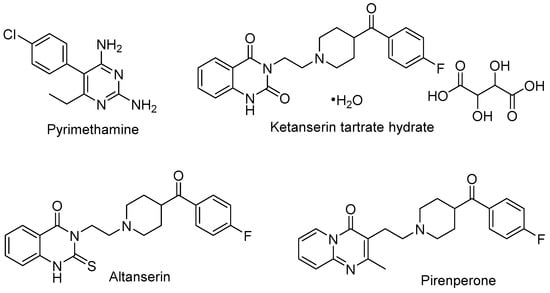
Figure 7.
Chemical structures of compounds that target VEEV nsP1.

Table 2.
In vitro data for compounds that target VEEV non-structural protein 1.
4.2. Non-Structural Protein 2 as a Target
The alphavirus nonstructural protein 2 (nsP2) is responsible for several key functions in the viral life cycle, making it an attractive therapeutic target [130,131,132,133]. Enzymatic activities accomplished by nsP2 include ATPase and GTPase activities [134] that are attributed to an N-terminal domain, helicase-mediated remodeling of viral RNA [135], proteolytic cleavage of the nonstructural polyprotein due to a cysteine protease domain [136], and RNA 5′-triphosphatase activity via an S-adenosyl-L-methionine-dependent RNA methyltransferase (SAM MTase) [137]. An X-ray crystal structure of the VEEV nsP2 protease has been resolved (5EZQ) [138], and a co-crystal structure with a protease inhibitor, E64d, has also been reported (PDB 5EZS) [139].
Zhang and co-workers [140] conducted a phenotypic screen using VEEV IC-SH3, a virulent strain isolated from humans during a VEE outbreak in the early 1990s, and a library of potential covalent binding compounds that might be expected to target the cysteine protease of VEEV nsP2. Based on an acrylate-linked 1,2-dihydroquinoline hit compound, a supporting cast of 15 analogs were generated to explore SAR in multiple cell types and with additional strains of VEEV. Key structural modifications of the hit scaffold included exchange of the acrylate warhead with an α,β-unsaturated methyl sulfone and saturation of the fused piperidine ring of the quinoline core. These efforts afforded compound 11 which addressed hydrolysis of the acrylate warhead and revealed VEEV EC50 values that were consistently around 2 µM across multiple cell lines using TC83 and TrD strains (Figure 8, Table 3). Some of the most active compounds from the study, including compound 11, were evaluated for their ability to inhibit the VEEV nsP2-dependent proteolytic cleavage of a FRET substrate in vitro. Complete inhibition of the VEEV nsP2 proteolytic activity was observed for these compounds at a concentration of 20 µM, while at 0.9 µM, compound 11 inhibited 39% of VEEV nsP2 proteolytic activity after 24 h, and related analog, compound 13, demonstrated an inhibition of 71% at that same concentration. In silico molecular docking studies were conducted with compound 11 to provide a rationale for the covalent inhibitor binding and key interactions with nsP2. The authors proposed future studies to improve metabolic stability and potency while also assessing the pharmacokinetic suitability of this dihydroquinoline scaffold ahead of in vivo efficacy determination [140].
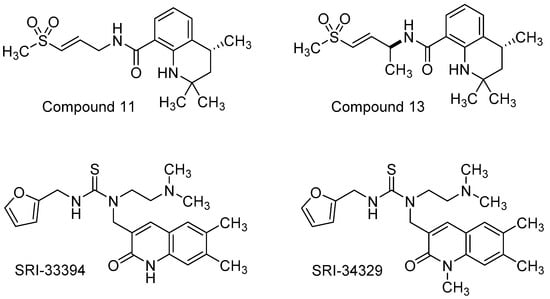
Figure 8.
Chemical structures of compounds that target nsP2.
Haese and co-workers reported the identification and SAR exploration of a 2-quinolinone hit compound following a CPE-based phenotypic screen using VEEV TC83 in Vero cells, followed by hit validation using normal human dermal fibroblasts (NHDFs) [141]. These efforts revealed SRI-33394 as a prime starting point for a systematic SAR investigation (Figure 8, Table 3). The team prepared approximately 81 analogs that surveyed five regions of the scaffold. Thiourea replacement with a urea was not tolerated, resulting in loss of potency; therefore, the thiourea was conserved. The presence of a basic nitrogen-containing functional group appended to the thiourea, along with a thiourea linked 2-methylfuranyl group, were also determined to be essential. Methylation of the SRI-33394 NH-quinazolinone core generated SRI-34329 which showed a 6-fold improvement in CPE antiviral activity but a 3 log loss in viral titer reduction compared to the parent hit. Plaque assays and northern blot assessments showed that quinolinone SRI-34329 inhibited an early viral replication step that affected viral RNA synthesis. Resistance mutations from passaged VEEV TC83 in the presence of SRI-34329 were sequenced, showing Y102C or Y102S mutations in nsP2. Additionally, the compound improved upon its antiviral spectrum compared to the hit compound, revealing low micromolar activity against ONNV, MAYV, and RRV. Ultimately, liabilities in solubility and microsomal stability dominated and could not be appreciably addressed through structural augmentation in the analog set; however, these compounds and the collective studies provided insight into potential VEEV nsP2 inhibition and a rationale as to why select compounds were not active against CHIKV. In silico docking of the analogs was performed using the crystal structure of CHIKV nsP2 for which VEEV nsP2 has high homology. Though select compounds did not inhibit CHIKV, the CHIKV nsP2 contains a lysine residue at position 102 in contrast to a tyrosine residue in VEEV that, when mutated at this position, rendered VEEV resistant to tested compounds in the set. These models, in tandem with resistance mutant analyses and reverse genetics, were used to rationalize the observed SAR for the series and provide evidence of nsP2 as the target of these compounds [141].
4.3. Non-Structural Protein 3–Host Protein Interactions as a Target
The alphavirus non-structural protein 3 (nsP3) is an integral component of the viral replication complex which is responsible for generating genomic, subgenomic, and negative-sense strands of viral RNA during infection [142,143,144]; however, physical attributes of its disordered C-terminal domain, nsP3 localization beyond that of the replication machinery, and association with different host proteins implicate additional functions that facilitate viral replication [145,146,147,148,149]. As such, identification of host proteins that interact with nsP3 may provide intervention points and therapeutic opportunities. While some VEEV nsP3–host protein interactions had been previously identified [102,107,144,150], a recent study sought to broaden the interactome while also identifying small molecule-based inhibitors. Bakovic and co-workers examined host protein interactions with VEEV nsP3 using an HA-tagged nsP3 mutant protein and mass spectroscopy and immunoprecipitation assays [151]. The study revealed 160 VEEV nsP3–host protein interactions and 42 potential inhibitors, of which nine compounds were selected for further evaluation. In vitro VEEV TC83 and cytotoxicity assays highlighted tomatidine, Z-VEID-FMK, and the selective serotonin reuptake inhibitor antidepressant, citalopram HBr, at 10 μM as inhibitors showing a >10-fold reduction in VEEV titer (Figure 9, Table 3). Subsequent testing of these compounds at 20 μM also showed antiviral activity against VEEV TrD and EEEV. Knockdown studies employing short-interfering RNA (siRNA) and VEEV TC83 stop codon mutants were conducted to confirm the relevance of host proteins putatively targeted by the inhibitors. Eukaryotic initiation factor 2 subunit 2 (eIF2S2), the putative target of tomatidine, was determined to be important to VEEV genomic RNA synthesis and independent of VEEV subgenomic RNA generation. Tomatidine only slightly impeded VEEV genomic RNA translation and more significantly abrogated viral subgenomic RNA synthesis, suggesting possible alternative mechanisms in play. Transcription factor AP-2 alpha (TFAP2A), putatively targeted by Z-VEID-FMK and citalopram HBr, was shown to not be directly involved in viral RNA synthesis, despite inhibitors of this protein still displaying antiviral activity [151].
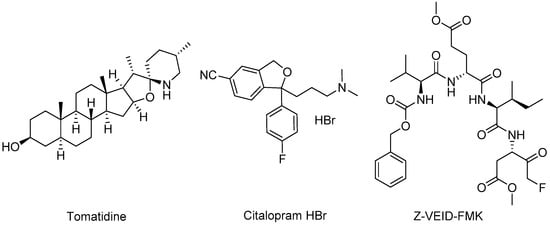
Figure 9.
Chemical structures of compounds that target nsP3.
4.4. Non-Structural Protein 4 as a Target
Alphavirus nonstructural protein 4 (nsP4) is a component of the viral replication complex that harbors RNA-dependent RNA polymerase (RdRp) and terminal adenylyltransferase (TAT) activities [152,153,154]. Due to these pivotal roles in viral replication and a high level of conservation across viral RdRps [155,156], its inhibition or exploitation is an attractive strategy for antiviral development, as the compounds discussed in this section exemplify.
Beta-d-N4–hydroxycytidine (NHC) is a ribonucleoside mimic featuring an N-4-hydroxyl group on a cytidine core and is the pre-phosphorylated active form of the ester prodrug, Molnupiravir, which can be used to treat patients infected with SARS-CoV-2. A unique architectural feature of NHC (Figure 10, Table 3) is that it can exist in tautomeric forms, thereby mimicking cytidine in the hydroxylamine form, or in the oxime tautomeric form, it better resembles uridine [157]. As a consequence, once NHC is converted into its triphosphate metabolite (NHC-TP) and incorporated by the viral RdRp into newly transcribed viral RNA, the NHC-containing template is read inconsistently as either uridine or cytidine, generating mutations that impair the viability of the virus [157,158]. Further, the mutations are not recognized by proofreading viral exonucleases as erroneous, thus the misinformation within the genome is propagated. NHC was evaluated against smallpox in the 1970s and was later found to be active against hepatitis viruses [159], norovirus [160], and chikungunya virus [161]. As a broad-spectrum antiviral agent, Urakova and co-workers [158] evaluated NHC against VEEV TC83, showing submicromolar potency without discernable cytotoxicity. The studies revealed that NHC treatment was most effective when it was applied in the first 4 h of infection, and the VEE virions produced from NHC-treated cells contained mutations that compromised replication capability. Monitoring of emerging resistance after NHC treatment showed that resistance was challenging to achieve after even 20 passages and appeared to require multiple coincident mutations in nsP4 [158]. Additional studies in mice were undertaken to assess the efficacy and safety profiles of NHC, later designated EIDD-1931 in preclinical development (Figure 10, Table 3) [162]. Painter and co-workers established the in vivo murine pharmacokinetic profile of EIDD-1931 and the triphosphate NHC derivative, EIDD-2061, after oral dosing, revealing distribution and exposure in plasma, spleen, and brain tissues and a good safety margin using up to 1000 mg/kg/day after repeat dosing for 7 days. Intranasally exposed, VEEV TrD-infected mice were used to assess the in vivo efficacy of EIDD-1931. Mice treated prior to viral exposure showed 90% survival in a 14-day study when dosed orally with 300 or 500 mg/kg twice daily for 6 days. Therapeutic treatment at 24 or 48 h post-infection in intranasally exposed, VEEV TrD-infected mice resulted in 90% and 40% survival, respectively, when dosed orally with EIDD-1931 at 500 mg/kg/day twice daily for 6 days [162]. These studies established the oral efficacy of EIDD-1931 against VEEV TrD in mice, providing a broader spectrum of activity for the compound and insights for further development.
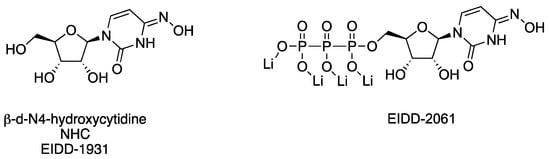
Figure 10.
Chemical structures of compounds that target nsP4.
4.5. Targeting Viral Replication through Alternative Modes of Action
ML336 is a benzamidine-based small molecule that demonstrates potent antiviral activity in VEEV TC83-infected mice (Figure 11, Table 3) [163]. Several studies have come into view since 2017 that explored various aspects of ML336. This includes an alternative formulation of ML336, examination of the breadth of ML336 antiviral activity in vitro and in vivo, assessment of ML336-associated, resistance-conferring VEEV mutations, or survey of benzamidine structure–activity and structure–property relationships.
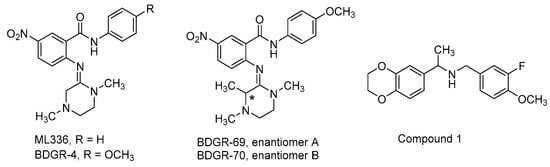
Figure 11.
Chemical structures for compounds that that target viral replication.
For instance, citing limitations in solubility and plasma stability, a study was undertaken to evaluate the use of lipid-coated mesoporous silica nanoparticles (LC-MSN) as a vehicle to deliver ML336 with anticipated increases in stability, solubility, tissue-targeting, and circulation time [164]. LC-MSNs contained about 20 μg ML336/mg LC-MSN and maintained colloidal stability for up to 4 days. In HeLa cells, ML336-loaded LC-MSNs inhibited VEEV TC83 replication up to a maximum of 6 log and without discernable cytotoxicity. In VEEV TC83-infected mice, ML336-loaded LC-MSNs resulted in a 10-fold reduction in brain viral load after 4 days compared to PBS-treated, infected mice [164], thus highlighting the potential of this delivery platform in the context of VEEV infection.
To better understand how ML336 exerts its antiviral effects, a more granular assessment of viral and host cell RNA synthesis was studied when ML336 treatment was applied [165]. Using nine structural analogs of ML336, it was shown that the most potent VEEV TC83 CPE assay inhibitors reduced viral RNA synthesis by the greatest amount. While ML336 did not appreciably affect host cell RNA synthesis, strand-specific qRT-PCR experiments showed that ML336 inhibited both positive and negative strand viral RNA synthesis. Further, ML336 inhibited the synthesis of both genomic and subgenomic viral RNA, and inhibition of all stages of viral RNA synthesis was dose dependent. These findings were also observed using the viral replicase enriched, membranous P15 fraction from VEEV-infected BHK cells. Collectively, these studies showed that benzamidine ML336 potently inhibited the synthesis of all VEEV RNA species through the intermediacy of the viral replication complex [165].
Previously, key in vitro mutations which rendered variant viruses inert to the effects of ML336 had been identified in both nsP2 (Y102C) and nsP4 (Q210K) [165,166]. In a follow-up study, the incidence and magnitude of resistance mutations was studied in nonhuman primate kidney epithelial cells and human astrocytes (Vero 76 and SVGA, respectively). The approach employed whole genome next-generation sequencing (NGS), thereby revealing single-nucleotide polymorphisms (SNPs) from passaged VEEV TC83 in the presence of the compound [167]. Notable outcomes included a common nsP4 Q210 mutation that dominated in both Vero 76 and SVGA cells. SNPs appeared more slowly in the SVGA cells, owing to their ability to produce type-1 IFNs. The major mutations were stable to additional passages in the absence of ML336 and maintained fitness. While RNA isolated from the brains of VEEV TC83-infected, ML336-treated mice were analyzed by NGS, depth of coverage was low, little to no overlap was observed between SNPs from in vivo and in vitro sources, and SNPs from mice were not ones known to confer resistance. A network analysis of the data showed that the microenvironment in these studies played a significant role in the evolution of mutations [167].
In a separate study, several analogs of ML336 were designed and synthesized to improve plasma stability and solubility without sacrificing VEEV potency [166]. The resulting benzamidine, BDGR-4 (Figure 11, Table 3), differs structurally from ML336 with the inclusion of a 4-methoxy group on the N-amide-like portion of the scaffold which enhanced solubilization by 2.6-fold and resulted in 11.6% more of the parent compound remaining after liver microsome incubation. This structural change was integrated into subsequent analogs such as BDGR-5 and enantiomeric benzamidines BDGR-69 and BDGR-70 which also featured a strategically placed methyl substituent to potentially thwart undesirable amidine hydrolysis in plasma. While all analogs were similar in potency, ultimately BDGR-4 delivered the best boost in solubility while maintaining plasma stability and protein binding characteristics like that of ML336. Testing of BDGR-4 in CPE assays against WEEV California and EEEV FL93-939 showed EC50 values in the 102–150 nM range. Prophylactic administration of BDGR-4, BDGR-69, or BDGR-70 to mice challenged intranasally with VEEV TC83 showed that BDGR-4 provided the best protection, resulting in 67% survival in a 21-day study after dosing BDGR-4 at 2.5 mg/kg b.i.d. for 5 days [166]. In a head-to-head comparison between BDGR-4 and ML336, mice challenged subcutaneously with VEEV TrD after being treated with either compound resulted in 100% protection while all untreated mice died by day 7. Delay of treatment studies in VEEV TrD-infected mice showed that BDGR-4 provided full protection when dosing was initiated at 24 h post infection and 88% survival when dosing was started at 48 h post infection [166]. In vivo efficacy was also established for BDGR-4 against EEEV FL93-939 infection in mice where prophylactic treatment resulted in 90% survival. Viral titers in brain tissue were determined and changes in weight for in study animals were catalogued for these studies. BDGR-4 did not induce type-1 IFNs, indicating that the antiviral activity of BDGR-4 was not a result of activation of the host immune response. Identification of resistance mutations from in vitro studies revealed mutations in nsP4 (several) and nsP2 (one) whose emergence was BDGR-4 concentration dependent. The results of these collective studies extended the in vivo efficacy profile of ML336 and, in comparison to structural analogs, showed that BDGR-4 offered a significant solubility improvement over ML336 that enabled in vivo assessments in VEEV and EEEV infected mice. Those experiments highlighted BDGR-4 as a potent inhibitor of VEEV and EEEV that offers significant protection in lethal murine models of infection [166].
Dibenzylamine hit compound 1, discovered in a CPE-based high throughput screen, potently inhibited a VEEV TC83 CPE with an EC90 value of 0.89 µM, without cytotoxic liability up to a concentration of 30 µM, and reduced VEEV titer at a concentration of 10 µM by 7.49 log (Figure 11, Table 3) [168]. Nguyen and coworkers generated 24 analogs of compound 1 that surveyed three structural scaffold regions to retain or improve the antiviral activity while addressing limitations in solubility and microsomal stability. Modifications to the ring fused 1,4-dioxane moiety and alterations of the fluoromethoxybenzene component rendered analogs less potent than compound 1 in the CPE assay and did not improve microsomal stability for those compounds that were assessed. Some modest gains in CPE potency were observed for a benzylic gem-dimethyl substitution of the monomethyl benzylic linker; however, microsomal stability remained similarly compromised. Compound 1 was inactive against a panel of Old World alphaviruses but did inhibit VEEV replication at an early stage beyond viral entry, resulting in a blockage of VEEV RNA and protein production. Further refinement of microsomal stability and mechanism of action studies were proposed for this structural series [168].

Table 3.
In vitro and in vivo anti-EEV data for direct acting antiviral small molecules.
Table 3.
In vitro and in vivo anti-EEV data for direct acting antiviral small molecules.
| Compound | Virus Strain | In Vitro Antiviral Data | In Vivo Antiviral Data | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|
| Cell Line | MOI | EC50 μM | CC50 μM | Titer Reduction (Concentration) | Mouse Challenge | Survival Dose | |||
| compound 11 | VEEV TC83 | BE(2)M17 | 1.2 | 1.4 | >25 | - | - | - | [140] |
| VEEV TC83 | Neuro-2a | 3.5 | 2.0 | >25 | - | - | - | [140] | |
| VEEV TrD | HeLa | 0.1 | 1.6 | >30 | - | - | - | [140] | |
| VEEV TrD | Vero | 0.01 | 2.4 | 30 | - | - | - | [140] | |
| compound 13 | VEEV TC83 | BE(2)M17 | 1.2 | 3.3 | >25 | - | - | - | [140] |
| VEEV TrD | Neuro-2a | 3.5 | 3.7 | >25 | - | - | - | [140] | |
| SRI-33394 | VEEV TC83 | NHDF | 1 | 0.77 | >30 | 8.98 log (10 µM) | - | - | [141] |
| SRI-34329 | VEEV TC83 | NHDF | 1 | 0.12 | >50 | 5.96 log (10 µM) | - | - | [141] |
| tomatidine | VEEV TC83 | U87MG | 0.1 | 2.5 | 175 | 11-fold (10 µM) | - | - | [151] |
| VEEV TrD | U87MG | 0.1 | - | 175 | 364-fold (20 µM) | - | - | [151] | |
| EEEV FL93-939 | U87MG | 0.1 | - | 175 | 314-fold (20 µM) | - | - | [151] | |
| Z-VEID-FMK | VEEV TC83 | U87MG | 0.1 | 0.5 | >150 | 128-fold (10 µM) | - | - | [151] |
| VEEV TrD | U87MG | 0.1 | - | 175 | 887-fold (20 µM) | - | - | [151] | |
| EEEV FL93-939 | U87MG | 0.1 | - | 175 | 100-fold (20 µM) | - | - | [151] | |
| citalopram HBr | VEEV TC83 | U87MG | 0.1 | 1 | >150 | 87-fold (10 µM) | - | - | [151] |
| VEEV TrD | U87MG | 0.1 | - | 175 | 19-fold (20 µM) | - | - | [151] | |
| EEEV FL93-939 | U87MG | 0.1 | - | 175 | 17-fold (20 µM) | - | - | [151] | |
| NHC or EIDD-1931 | VEEV TC83 | Vero | 0.5 | 0.426 | >200 | 2 log (1 µM) 4 log (2.5 µM) | - | - | [158] |
| VEEV TrD | - | - | - | - | - | CD-1 intranasal 100 LD50 | 90% a,b 300 mg/kg, BID, 6 d, PO | [162] | |
| VEEV TrD | CD-1 intranasal 100 LD50 | 90% a (+24 h PI) c 500 mg/kg, BID, 6 d, PO | [162] | ||||||
| VEEV TrD | CD-1 intranasal 100 LD50 | 40% a (+48 h PI)c 500 mg/kg, BID, 6 d, PO | [162] | ||||||
| ML336 | VEEV TC83 | Vero76 | 0.05 | 0.032 | >50 | 7.0 log (5 µM) | C3H/HeN intranasal 10LD50 | 71% b,d 5 mg/kg, BID, 4 d, IP | [163] |
| VEEV TrD | Vero76 | 0.05 | 0.04 | >50 | BLD (1 µM) BLD (0.5 µM) | BALB/c SC challenge 10LD50 | 100% a,b 25 mg/kg, BID, 8 d, IP | [163] [166] | |
| ML336/LC-MSN | VEEV TC83 | Vero | 0.1 | - | - | 6 log (2.5µg/mL) | C3H/HeN intranasal 108 PFU | reduced viral brain titer by 10-fold | [164] |
| BDGR-4 | VEEV TC83 | Vero76 | 0.05 | 0.047 | >50 | 7.0 log (5 µM) | C3H/HeN SC challenge 1 × 107 PFU | 100% b,e 5 mg/kg, BID, 8 d, IP | [166] |
| VEEV TrD | - | - | - | - | - | BALB/c SC challenge 10LD50 | 100% a,b 25 mg/kg, BID, 8 d, IP | [166] | |
| VEEV TrD | - | - | - | - | - | BALB/c SC challenge 10LD50 | 100% a (+24 h PI) c 25 mg/kg, BID, 8 d, IP | [166] | |
| VEEV TrD | - | - | - | - | - | BALB/c SC challenge 10LD50 | 88% a (+48 h PI) c 25 mg/kg, BID, 8 d, IP | [166] | |
| WEEV California | Vero76 | 0.05 | 0.102 | >50 | >6.2 log (5 µM) | - | - | [166] | |
| EEEV FL93-939 | Vero76 | 0.05 | 0.149 | >50 | - | C57BL/6 SC challenge 104.3 CCID50 | 90% a,b 50 mg/kg, BID, 8 d, IP | [166] | |
| BDGR-69 | VEEV TC83 | Vero76 | 0.05 | 0.028 | >50 | 7.2 log (5 µM) | C3H/HeN intranasal 1 × 107 PFU | 50% a,b 25 mg/kg, BID, 5 d, IP | [166] |
| BDGR-70 | VEEV TC83 | Vero76 | 0.05 | 0.025 | >50 | 7.2 log (5 µM) | C3H/HeN intranasal 1 × 107 PFU | 67% a,b 25 mg/kg, BID, 5 d, IP | [166] |
| VEEV TC83 | Vero76 | 0.05 | 0.025 | >50 | 7.2 log (5 µM) | C3H/HeN intranasal 1 × 107 PFU | 100% b,e 2.5 mg/kg, BID, 8 d, IP | [166] | |
| compound 1 | VEEV TC83 | THFF | 1 | 0.89 f | >30 | 7.5 log (10 µM) | - | - | [168] |
Only compounds with in vitro or in vivo data for any of the EEVs is tabulated; EC50, effective concentration required to inhibit 50% of the treated cell population; CC50, cytotoxic concentration that results in death of 50% of the treated cell population; BID, twice daily; PO, oral administration; SC, subcutaneous administration; IP, intraperitoneal administration; BLD, below limits of detection; a compared to 0% survival in untreated controls; b prophylactic study with dosing initiated two hours prior to viral challenge; c delay of treatment study with dosing initiated at the indicated time point post-infection (PI); d survival in untreated control group was 14%. e survival in untreated control group was 17%; f EC90 value.
5. Additional Small Molecule Inhibitors of Encephalitic Alphaviruses
Compounds were published with activity against VEEV, WEEV, and/or EEEV that are not discussed in the preceding sections. Generally, these compounds have yet to be subjected to in-depth mechanistic studies or divergent mechanisms are suggested to account for the observed activity, thereby complicating a straightforward categorization. In some cases, the compounds were identified from studies centered on other viruses which reported anti-EEV activity as part of the antiviral spectrum. Nonetheless, these compounds are important to track in the development pipeline as they may hold insights into unique or broader approaches to preventing or treating encephalitic alphavirus infection.
The synthetic chemistry that led to the discovery and development of anti-VEEV benzamidines ML336 and BDGR-4 [163,166] was recently modified, leading to the formation of a new class of benzodiazepinones [169]. A structural model overlaying the benzodiazepinone framework with BDGR-4 showed remarkable alignment. As such, a library of 17 benzodiazepinones was prepared bearing substituents known to impart anti-VEEV activity on the benzamidine core. The resulting benzodiazepinones were assessed in CPE and titer reduction assays using VEEV INH9813 or EEEV V105 in Vero 76 cells. The most potent compounds (EC50 = 27–48 nM for VEEV and EEEV) featured a C8 nitro group, though replacement with a nitrile moiety was possible in combination with other structural changes without significant potency loss. At a concentration of 5 µM, VEEV and EEEV titers were reduced by >5 log for many examples. Compounds 7a, 7b, 7o and the separated enantiomers of 7o were evaluated for VEEV and EEEV yield reduction in human brain primary neuronal cells (Figure 12, Table 4). At a concentration of 5 µM, viral titers were reduced to the limits of assay detection. Compound 7o and its individual isomers, assessed at 1 µM, were not significantly different from each other and resulted in at least a 2 log reduction in VEEV and EEEV titers. Aqueous solubility was modest for several compounds, and microsomal stability was marginal for compounds 7a and 7b (t1/2 = 10 min) but was improved for 7o (t1/2 = 45 min). The need for mechanism of action insights and structural modifications to improve microsomal stability were discussed to advance this new chemotype with potent antiviral cell activity against both VEEV and EEEV [169].
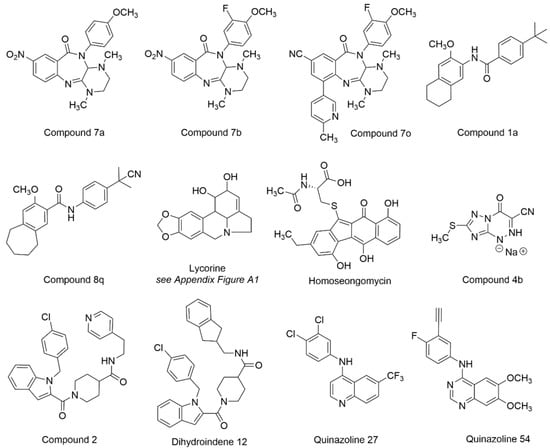
Figure 12.
Chemical structures for compounds intervening in the EEV life cycle.
In a study aimed at developing inhibitors of CHIKV, select compounds that had been originally tested against VEEV were evaluated against CHIKV for new medicinal chemistry opportunities [170]. Compound 1a (Figure 12, Table 4) modestly inhibited VEEV TC83 (EC50 = 13.2 µM, Vero cells) [163], but 1a demonstrated improved CHIKV potency and was selected as an optimization point, leading to the development of compound 8q. In normal human dermal fibroblast (NHDF) cells, compound 8q inhibited several alphaviruses, including VEEV TC83, with an EC90 value of 0.40 µM without cytotoxicity at the highest concentration tested (CC50 > 30 µM). At a concentration of 10 µM, VEEV viral titer was reduced by 3.1 log. Pharmacokinetic parameters for compound 8q were determined in mice, which may inform potential VEEV-centered studies. Resistant mutations in CHIKV implicated involvement of the nsP3 macrodomain in the compound’s mechanism of action. Compound 8q also inhibited human dihydroorotate dehydrogenase (IC50 = 0.31 µM), a host target whose inhibition has been associated with in vitro antiviral activity due to the role of the enzyme in nucleotide biosynthesis [170,171,172]. In sum, these efforts may help guide additional work with this scaffold on encephalitic alphaviruses.
In a separate CHIKV-focused study, lycorine (Figure 12, Table 4) was assessed for activity in cells infected with various alphaviruses [173]. Lycorine is an alkaloid derived from plants that has been studied for a range of pharmacological effects, including antiviral activity [173,174] (see Appendix A Figure A1 for structural note). The compound was found to inhibit CHIKV infection in various cell types with submicromolar IC50 values and without apparent cytotoxicity. Lycorine was tested against a panel of alphaviruses, including VEEV, which revealed an IC50 of 0.31 µM. Several mechanisms have been proposed to account for the observed effects of lycorine on other viruses [175,176,177,178] and in this study, the time of addition studies and subsequent experiments with an nsP4-inactivated viral mutant suggest that lycorine inhibited CHIKV replication by interrupting viral RNA translation [162].
Homoseongomycin is a marine natural product that was found during a high throughput screen [179] aimed at identifying compounds with activity against VEEV (Figure 12, Table 4). In Vero cells, homoseongomycin inhibited VEEV with EC50 values of 8.6 µM (TC83-luc assay) and 9.1 µM (ZPC738 strain). At a compound concentration of 50 µM, viral titers were reduced by 8 log (TC83, Vero cells) and 4 log (ZPC738, U87MG cells) with no observable toxicity up to 50 µM in either cell line. A nano luciferase reporter virus of EEEV FL93-939 was also inhibited by homoseongomycin (EC50 = 1.2 µM). Time of addition studies showed that homoseongomycin significantly inhibited VEEV entry as well as later stages of viral infection. The intermediacy of host factors in the observed antiviral effects of homoseongomycin is undetermined at this time but these experiments and assessments in animal models are proposed [179].
Triazavirin (riamilovir) is a non-nucleoside triazolotriazine-based analog that has been investigated for its antiviral activity against tick-borne encephalitis and influenza [180,181]. To assess if derivatives of triazavirin would be effective in VEEV and EEEV cell culture and in infected mice, analogs were generated as sodium salts that incorporated a nitrile group in place of the nitro group of the parent structure, and then modifications were made to the triazine thiomethyl substituent or in the triazine ring itself [181]. At a concentration of 100 µg/mL, a 1.9 and 2.6 log reduction in VEEV and EEEV titers, respectively, was observed for compound 4b (Figure 12, Table 4). In VEEV strain 230-infected mice (parental challenge), compound 4 was dosed orally at 50 or 100 mg/kg at 2 h post infection and continued for 5 days, resulting in 60% and 80% survival, respectively. Oral dosing initiated at 24 h post infection in VEEV-infected mice and continued for 4 days resulted in 40% and 70% survival at 50 or 100 mg/kg doses, respectively. Similar protocols employed for EEEV strain 463 efficacy studies showed 50% and 70% survival in mice dosed with 50 or 100 mg/kg/d at 2 h post-infection, respectively. For mice dosed at 24 h post infection, 40% and 70% survival were observed with 50 or 100 mg/kg/d of compound 4b, respectively. Mouse toxicology experiments did not reveal behavioral or morphological changes in mice up to 28 days with daily injections of 375 mg/kg [181].
A series of indole 2-carboxamides, represented by compound 2 (Figure 12, Table 4), was described previously [182] with activity against WEEV in a replicon assay (IC50 = 0.5 µM, CC50 = 65 µM); however, the precise mechanism of action regarding the target and how the compounds may bind was unknown. Conformationally restricted indole 2-carboxamides were synthesized to improve potency against WEEV and define a pharmacophoric model [183]. Though significant gains in potency were not achieved compared to compound 2, dihydroindene 12 showed comparable potency against WEEV without notable cytotoxicity (IC50 = 0.53 µM, CC50 > 100 µM). Scaffold rigidification and SAR, with computational analysis, refined a pharmacophoric model for this chemotype [183].
A collection of quinazolines and quinolines featuring a 4-aminoaryl group was designed based on structural similarities between the anti-DENV 4-aminoquinazoline, erlotinib, and a 4-aminoquinoline that demonstrated antiviral activity against both DENV and VEEV [184]. Following a systematic SAR effort, quinoline 27 and quinazoline 54 (Figure 12, Table 4) showed activity in human U87MG astrocytes infected with VEEV TC83 (EC50 = 0.50–0.60 µM, CC50 > 10 µM). In a separate study, structurally related compounds that more deeply assessed substitutions of the 4-aminoaryl substituent were assessed against these viruses [185], resulting in several analogs with single digit TC83 micromolar activity.

Table 4.
In vitro and in vivo anti-EEV data for antiviral small molecules.
Table 4.
In vitro and in vivo anti-EEV data for antiviral small molecules.
| Compound | Virus Strain | In Vitro Antiviral Data | In Vivo Antiviral Data | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|
| Cell Line | MOI | EC50 μM | CC50 μM | Titer Reduction (Concentration) | Mouse Challenge | Survival Dose | |||
| compound 7a | VEEV INH9813 | Vero76 | 0.05 | 0.041 | >30 | 5.5 log (5 µM) | - | - | [169] |
| EEEV V105 | Vero76 | 0.05 | 0.033 | >30 | 7.9 log (5 µM) | - | - | [169] | |
| compound 7o | VEEV INH9813 | Vero76 | 0.05 | 0.24 | 23.3 | 5.2 log (5 µM) | - | - | [169] |
| EEEV V105 | Vero76 | 0.05 | 0.16 | 23.3 | 6.9 log (5 µM) | - | - | [169] | |
| compound 1a | VEEV TC83 | NHDF | 1 | 2.1 c | >30 | - | - | - | [170] |
| compound 8q | VEEV TC83 | NHDF | 1 | 0.4 c | >30 | 3.1 log (10 µM) | - | - | [170] |
| lycorine | VEEV TC83 | Vero | 0.01 | 0.31 | >10 | - | - | - | [173] |
| homoseon- gomycin | VEEV TC83-luc | Vero | 0.1 | 8.6 | >50 | 8 log (50 µM) | - | - | [179] |
| VEEV ZPC738-luc | U87MG | 0.1 | 9.1 a | >50 | 4 log (50 µM) | - | - | [179] | |
| EEEV FL93-939-luc | Vero | 0.1 | 1.2 | >50 | - | - | - | [179] | |
| compound 4b | VEEV 230 | PEKC | 0.02 | - | - | 1.9 log PFU (100 µg/mL) | outbred albino SC challenge 10LD50 | 60% b (+2 h PI) 50 mg/kg, QD, 5 d, PO | [181] |
| VEEV 230 | PEKC | 0.02 | - | - | 1.9 log PFU (100 µg/mL) | outbred albino SC challenge 10LD50 | 80% b (+24 h PI) 100 mg/kg, QD, 5 d, PO | [181] | |
| EEEV 463 | PEKC | 0.02 | - | - | 2.6 log PFU (100 µg/mL) | outbred albino SC challenge 10LD50 | 50% b (+2 h PI) 50 mg/kg, QD, 5 d, PO | [181] | |
| EEEV 463 | PEKC | 0.02 | - | - | 2.6 log PFU (100 µg/mL) | outbred albino SC challenge 10LD50 | 70% b (+24 h PI) 100 mg/kg, QD, 5 d, PO | [181] | |
| compound 2 | WEEV Cba-87 | BSR-Z7/C3 | 0.1 | 0.53 | 65 | - | - | - | [183] |
| dihydro- indene 12 | WEEV Cba-87 | BSR-Z7/C3 | 0.1 | 0.53 | >100 | - | - | - | [183] |
| quinoline 27 | VEEV TC83 | U87MG | 0.1 | 0.50 | >10 | - | - | - | [184] |
| quinazoline 54 | VEEV TC83 | U87MG | 0.1 | 0.60 | >10 | - | - | - | [184] |
Only compounds with in vitro or in vivo data for any of the EEVs is tabulated; EC50, effective concentration required to inhibit 50% of the treated cell population; CC50, cytotoxic concentration that results in death of 50% of the treated cell population; PO, oral administration; SC, subcutaneous administration; a In Vero cells; b delay of treatment study with dosing initiated at the indicated time point post-infection (PI); c EC90 value.
6. Discussion
Alphavirus infection is a serious public health concern due to a constellation of factors. For instance, the absence of approved, effective, and safe prophylactic or therapeutic modalities creates vulnerabilities for which we are unprepared. This is exacerbated by the unpredictable nature of outbreaks and global climate changes that expand the geographic regions affected by mosquito-borne transmission. In addition, VEEV, WEEV and EEEV are classified by the National Institute of Allergy and Infectious Diseases (NIAID) as category B pathogens due to their biowarfare potential via aerosolization and deliberate distribution [186]. Further, human acquired infections caused by these viruses have been shown to place a substantial physical and economic burden on survivors [187,188]. Taken together, the need for translational research and outcomes in this area are underscored.
Herein, small molecules affecting EEVs were highlighted that have been discovered or advanced in development since a thorough review was done in 2017 [34]. In this report, assessment of compounds against specific targets, determination of cellular antiviral activity and spectrum, exploration of SAR and establishment of ADME, pharmacokinetic and toxicologic parameters, and examination of formulation were showcased. Additionally, several compounds were evaluated in mouse infection models for VEEV (8 compounds, 5 distinct chemotypes), WEEV (1 compound/chemotype), and EEEV (2 compounds/chemotypes), revealing significant survival outcomes in mice across these viruses. Collectively, these studies provide benchmarks and granularity on mechanisms of action that may be useful in the optimization and study of future analogs and chemotypes.
As new compounds are identified and existing antivirals are advanced for drug development purposes, it is important to define the target product profile (TPP). The TPP provides a roadmap to the experiments and data that will be required to demonstrate safety and efficacy and accounts for pipeline challenges that need to be addressed. Ideally, an EEV antiviral is orally administered, quickly distributed to the brain and other tissues with once-a-day dosing, efficacious and safe for the broadest patient population, inhibits all three EEVs in prophylactic and therapeutic scenarios and is independent of route of exposure, avoids the development of resistance, and is shelf stable without a cold chain requirement. To achieve these goals, long range planning is essential. For instance, the choice of cell lines, viral strain alignment from in vitro to in vivo assessment, the readiness of animal models that recapitulate human disease, biomarker identification, and even the outlook for clinical trial patient recruitment should be examined, as these factors may require adjustment of the development plan and experiments needed along the way.
Considerations for early-stage compounds include an understanding of mechanism, demonstration of antiviral activity and selectivity indices in relevant cell lines, and with wild-type strains of alphaviruses that will likely be used in downstream efficacy models. Establishment of structure–activity and structure–property relationships are important to indicate that the scaffold can tolerate structural modifications with improvement in multiple profile parameters. Physiochemical properties should be evaluated once compound hits are validated, as solubility and microsomal stability are critical guideposts for optimization. Brain exposure is also an important aspect of the compound profile given the need to penetrate the blood brain barrier (BBB) to inhibit EEV replication in neuronal cells. In vitro assays are available to assess BBB permeability and can be useful in the down-selection of compounds for advanced in vivo studies. Tiered ADME studies assist in compound prioritization for PK and efficacy studies, while also pinpointing potential liabilities that may be remedied through medicinal chemistry efforts. PK studies determine plasma and tissue exposure as a function of dose and route of administration and help guide dosing in animal models. Demonstration of efficacy and safety in validated animal models with viral strains that are clinically relevant are also critical milestones.
Drug discovery and development activities centered on EEVs are impeded by unique challenges, in addition to the traditional drug pipeline bottlenecks, risks, triage or failure observed during hit-to-lead, lead advancement, and preclinical studies. While very early activities may avoid the use of highly pathogenic viruses or select agents, at some point, specialized biosafety facilities, regulatory oversight, and trained personnel are required to execute studies with relevant virus strains. VEEV and EEEV are categorized as select agents, except for the attenuated VEEV TC83 strain that can be used at a BSL2 level. Therefore, highly pathogenic, wild-type strains of VEEV, WEEV, and EEEV are restricted to use within a limited number of BSL3 facilities. Additionally, our understanding of alphavirus pathogenesis and the intermediacy of host proteins continues to grow, hampered in part by the absence of X-ray crystal structures of some targets such as the nsP4 viral polymerase that would help guide compound optimization. Priming the pipeline with high-quality, drug-like chemical matter from the start is also imperative to enhance the success of translation. Data from validated animal models of EEV infection that strongly mirror the pathogenesis and hallmarks of the human disease and provide clear clinical endpoints are also critical. This point is especially poignant due to the applicability of the animal rule [189,190], as patient recruitment for an EEV human clinical efficacy study is more difficult given the unpredictable nature of EEV outbreaks and coincident infections that obscure straightforward diagnoses of encephalitic alphavirus infections.
Despite these hurdles, the outlook for finding small molecule derived drugs for EEVs is promising. Technological innovation in cryo-EM and genetic and analytical tools may enable examination of compound-target interactions that to date have been challenging to study. Compounds in the EEV pipeline have advanced further and with greater characterization than in previous years, as exemplified by EIDD-1931, which progressed within the EEV pipeline until a strategic pivot to SARS-CoV-2 was implemented, resulting in the emergency use authorization of Molnupiravir [191]. In fact, the global impact of COVID-19 has highlighted a gap in the availability and development of anti-infective agents for viruses that pose a significant threat or have pandemic potential [192,193]. Consequently, funding initiatives have been established to support efforts in this area which includes the Togaviridae EEVs, VEEV and EEEV, as viruses of concern. In addition to EIDD-1931, the benzamidine BDGR-4 is notable due to its in vitro activity against all three wild-type EEVs and demonstrated in vivo efficacy against VEEV and EEEV. These and other compounds that advance into higher species for efficacy and safety may serve as informative benchmarks by which better antivirals will be designed. Ultimately, this momentum in the field enhances the likelihood that safe and effective small molecule-based drugs can be developed to address gaps in the EEV pipeline.
Author Contributions
T.J.O. conducted the literature review; T.J.O. and J.E.G. reviewed the articles, wrote the manuscript, and read and agreed to the published version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge support from the National Institutes of Health (U19AI142762, J.E.G.) for time to write this review on compounds directly related to and/or stemming from that funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
J.E.G. is a co-inventor of ML336, BDGR-4, BDGR-5, BDGR-69-70, and compounds 7a, 7b, and 7o which are included in this review.
Appendix A
The structure of lycorine was published [173] as shown in Figure A1, panel A. Commercial vendors indicate the presence of a double bond (shaded yellow region) that is not present in the structure published by Li et al. [173]. The authors cite the source of their material as Sichuan Victory Biological Technology Co., Ltd., China. This vendor also depicts the presence of the double bond, along with at least three of four designated stereocenters (differences in one stereocenter shaded in green), as compounds available for purchase (see Figure A1, panels B and C). In this review, we depict the structure of lycorine (see in Figure 12) with the double bond to accurately reflect the correct structural skeleton but the stereocenters are not designated given that the authors did not do so, and we cannot account for the possibility of a compound purchased with different stereoisomers that may no longer be available from this vendor.
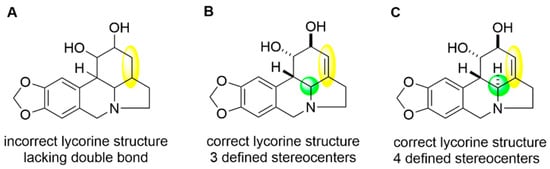
Figure A1.
Lycorine structural inconsistencies. (A) is the published incorrect lycorine structure [173] that lacks a double bond, highlighted in yellow. (B,C) depict lycorine structures from the referenced vendor that contain the required double bond, but also show stereochemical assignments that were not explicitly detailed in the reference.
References
- Zacks, M.A.; Paessler, S. Encephalitic alphaviruses. Vet. Microbiol. 2010, 140, 281–286. [Google Scholar] [CrossRef]
- Johnson, K.M.; Martin, D.H. Venezuelan equine encephalitis. Adv. Vet. Sci. Comp. Med. 1974, 18, 79–116. [Google Scholar]
- Suhrbier, A.; Jaffar-Bandjee, M.C.; Gasque, P. Arthritogenic alphaviruses—An overview. Nat. Rev. Rheumatol. 2012, 8, 420–429. [Google Scholar] [CrossRef]
- Chen, W.; Foo, S.S.; Sims, N.A.; Herrero, L.J.; Walsh, N.C.; Mahalingam, S. Arthritogenic alphaviruses: New insights into arthritis and bone pathology. Trends Microbiol. 2015, 23, 35–43. [Google Scholar] [CrossRef]
- Forrester, N.L.; Wertheim, J.O.; Dugan, V.G.; Auguste, A.J.; Lin, D.; Adams, A.P.; Chen, R.; Gorchakov, R.; Leal, G.; Estrada-Franco, J.G.; et al. Evolution and spread of Venezuelan equine encephalitis complex alphavirus in the Americas. PLoS Negl. Trop. Dis. 2017, 11, e0005693. [Google Scholar] [CrossRef]
- Aguilar, P.V.; Estrada-Franco, J.G.; Navarro-Lopez, R.; Ferro, C.; Haddow, A.D.; Weaver, S.C. Endemic Venezuelan equine encephalitis in the Americas: Hidden under the dengue umbrella. Future Virol. 2011, 6, 721–740. [Google Scholar] [CrossRef]
- Guzmán-Terán, C.; Calderón-Rangel, A.; Rodriguez-Morales, A.; Mattar, S. Venezuelan equine encephalitis virus: The problem is not over for tropical America. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Phelps, A.L.; O’Brien, L.M.; Eastaugh, L.S.; Davies, C.; Lever, M.S.; Ennis, J.; Zeitlin, L.; Nunez, A.; Ulaeto, D.O. Susceptibility and Lethality of Western Equine Encephalitis Virus in Balb/c Mice When Infected by the Aerosol Route. Viruses 2017, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Reisen, W.K. Western Equine Encephalomyelitis. In The Arboviruses, 1st ed.; Monath, T.P., Ed.; CRC Press: Boca Raton, FL, USA, 1988; Volume 5, pp. 89–137. [Google Scholar]
- Calisher, C.H. Medically important arboviruses of the United States and Canada. Clin. Microbiol. Rev. 1994, 7, 89–116. [Google Scholar] [CrossRef] [PubMed]
- Millet, N.; Faiek, S.; Gurrieri, D.; Kals, K.; Adams, W.; Hamaty, E.; Trivedi, M.; Zeidwerg, D. Deadly Neuroinvasive Mosquito-Borne Virus: A Case of Eastern Equine Encephalitis. Perm. J. 2021, 25, 20.288. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, N.P.; Martin, S.W.; Staples, J.E.; Fischer, M. Notes from the Field: Multistate Outbreak of Eastern Equine Encephalitis Virus—United States, 2019. MMWR Morb. Mortal. Wkly Rep. 2020, 69, 50–51. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention | Eastern Equine Encephalitis. Available online: https://www.cdc.gov/easternequineencephalitis/statistics-maps/index.html (accessed on 1 December 2022).
- Ronca, S.E.; Dineley, K.T.; Paessler, S. Neurological Sequelae Resulting from Encephalitic Alphavirus Infection. Front. Microbiol. 2016, 7, 959. [Google Scholar] [CrossRef]
- Bowen, G.S.; Fashinell, T.R.; Dean, P.B.; Gregg, M.B. Clinical aspects of human Venezuelan equine encephalitis in Texas. Bull. Pan Am. Health Organ 1976, 10, 46–57. [Google Scholar] [PubMed]
- Hayes, E.B.; Staples, J.E. Principles and Practice of Pediatric Infectious Diseases, 4th ed.; Elsevier: London, UK, 2012; pp. 1097–1099. [Google Scholar]
- Reichert, E.; Clase, A.; Bacetty, A.; Larsen, J. Alphavirus antiviral drug development: Scientific gap analysis and prospective research areas. Biosecur. Bioterror. 2009, 7, 413–427. [Google Scholar] [CrossRef]
- Sidwell, R.W.; Smee, D.F. Viruses of the Bunya- and Togaviridae families: Potential as bioterrorism agents and means of control. Antivir. Res. 2003, 57, 101–111. [Google Scholar] [CrossRef]
- Christopher, G.W.; Cieslak, T.J.; Pavlin, J.A.; Eitzen, E.M., Jr. Biological warfare. A historical perspective. JAMA 1997, 278, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Alevizatos, A.C.; McKinney, R.W.; Feigin, R.D. Live, attenuated Venezuelan equine encephalomyelitis virus vaccine. I. Clinical effects in man. Am. J. Trop. Med. Hyg. 1967, 16, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Pittman, P.R.; Makuch, R.S.; Mangiafico, J.A.; Cannon, T.L.; Gibbs, P.H.; Peters, C.J. Long-term duration of detectable neutralizing antibodies after administration of live-attenuated VEE vaccine and following booster vaccination with inactivated VEE vaccine. Vaccine 1996, 14, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention | Treatment & Prevention | Eastern Equine Encephalitis. Available online: https://www.cdc.gov/easternequineencephalitis/healthcare-providers/treatment-prevention.html (accessed on 2 October 2022).
- Stromberg, Z.R.; Fischer, W.; Bradfute, S.B.; Kubicek-Sutherland, J.Z.; Hraber, P. Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines 2020, 8, 273. [Google Scholar] [CrossRef] [PubMed]
- Paessler, S.; Weaver, S.C. Vaccines for Venezuelan equine encephalitis. Vaccine 2009, 27 (Suppl. S4), D80–D85. [Google Scholar] [CrossRef]
- Powers, A.M. Resurgence of Interest in Eastern Equine Encephalitis Virus Vaccine Development. J. Med. Entom. 2021, 59, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Burke, C.W.; Froude, J.W.; Rossi, F.; White, C.E.; Moyer, C.L.; Ennis, J.; Pitt, M.L.; Streatfield, S.; Jones, R.M.; Musiychuk, K.; et al. Therapeutic monoclonal antibody treatment protects nonhuman primates from severe Venezuelan equine encephalitis virus disease after aerosol exposure. PLoS Pathog. 2019, 15, e1008157. [Google Scholar] [CrossRef] [PubMed]
- Williamson, L.E.; Gilliland, T., Jr.; Yadav, P.K.; Binshtein, E.; Bombardi, R.; Kose, N.; Nargi, R.S.; Sutton, R.E.; Durie, C.L.; Armstrong, E.; et al. Human Antibodies Protect against Aerosolized Eastern Equine Encephalitis Virus Infection. Cell 2020, 183, 1884–1900.e1823. [Google Scholar] [CrossRef]
- Kim, A.S.; Austin, S.K.; Gardner, C.L.; Zuiani, A.; Reed, D.S.; Trobaugh, D.W.; Sun, C.; Basore, K.; Williamson, L.E.; Crowe, J.E., Jr.; et al. Protective antibodies against Eastern equine encephalitis virus bind to epitopes in domains A and B of the E2 glycoprotein. Nat. Microbiol. 2019, 4, 187–197. [Google Scholar] [CrossRef]
- ChemicalProbes.Org. Chemical Probes | Information Centre | Probes Criteria. Available online: https://www.chemicalprobes.org/information-centre#probes-criteria (accessed on 31 January 2023).
- Antolin, A.A.; Sanfelice, D.; Crisp, A.; Villasclaras Fernandez, E.; Mica, I.L.; Chen, Y.; Collins, I.; Edwards, A.; Müller, S.; Al-Lazikani, B.; et al. The Chemical Probes Portal: An expert review-based public resource to empower chemical probe assessment, selection and use. Nucleic Acids Res. 2022, 51, D1492–D1502. [Google Scholar] [CrossRef] [PubMed]
- Kehn-Hall, K.; Bradfute, S.B. Understanding host responses to equine encephalitis virus infection: Implications for therapeutic development. Expert Rev. Anti-Infect Ther. 2022, 20, 1551–1566. [Google Scholar] [CrossRef]
- Bassetto, M.; Brancale, A. Chapter Four—The search for antivirals to treat alphavirus infections. Annu. Rep. Med. Chem. 2021, 57, 133–151. [Google Scholar]
- Nagata, L.P.; Wong, J.P.; Hu, W.-g.; Wu, J.Q. Vaccines and therapeutics for the encephalitic alphaviruses. Future Virol. 2013, 8, 661–674. [Google Scholar] [CrossRef]
- Ching, K.C.; FP Ng, L.; Chai, C.L.L. A compendium of small molecule direct-acting and host-targeting inhibitors as therapies against alphaviruses. J. Antimicrob. Chemother. 2017, 72, 2973–2989. [Google Scholar] [CrossRef]
- Snyder, J.E.; Kulcsar, K.A.; Schultz, K.L.W.; Riley, C.P.; Neary, J.T.; Marr, S.; Jose, J.; Griffin, D.E.; Kuhn, R.J. Functional Characterization of the Alphavirus TF Protein. J. Virol. 2013, 87, 8511–8523. [Google Scholar] [CrossRef]
- Sharma, A.; Knollmann-Ritschel, B. Current Understanding of the Molecular Basis of Venezuelan Equine Encephalitis Virus Pathogenesis and Vaccine Development. Viruses 2019, 11, 164. [Google Scholar] [CrossRef]
- Ramsey, J.; Mukhopadhyay, S. Disentangling the Frames, the State of Research on the Alphavirus 6K and TF Proteins. Viruses 2017, 9, 228. [Google Scholar] [CrossRef]
- Hardy, W.R.; Strauss, J.H. Processing the nonstructural polyproteins of sindbis virus: Nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 1989, 63, 4653–4664. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.Y.-S.; Ng, M.M.-L.; Chu, J.J.H.C. Replication of Alphaviruses: A Review on the Entry Process of Alphaviruses into Cells. Adv. Virol. 2011, 2011, 249640. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Snyder, J.E.; Kuhn, R.J. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009, 4, 837–856. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Zhang, W.; Gabler, S.; Chipman, P.R.; Strauss, E.G.; Strauss, J.H.; Baker, T.S.; Kuhn, R.J.; Rossmann, M.G. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 2006, 14, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.S.; Diamond, M.S. A molecular understanding of alphavirus entry and antibody protection. Nat. Rev. Microbiol. 2022, 1–12. [Google Scholar] [CrossRef]
- Gardner, C.L.; Ebel, G.D.; Ryman, K.D.; Klimstra, W.B. Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc. Natl. Acad. Sci. USA 2011, 108, 16026–16031. [Google Scholar] [CrossRef]
- Mudhakir, D.; Harashima, H. Learning from the viral journey: How to enter cells and how to overcome intracellular barriers to reach the nucleus. AAPS J. 2009, 11, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.A.M.; Silva, J.L.; Oliveira, A.C.; Gomes, A.M.O. On the entry of an emerging arbovirus into host cells: Mayaro virus takes the highway to the cytoplasm through fusion with early endosomes and caveolae-derived vesicles. PeerJ 2017, 5, e3245. [Google Scholar] [CrossRef]
- Lee, C.H.R.; Mohamed Hussain, K.; Chu, J.J.H. Macropinocytosis dependent entry of Chikungunya virus into human muscle cells. PLoS Negl. Trop. Dis. 2019, 13, e0007610. [Google Scholar] [CrossRef] [PubMed]
- Paredes, A.M.; Ferreira, D.; Horton, M.; Saad, A.; Tsuruta, H.; Johnston, R.; Klimstra, W.; Ryman, K.; Hernandez, R.; Chiu, W.; et al. Conformational changes in Sindbis virions resulting from exposure to low pH and interactions with cells suggest that cell penetration may occur at the cell surface in the absence of membrane fusion. Virology 2004, 324, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Vancini, R.; Wang, G.; Ferreira, D.; Hernandez, R.; Brown, D.T. Alphavirus Genome Delivery Occurs Directly at the Plasma Membrane in a Time- and Temperature-Dependent Process. J. Virol. 2013, 87, 4352–4359. [Google Scholar] [CrossRef] [PubMed]
- Cain, M.D.; Salimi, H.; Gong, Y.; Yang, L.; Hamilton, S.L.; Heffernan, J.R.; Hou, J.; Miller, M.J.; Klein, R.S. Virus entry and replication in the brain precedes blood-brain barrier disruption during intranasal alphavirus infection. J. Neuroimmunol. 2017, 308, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Bhomia, M.; Honnold, S.P.; Maheshwari, R.K. Role of adhesion molecules and inflammation in Venezuelan equine encephalitis virus infected mouse brain. Virology J. 2011, 8, 197. [Google Scholar] [CrossRef]
- Jackson, A.C.; Rossiter, J.P. Apoptotic cell death is an important cause of neuronal injury in experimental Venezuelan equine encephalitis virus infection of mice. Acta Neuropathol. 1997, 93, 349–353. [Google Scholar] [CrossRef]
- Schoneboom, B.A.; Catlin, K.M.; Marty, A.M.; Grieder, F.B. Inflammation is a component of neurodegeneration in response to Venezuelan equine encephalitis virus infection in mice. J. Neuroimmunol. 2000, 109, 132–146. [Google Scholar] [CrossRef]
- Risner, K.; Ahmed, A.; Bakovic, A.; Kortchak, S.; Bhalla, N.; Narayanan, A. Efficacy of FDA-Approved Anti-Inflammatory Drugs Against Venezuelan Equine Encephalitis Virus Infection. Viruses 2019, 11, 1151. [Google Scholar] [CrossRef]
- MacMicking, J.D. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012, 12, 367–382. [Google Scholar] [CrossRef]
- Crosse, K.M.; Monson, E.A.; Beard, M.R.; Helbig, K.J. Interferon-Stimulated Genes as Enhancers of Antiviral Innate Immune Signaling. J. Innate Immun. 2018, 10, 85–93. [Google Scholar] [CrossRef]
- Zhang, Y.; Burke, C.W.; Ryman, K.D.; Klimstra, W.B. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 2007, 81, 11246–11255. [Google Scholar] [CrossRef]
- Gall, B.; Pryke, K.; Abraham, J.; Mizuno, N.; Botto, S.; Sali, T.M.; Broeckel, R.; Haese, N.; Nilsen, A.; Placzek, A.; et al. Emerging Alphaviruses Are Sensitive to Cellular States Induced by a Novel Small-Molecule Agonist of the STING Pathway. J. Virol. 2018, 92, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Cavlar, T.; Deimling, T.; Ablasser, A.; Hopfner, K.P.; Hornung, V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013, 32, 1440–1450. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, X.; Qin, N.; Qiao, Y.; Xing, S.; Liu, W.; Feng, F.; Liu, Z.; Sun, H. STING, a promising target for small molecular immune modulator: A review. Eur. J. Med. Chem. 2021, 211, 113113. [Google Scholar] [CrossRef] [PubMed]
- Ekins, S.; Lane, T.R.; Madrid, P.B. Tilorone: A Broad-Spectrum Antiviral Invented in the USA and Commercialized in Russia and beyond. Pharm. Res. 2020, 37, 71. [Google Scholar] [CrossRef]
- Ekins, S.; Lingerfelt, M.A.; Comer, J.E.; Freiberg, A.N.; Mirsalis, J.C.; O’Loughlin, K.; Harutyunyan, A.; McFarlane, C.; Green, C.E.; Madrid, P.B. Efficacy of Tilorone Dihydrochloride against Ebola Virus Infection. Antimicrob. Agents Chemother. 2018, 62, 1–13. [Google Scholar] [CrossRef]
- Ekins, S.; Madrid, P.B. Tilorone, a Broad-Spectrum Antiviral for Emerging Viruses. Antimicrob. Agents Chemother. 2020, 64, 1–3. [Google Scholar] [CrossRef]
- Keyer, V.; Syzdykova, L.; Zauatbayeva, G.; Zhulikeyeva, A.; Ramanculov, Y.; Shustov, A.V.; Shulgau, Z. Tilorone and Cridanimod Protect Mice and Show Antiviral Activity in Rats despite Absence of the Interferon-Inducing Effect in Rats. Pharmaceuticals 2022, 15, 617. [Google Scholar] [CrossRef]
- Alam, S.; Javor, S.; Degardin, M.; Ajami, D.; Rebek, M.; Kissner, T.L.; Waag, D.M.; Rebek, J., Jr.; Saikh, K.U. Structure-Based Design and Synthesis of a Small Molecule that Exhibits Anti-inflammatory Activity by Inhibition of MyD88-mediated Signaling to Bacterial Toxin Exposure. Chem. Biol. Drug Des. 2015, 86, 200–209. [Google Scholar] [CrossRef]
- Sharma, A.; Maheshwari, R.K. Oligonucleotide array analysis of Toll-like receptors and associated signalling genes in Venezuelan equine encephalitis virus-infected mouse brain. J. Gen. Virol. 2009, 90, 1836–1847. [Google Scholar] [CrossRef]
- Fuse, K.; Chan, G.; Liu, Y.; Gudgeon, P.; Husain, M.; Chen, M.; Yeh, W.C.; Akira, S.; Liu, P.P. Myeloid differentiation factor-88 plays a crucial role in the pathogenesis of Coxsackievirus B3-induced myocarditis and influences type I interferon production. Circulation 2005, 112, 2276–2285. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, P.V.; Adams, A.P.; Wang, E.; Kang, W.; Carrara, A.S.; Anishchenko, M.; Frolov, I.; Weaver, S.C. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J. Virol. 2008, 82, 4920–4930. [Google Scholar] [CrossRef]
- Saikh, K.U.; Morazzani, E.M.; Piper, A.E.; Bakken, R.R.; Glass, P.J. A small molecule inhibitor of MyD88 exhibits broad spectrum antiviral activity by up regulation of type I interferon. Antivir. Res. 2020, 181, 104854. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, A.J.; Jans, D.A. Regulation of nucleocytoplasmic trafficking of viral proteins: An integral role in pathogenesis? Biochim. Biophys. Acta 2011, 1813, 2176–2190. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Wagstaff, K.M.; Jans, D.A. Nuclear trafficking of proteins from RNA viruses: Potential target for antivirals? Antivir. Res. 2012, 95, 202–206. [Google Scholar] [CrossRef]
- Atasheva, S.; Fish, A.; Fornerod, M.; Frolova, E.I. Venezuelan equine Encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin alpha/beta that obstructs nuclear pore complex function. J. Virol. 2010, 84, 4158–4171. [Google Scholar] [CrossRef]
- Ryman, K.D.; Klimstra, W.B. Host responses to alphavirus infection. Immunol. Rev. 2008, 225, 27–45. [Google Scholar] [CrossRef]
- Shechter, S.; Thomas, D.R.; Lundberg, L.; Pinkham, C.; Lin, S.-C.; Wagstaff, K.M.; Debono, A.; Kehn-Hall, K.; Jans, D.A. Novel inhibitors targeting Venezuelan equine encephalitis virus capsid protein identified using In Silico Structure-Based-Drug-Design. Sci. Rep. 2017, 7, 17705. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Rawlinson, S.M.; Hearps, A.C.; Jans, D.A. An AlphaScreen®-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen 2011, 16, 192–200. [Google Scholar] [CrossRef]
- Wagstaff, K.M.; Sivakumaran, H.; Heaton, S.M.; Harrich, D.; Jans, D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012, 443, 851–856. [Google Scholar] [CrossRef]
- Thomas, D.R.; Lundberg, L.; Pinkham, C.; Shechter, S.; DeBono, A.; Baell, J.; Wagstaff, K.M.; Hick, C.A.; Kehn-Hall, K.; Jans, D.A. Identification of novel antivirals inhibiting recognition of Venezuelan equine encephalitis virus capsid protein by the Importin α/β1 heterodimer through high-throughput screening. Antivir. Res. 2018, 151, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, L.; Fontenot, J.; Lin, S.C.; Pinkham, C.; Carey, B.D.; Campbell, C.E.; Kehn-Hall, K. Venezuelan Equine Encephalitis Virus Capsid Implicated in Infection-Induced Cell Cycle Delay in vitro. Front. Microbiol. 2018, 9, 3126. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.T.; Hwang, A.C.; Harper, C.C.; Stewart, F.H. Safety of mifepristone abortions in clinical use. Contraception 2005, 72, 175–178. [Google Scholar] [CrossRef]
- Schaff, E.A. Mifepristone: Ten years later. Contraception 2010, 81, 1–7. [Google Scholar] [CrossRef]
- Lundberg, L.; Pinkham, C.; Baer, A.; Amaya, M.; Narayanan, A.; Wagstaff, K.M.; Jans, D.A.; Kehn-Hall, K. Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antivir. Res. 2013, 100, 662–672. [Google Scholar] [CrossRef] [PubMed]
- DeBono, A.; Thomas, D.R.; Lundberg, L.; Pinkham, C.; Cao, Y.; Graham, J.D.; Clarke, C.L.; Wagstaff, K.M.; Shechter, S.; Kehn-Hall, K.; et al. Novel RU486 (mifepristone) analogues with increased activity against Venezuelan Equine Encephalitis Virus but reduced progesterone receptor antagonistic activity. Sci. Rep. 2019, 9, 2634. [Google Scholar] [CrossRef]
- Schor, S.; Einav, S. Repurposing of Kinase Inhibitors as Broad-Spectrum Antiviral Drugs. DNA Cell Biol. 2018, 37, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Laufer, S. Kinases as Potential Therapeutic Targets for Anti-coronaviral Therapy. J. Med. Chem. 2022, 65, 955–982. [Google Scholar] [CrossRef]
- García-Cárceles, J.; Caballero, E.; Gil, C.; Martínez, A. Kinase Inhibitors as Underexplored Antiviral Agents. J. Med. Chem. 2022, 65, 935–954. [Google Scholar] [CrossRef]
- Yi, F.; Guo, J.; Dabbagh, D.; Spear, M.; He, S.; Kehn-Hall, K.; Fontenot, J.; Yin, Y.; Bibian, M.; Park, C.M.; et al. Discovery of Novel Small-Molecule Inhibitors of LIM Domain Kinase for Inhibiting HIV-1. J. Virol. 2017, 91, 1–21. [Google Scholar] [CrossRef]
- Spear, M.; Wu, Y. Viral exploitation of actin: Force-generation and scaffolding functions in viral infection. Virol. Sin. 2014, 29, 139–147. [Google Scholar] [CrossRef]
- Vorster, P.J.; Guo, J.; Yoder, A.; Wang, W.; Zheng, Y.; Xu, X.; Yu, D.; Spear, M.; Wu, Y. LIM kinase 1 modulates cortical actin and CXCR4 cycling and is activated by HIV-1 to initiate viral infection. J. Biol. Chem. 2011, 286, 12554–12564. [Google Scholar] [CrossRef]
- Wen, X.; Ding, L.; Wang, J.-J.; Qi, M.; Hammonds, J.; Chu, H.; Chen, X.; Hunter, E.; Spearman, P. ROCK1 and LIM Kinase Modulate Retrovirus Particle Release and Cell-Cell Transmission Events. J. Virol. 2014, 88, 6906–6921. [Google Scholar] [CrossRef]
- Wilhelm, S.; Carter, C.; Lynch, M.; Lowinger, T.; Dumas, J.; Smith, R.A.; Schwartz, B.; Simantov, R.; Kelley, S. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat. Rev. Drug Discov. 2006, 5, 835–844. [Google Scholar] [CrossRef]
- Ibrahim, N.; Yu, Y.; Walsh, W.R.; Yang, J.L. Molecular targeted therapies for cancer: Sorafenib mono-therapy and its combination with other therapies (review). Oncol. Rep. 2012, 27, 1303–1311. [Google Scholar] [CrossRef]
- Wilhelm, S.M.; Adnane, L.; Newell, P.; Villanueva, A.; Llovet, J.M.; Lynch, M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol. Cancer Ther. 2008, 7, 3129–3140. [Google Scholar] [CrossRef]
- Lundberg, L.; Brahms, A.; Hooper, I.; Carey, B.; Lin, S.C.; Dahal, B.; Narayanan, A.; Kehn-Hall, K. Repurposed FDA-Approved drug sorafenib reduces replication of Venezuelan equine encephalitis virus and other alphaviruses. Antivir. Res. 2018, 157, 57–67. [Google Scholar] [CrossRef]
- Chen, K.F.; Tai, W.T.; Huang, J.W.; Hsu, C.Y.; Chen, W.L.; Cheng, A.L.; Chen, P.J.; Shiau, C.W. Sorafenib derivatives induce apoptosis through inhibition of STAT3 independent of Raf. Eur. J. Med. Chem. 2011, 46, 2845–2851. [Google Scholar] [CrossRef]
- Su, T.H.; Shiau, C.W.; Jao, P.; Liu, C.H.; Liu, C.J.; Tai, W.T.; Jeng, Y.M.; Yang, H.C.; Tseng, T.C.; Huang, H.P.; et al. Sorafenib and its derivative SC-1 exhibit antifibrotic effects through signal transducer and activator of transcription 3 inhibition. Proc. Natl. Acad. Sci. USA 2015, 112, 7243–7248. [Google Scholar] [CrossRef]
- Benedict, A.; Bansal, N.; Senina, S.; Hooper, I.; Lundberg, L.; de la Fuente, C.; Narayanan, A.; Gutting, B.; Kehn-Hall, K. Repurposing FDA-approved drugs as therapeutics to treat Rift Valley fever virus infection. Front. Microbiol. 2015, 6, 676. [Google Scholar] [CrossRef]
- Brahms, A.; Mudhasani, R.; Pinkham, C.; Kota, K.; Nasar, F.; Zamani, R.; Bavari, S.; Kehn-Hall, K. Sorafenib Impedes Rift Valley Fever Virus Egress by Inhibiting Valosin-Containing Protein Function in the Cellular Secretory Pathway. J. Virol. 2017, 91, 1–17. [Google Scholar] [CrossRef]
- Himmelsbach, K.; Sauter, D.; Baumert, T.F.; Ludwig, L.; Blum, H.E.; Hildt, E. New aspects of an anti-tumour drug: Sorafenib efficiently inhibits HCV replication. Gut 2009, 58, 1644–1653. [Google Scholar] [CrossRef]
- Lehman, C.W.; Kehn-Hall, K.; Aggarwal, M.; Bracci, N.R.; Pan, H.C.; Panny, L.; Lamb, R.A.; Lin, S.C. Resveratrol Inhibits Venezuelan Equine Encephalitis Virus Infection by Interfering with the AKT/GSK Pathway. Plants 2021, 10, 346. [Google Scholar] [CrossRef]
- Thaa, B.; Biasiotto, R.; Eng, K.; Neuvonen, M.; Götte, B.; Rheinemann, L.; Mutso, M.; Utt, A.; Varghese, F.; Balistreri, G.; et al. Differential Phosphatidylinositol-3-Kinase-Akt-mTOR Activation by Semliki Forest and Chikungunya Viruses Is Dependent on nsP3 and Connected to Replication Complex Internalization. J. Virol. 2015, 89, 11420–11437. [Google Scholar] [CrossRef]
- Mazzon, M.; Castro, C.; Thaa, B.; Liu, L.; Mutso, M.; Liu, X.; Mahalingam, S.; Griffin, J.L.; Marsh, M.; McInerney, G.M. Alphavirus-induced hyperactivation of PI3K/AKT directs pro-viral metabolic changes. PLoS Pathog. 2018, 14, e1006835. [Google Scholar] [CrossRef]
- Saqib, U.; Kelley, T.T.; Panguluri, S.K.; Liu, D.; Savai, R.; Baig, M.S.; Schürer, S.C. Polypharmacology or Promiscuity? Structural Interactions of Resveratrol With Its Bandwagon of Targets. Front. Pharmacol. 2018, 9, 1201. [Google Scholar] [CrossRef]
- Amaya, M.; Voss, K.; Sampey, G.; Senina, S.; de la Fuente, C.; Mueller, C.; Calvert, V.; Kehn-Hall, K.; Carpenter, C.; Kashanchi, F.; et al. The role of IKKβ in Venezuelan equine encephalitis virus infection. PLoS ONE 2014, 9, e86745. [Google Scholar] [CrossRef]
- Le Negrate, G. Viral interference with innate immunity by preventing NF-κB activity. Cell Microbiol. 2012, 14, 168–181. [Google Scholar] [CrossRef]
- Karin, M. The beginning of the end: IkappaB kinase (IKK) and NF-kappaB activation. J. Biol. Chem. 1999, 274, 27339–27342. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Signaling to NF-kappaB. Genes Dev. 2004, 18, 2195–2224. [Google Scholar] [CrossRef]
- Santoro, M.G.; Rossi, A.; Amici, C. NF-kappaB and virus infection: Who controls whom. EMBO J. 2003, 22, 2552–2560. [Google Scholar] [CrossRef]
- Bakovic, A.; Bhalla, N.; Kortchak, S.; Sun, C.; Zhou, W.; Ahmed, A.; Risner, K.; Klimstra, W.B.; Narayanan, A. Venezuelan Equine Encephalitis Virus nsP3 Phosphorylation Can Be Mediated by IKKβ Kinase Activity and Abrogation of Phosphorylation Inhibits Negative-Strand Synthesis. Viruses 2020, 12, 1021. [Google Scholar] [CrossRef]
- Ammosova, T.; Platonov, M.; Ivanov, A.; Kont, Y.S.; Kumari, N.; Kehn-Hall, K.; Jerebtsova, M.; Kulkarni, A.A.; Uren, A.; Kovalskyy, D.; et al. 1E7-03, a low MW compound targeting host protein phosphatase-1, inhibits HIV-1 transcription. Br. J. Pharmacol. 2014, 171, 5059–5075. [Google Scholar] [CrossRef]
- Lin, X.; Kumari, N.; DeMarino, C.; Kont, Y.S.; Ammosova, T.; Kulkarni, A.; Jerebtsova, M.; Vazquez-Meves, G.; Ivanov, A.; Dmytro, K.; et al. Inhibition of HIV-1 infection in humanized mice and metabolic stability of protein phosphatase-1-targeting small molecule 1E7-03. Oncotarget 2017, 8, 76749–76769. [Google Scholar] [CrossRef]
- Carey, B.D.; Ammosova, T.; Pinkham, C.; Lin, X.; Zhou, W.; Liotta, L.A.; Nekhai, S.; Kehn-Hall, K. Protein Phosphatase 1α Interacts with Venezuelan Equine Encephalitis Virus Capsid Protein and Regulates Viral Replication through Modulation of Capsid Phosphorylation. J. Virol. 2018, 92, 1–18. [Google Scholar] [CrossRef]
- Ellgaard, L.; Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003, 4, 181–191. [Google Scholar] [CrossRef]
- Singh, I.; Doms, R.W.; Wagner, K.R.; Helenius, A. Intracellular transport of soluble and membrane-bound glycoproteins: Folding, assembly and secretion of anchor-free influenza hemagglutinin. Embo J. 1990, 9, 631–639. [Google Scholar] [CrossRef]
- Mulvey, M.; Brown, D.T. Involvement of the molecular chaperone BiP in maturation of Sindbis virus envelope glycoproteins. J. Virol. 1995, 69, 1621–1627. [Google Scholar] [CrossRef]
- Choukhi, A.; Ung, S.; Wychowski, C.; Dubuisson, J. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 1998, 72, 3851–3858. [Google Scholar] [CrossRef]
- Little, E.; Ramakrishnan, M.; Roy, B.; Gazit, G.; Lee, A.S. The glucose-regulated proteins (GRP78 and GRP94): Functions, gene regulation, and applications. Crit. Rev. Eukaryot. Gene Expr. 1994, 4, 1–18. [Google Scholar] [CrossRef]
- Pfaffenbach, K.T.; Lee, A.S. The critical role of GRP78 in physiologic and pathologic stress. Curr. Opin. Cell Biol. 2011, 23, 150–156. [Google Scholar] [CrossRef]
- Barrera, M.D.; Callahan, V.; Akhrymuk, I.; Bhalla, N.; Zhou, W.; Campbell, C.; Narayanan, A.; Kehn-Hall, K. Proteomic Discovery of VEEV E2-Host Partner Interactions Identifies GRP78 Inhibitor HA15 as a Potential Therapeutic for Alphavirus Infections. Pathogens 2021, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Barke, J.; Brearley, C.; Hill, L.; Yu, D.W.; Goss, R.J.M.; Hutchings, M.I. A Single Streptomyces Symbiont Makes Multiple Antifungals to Support the Fungus Farming Ant Acromyrmex octospinosus. PLoS ONE 2011, 6, e22028. [Google Scholar] [CrossRef] [PubMed]
- Shum, D.; Smith, J.L.; Hirsch, A.J.; Bhinder, B.; Radu, C.; Stein, D.A.; Nelson, J.A.; Früh, K.; Djaballah, H. High-content assay to identify inhibitors of dengue virus infection. Assay Drug Dev. Technol. 2010, 8, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Hui, E.K.-W.; Nayak, D.P. Role of ATP in Influenza Virus Budding. Virology 2001, 290, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Raveh, A.; Delekta, P.C.; Dobry, C.J.; Peng, W.; Schultz, P.J.; Blakely, P.K.; Tai, A.W.; Matainaho, T.; Irani, D.N.; Sherman, D.H.; et al. Discovery of Potent Broad Spectrum Antivirals Derived from Marine Actinobacteria. PLoS ONE 2013, 8, e82318. [Google Scholar] [CrossRef]
- Hamilton, P.B.; Carroll, F.I.; Rutledge, J.H.; Mason, J.E.; Harris, B.S.; Fenske, C.S.; Wall, M.E. Simple isolation of antimycin A1 and some of its toxicological properties. Appl. Microbiol. 1969, 17, 102–105. [Google Scholar] [CrossRef]
- Kasprzyk, R.; Jemielity, J. Enzymatic Assays to Explore Viral mRNA Capping Machinery. ChemBioChem 2021, 22, 3236–3253. [Google Scholar] [CrossRef]
- Ferreira-Ramos, A.S.; Li, C.; Eydoux, C.; Contreras, J.M.; Morice, C.; Quérat, G.; Gigante, A.; Pérez Pérez, M.J.; Jung, M.L.; Canard, B.; et al. Approved drugs screening against the nsP1 capping enzyme of Venezuelan equine encephalitis virus using an immuno-based assay. Antivir. Res. 2019, 163, 59–69. [Google Scholar] [CrossRef]
- Li, C.; Guillén, J.; Rabah, N.; Blanjoie, A.; Debart, F.; Vasseur, J.J.; Canard, B.; Decroly, E.; Coutard, B. mRNA Capping by Venezuelan Equine Encephalitis Virus nsP1: Functional Characterization and Implications for Antiviral Res.earch. J. Virol. 2015, 89, 8292–8303. [Google Scholar] [CrossRef]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2012, 10, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Ferron, F.; Decroly, E.; Selisko, B.; Canard, B. The viral RNA capping machinery as a target for antiviral drugs. Antivir. Res. 2012, 96, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Rabah, N.; Ortega Granda, O.; Quérat, G.; Canard, B.; Decroly, E.; Coutard, B. Mutations on VEEV nsP1 relate RNA capping efficiency to ribavirin susceptibility. Antivir. Res. 2020, 182, 104883. [Google Scholar] [CrossRef] [PubMed]
- Delang, L.; Li, C.; Tas, A.; Quérat, G.; Albulescu, I.C.; De Burghgraeve, T.; Guerrero, N.A.; Gigante, A.; Piorkowski, G.; Decroly, E.; et al. The viral capping enzyme nsP1: A novel target for the inhibition of chikungunya virus infection. Sci. Rep. 2016, 6, 31819. [Google Scholar] [CrossRef] [PubMed]
- Rupp, J.C.; Sokoloski, K.J.; Gebhart, N.N.; Hardy, R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015, 96, 2483–2500. [Google Scholar] [CrossRef] [PubMed]
- Abu Bakar, F.; Ng, L.F.P. Nonstructural Proteins of Alphavirus-Potential Targets for Drug Development. Viruses 2018, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Bozóki, B.; Mótyán, J.A.; Hoffka, G.; Waugh, D.S.; Tőzsér, J. Specificity Studies of the Venezuelan Equine Encephalitis Virus Non-Structural Protein 2 Protease Using Recombinant Fluorescent Substrates. Int. J. Mol. Sci. 2020, 21, 7686. [Google Scholar] [CrossRef]
- Morazzani, E.M.; Compton, J.R.; Leary, D.H.; Berry, A.V.; Hu, X.; Marugan, J.J.; Glass, P.J.; Legler, P.M. Proteolytic cleavage of host proteins by the Group IV viral proteases of Venezuelan equine encephalitis virus and Zika virus. Antivir. Res. 2019, 164, 106–122. [Google Scholar] [CrossRef]
- Rikkonen, M.; Peränen, J.; Kääriäinen, L. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 1994, 68, 5804–5810. [Google Scholar] [CrossRef]
- Gomez de Cedrón, M.; Ehsani, N.; Mikkola, M.L.; García, J.A.; Kääriäinen, L. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 1999, 448, 19–22. [Google Scholar] [CrossRef]
- Vasiljeva, L.; Valmu, L.; Kääriäinen, L.; Merits, A. Site-specific protease activity of the carboxyl-terminal domain of Semliki Forest virus replicase protein nsP2. J. Biol. Chem. 2001, 276, 30786–30793. [Google Scholar] [CrossRef]
- Vasiljeva, L.; Merits, A.; Auvinen, P.; Kääriäinen, L. Identification of a novel function of the alphavirus capping apparatus. RNA 5’-triphosphatase activity of Nsp2. J. Biol. Chem. 2000, 275, 17281–17287. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.T.; White, M.A.; Watowich, S.J. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 2006, 14, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Compton, J.R.; Leary, D.H.; Olson, M.A.; Lee, M.S.; Cheung, J.; Ye, W.; Ferrer, M.; Southall, N.; Jadhav, A.; et al. Kinetic, Mutational, and Structural Studies of the Venezuelan Equine Encephalitis Virus Nonstructural Protein 2 Cysteine Protease. Biochemistry 2016, 55, 3007–3019. [Google Scholar] [CrossRef]
- Zhang, H.; Harmon, M.; Radoshitzky, S.R.; Soloveva, V.; Kane, C.D.; Duplantier, A.J.; Ogungbe, I.V. Vinyl Sulfone-Based Inhibitors of Nonstructural Protein 2 Block the Replication of Venezuelan Equine Encephalitis Virus. ACS MedChem Lett. 2020, 11, 2139–2145. [Google Scholar] [CrossRef]
- Haese, N.N.; May, N.A.; Taft-Benz, S.; Moukha-Chafiq, O.; Madadi, N.; Zhang, S.; Karyakarte, S.D.; Rodzinak, K.J.; Nguyen, T.H.; Denton, M.; et al. Identification of Quinolinones as Antivirals against Venezuelan Equine Encephalitis Virus. Antimicrob. Agents Chemother. 2021, 65, e0024421. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef]
- Wang, Y.F.; Sawicki, S.G.; Sawicki, D.L. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 1994, 68, 6466–6475. [Google Scholar] [CrossRef]
- Götte, B.; Liu, L.; McInerney, G.M. The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last. Viruses 2018, 10, 105. [Google Scholar] [CrossRef]
- Frolov, I.; Kim, D.Y.; Akhrymuk, M.; Mobley, J.A.; Frolova, E.I. Hypervariable Domain of Eastern Equine Encephalitis Virus nsP3 Redundantly Utilizes Multiple Cellular Proteins for Replication Complex Assembly. J. Virol. 2017, 91, 1–22. [Google Scholar] [CrossRef]
- Kim, D.Y.; Reynaud, J.M.; Rasalouskaya, A.; Akhrymuk, I.; Mobley, J.A.; Frolov, I.; Frolova, E.I. New World and Old World Alphaviruses Have Evolved to Exploit Different Components of Stress Granules, FXR and G3BP Proteins, for Assembly of Viral Replication Complexes. PLoS Pathog. 2016, 12, e1005810. [Google Scholar] [CrossRef]
- Lark, T.; Keck, F.; Narayanan, A. Interactions of Alphavirus nsP3 Protein with Host Proteins. Front. Microbiol. 2018, 8, 2652. [Google Scholar] [CrossRef]
- Foy, N.J.; Akhrymuk, M.; Akhrymuk, I.; Atasheva, S.; Bopda-Waffo, A.; Frolov, I.; Frolova, E.I. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J. Virol. 2013, 87, 1997–2010. [Google Scholar] [CrossRef]
- Nowee, G.; Bakker, J.W.; Geertsema, C.; Ros, V.I.D.; Göertz, G.P.; Fros, J.J.; Pijlman, G.P. A Tale of 20 Alphaviruses; Inter-species Diversity and Conserved Interactions Between Viral Non-structural Protein 3 and Stress Granule Proteins. Front. Cell Dev. Biol. 2021, 9, 625711. [Google Scholar] [CrossRef]
- Amaya, M.; Brooks-Faulconer, T.; Lark, T.; Keck, F.; Bailey, C.; Raman, V.; Narayanan, A. Venezuelan equine encephalitis virus non-structural protein 3 (nsP3) interacts with RNA helicases DDX1 and DDX3 in infected cells. Antivir. Res. 2016, 131, 49–60. [Google Scholar] [CrossRef]
- Bakovic, A.; Bhalla, N.; Alem, F.; Campbell, C.; Zhou, W.; Narayanan, A. Inhibitors of Venezuelan Equine Encephalitis Virus Identified Based on Host Interaction Partners of Viral Non-Structural Protein 3. Viruses 2021, 13, 1533. [Google Scholar] [CrossRef]
- Tomar, S.; Hardy, R.W.; Smith, J.L.; Kuhn, R.J. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 2006, 80, 9962–9969. [Google Scholar] [CrossRef]
- Chen, M.W.; Tan, Y.B.; Zheng, J.; Zhao, Y.; Lim, B.T.; Cornvik, T.; Lescar, J.; Ng, L.F.P.; Luo, D. Chikungunya virus nsP4 RNA-dependent RNA polymerase core domain displays detergent-sensitive primer extension and terminal adenylyltransferase activities. Antivir. Res. 2017, 143, 38–47. [Google Scholar] [CrossRef]
- Rubach, J.K.; Wasik, B.R.; Rupp, J.C.; Kuhn, R.J.; Hardy, R.W.; Smith, J.L. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 2009, 384, 201–208. [Google Scholar] [CrossRef]
- Ollis, D.L.; Brick, P.; Hamlin, R.; Xuong, N.G.; Steitz, T.A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 1985, 313, 762–766. [Google Scholar] [CrossRef]
- van der Heijden, M.W.; Bol, J.F. Composition of alphavirus-like replication complexes: Involvement of virus and host encoded proteins. Arch. Virol. 2002, 147, 875–898. [Google Scholar] [CrossRef] [PubMed]
- Malone, B.; Campbell, E.A. Molnupiravir: Coding for catastrophe. Nat. Struct. Mol. Biol. 2021, 28, 706–708. [Google Scholar] [CrossRef] [PubMed]
- Urakova, N.; Kuznetsova, V.; Crossman, D.K.; Sokratian, A.; Guthrie, D.B.; Kolykhalov, A.A.; Lockwood, M.A.; Natchus, M.G.; Crowley, M.R.; Painter, G.R.; et al. β-d-N(4)-Hydroxycytidine Is a Potent Anti-alphavirus Compound That Induces a High Level of Mutations in the Viral Genome. J. Virol. 2018, 92, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Stuyver, L.J.; Whitaker, T.; McBrayer, T.R.; Hernandez-Santiago, B.I.; Lostia, S.; Tharnish, P.M.; Ramesh, M.; Chu, C.K.; Jordan, R.; Shi, J.; et al. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 2003, 47, 244–254. [Google Scholar] [CrossRef]
- Costantini, V.P.; Whitaker, T.; Barclay, L.; Lee, D.; McBrayer, T.R.; Schinazi, R.F.; Vinjé, J. Antiviral activity of nucleoside analogues against norovirus. Antivir. Ther. 2012, 17, 981–991. [Google Scholar] [CrossRef]
- Ehteshami, M.; Tao, S.; Zandi, K.; Hsiao, H.M.; Jiang, Y.; Hammond, E.; Amblard, F.; Russell, O.O.; Merits, A.; Schinazi, R.F. Characterization of β-d-N(4)-Hydroxycytidine as a Novel Inhibitor of Chikungunya Virus. Antimicrob. Agents Chemother. 2017, 61, 1–6. [Google Scholar] [CrossRef]
- Painter, G.R.; Bowen, R.A.; Bluemling, G.R.; DeBergh, J.; Edpuganti, V.; Gruddanti, P.R.; Guthrie, D.B.; Hager, M.; Kuiper, D.L.; Lockwood, M.A.; et al. The prophylactic and therapeutic activity of a broadly active ribonucleoside analog in a murine model of intranasal venezuelan equine encephalitis virus infection. Antivir. Res. 2019, 171, 104597. [Google Scholar] [CrossRef]
- Schroeder, C.E.; Yao, T.; Sotsky, J.; Smith, R.A.; Roy, S.; Chu, Y.K.; Guo, H.; Tower, N.A.; Noah, J.W.; McKellip, S.; et al. Development of (E)-2-((1,4-dimethylpiperazin-2-ylidene)amino)-5-nitro-N-phenylbenzamide, ML336: Novel 2-amidinophenylbenzamides as potent inhibitors of venezuelan equine encephalitis virus. J. Med. Chem. 2014, 57, 8608–8621. [Google Scholar] [CrossRef]
- LaBauve, A.E.; Rinker, T.E.; Noureddine, A.; Serda, R.E.; Howe, J.Y.; Sherman, M.B.; Rasley, A.; Brinker, C.J.; Sasaki, D.Y.; Negrete, O.A. Lipid-Coated Mesoporous Silica Nanoparticles for the Delivery of the ML336 Antiviral to Inhibit Encephalitic Alphavirus Infection. Sci. Rep. 2018, 8, 13990. [Google Scholar] [CrossRef]
- Skidmore, A.M.; Adcock, R.S.; Jonsson, C.B.; Golden, J.E.; Chung, D.H. Benzamidine ML336 inhibits plus and minus strand RNA synthesis of Venezuelan equine encephalitis virus without affecting host RNA production. Antivir. Res. 2020, 174, 104674. [Google Scholar] [CrossRef]
- Jonsson, C.B.; Cao, X.; Lee, J.; Gabbard, J.D.; Chu, Y.K.; Fitzpatrick, E.A.; Julander, J.; Chung, D.H.; Stabenow, J.; Golden, J.E. Efficacy of a ML336 derivative against Venezuelan and eastern equine encephalitis viruses. Antivir. Res. 2019, 167, 25–34. [Google Scholar] [CrossRef]
- Lee, J.; Parvathareddy, J.; Yang, D.; Bansal, S.; O’Connell, K.; Golden, J.E.; Jonsson, C.B. Emergence and Magnitude of ML336 Resistance in Venezuelan Equine Encephalitis Virus Depend on the Microenvironment. J. Virol. 2020, 94, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Haese, N.N.; Madadi, N.; Sarkar, S.; Bonin, K.; Streblow, C.E.; Taft-Benz, S.; Tower, N.A.; Rasmussen, L.; Bostwick, R.; et al. Studies on Dibenzylamines as Inhibitors of Venezuelan Equine Encephalitis Virus. ACS Infect. Dis. 2019, 5, 2014–2028. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.C.; Kim, E.; Cao, X.; Reichard, W.; Ogorek, T.J.; Das, P.; Jonsson, C.B.; Baudry, J.; Chung, D.; Golden, J.E. Piperazinobenzodiazepinones: New Encephalitic Alphavirus Inhibitors via Ring Expansion of 2-Dichloromethylquinazolinones. ACS Med. Chem. Lett. 2022, 13, 546–553. [Google Scholar] [CrossRef]
- Ahmed, S.K.; Haese, N.N.; Cowan, J.T.; Pathak, V.; Moukha-Chafiq, O.; Smith, V.J.; Rodzinak, K.J.; Ahmad, F.; Zhang, S.; Bonin, K.M.; et al. Targeting Chikungunya Virus Replication by Benzoannulene Inhibitors. J. Med. Chem. 2021, 64, 4762–4786. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Bushell, S.; Qing, M.; Xu, H.Y.; Bonavia, A.; Nunes, S.; Zhou, J.; Poh, M.K.; Florez de Sessions, P.; Niyomrattanakit, P.; et al. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J. Virol. 2011, 85, 6548–6556. [Google Scholar] [CrossRef] [PubMed]
- Luthra, P.; Naidoo, J.; Pietzsch, C.A.; De, S.; Khadka, S.; Anantpadma, M.; Williams, C.G.; Edwards, M.R.; Davey, R.A.; Bukreyev, A.; et al. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antivir. Res. 2018, 158, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Z.; Wang, R.; Zhang, Z.R.; Zhang, Y.N.; Deng, C.L.; Zhang, B.; Shang, L.Q.; Ye, H.Q. In Vitro Inhibition of Alphaviruses by Lycorine. Virol. Sin. 2021, 36, 1465–1474. [Google Scholar] [CrossRef]
- Roy, M.; Liang, L.; Xiao, X.; Feng, P.; Ye, M.; Liu, J. Lycorine: A prospective natural lead for anticancer drug discovery. Biomed. Pharmacother. 2018, 107, 615–624. [Google Scholar] [CrossRef]
- Wang, H.; Guo, T.; Yang, Y.; Yu, L.; Pan, X.; Li, Y. Lycorine Derivative LY-55 Inhibits EV71 and CVA16 Replication Through Downregulating Autophagy. Front. Cell Infect. Microbiol. 2019, 9, 277. [Google Scholar] [CrossRef]
- He, J.; Qi, W.B.; Wang, L.; Tian, J.; Jiao, P.R.; Liu, G.Q.; Ye, W.C.; Liao, M. Amaryllidaceae alkaloids inhibit nuclear-to-cytoplasmic export of ribonucleoprotein (RNP) complex of highly pathogenic avian influenza virus H5N1. Influenza Other Respir. Viruses 2013, 7, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, Y.; Xu, Y.; Ma, C.; Qin, C.; Zhang, L. Lycorine reduces mortality of human enterovirus 71-infected mice by inhibiting virus replication. Virol. J. 2011, 8, 483. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, Y.; Cao, L.; Wang, P.; Qing, J.; Zheng, Q.; Shang, L.; Yin, Z.; Sun, Y. A Conserved Inhibitory Mechanism of a Lycorine Derivative against Enterovirus and Hepatitis C Virus. Antimicrob. Agents Chemother. 2016, 60, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.C.; Lehman, C.W.; Stewart, A.K.; Panny, L.; Bracci, N.; Wright, J.L.C.; Paige, M.; Strangman, W.K.; Kehn-Hall, K. Homoseongomycin, a compound isolated from marine actinomycete bacteria K3-1, is a potent inhibitor of encephalitic alphaviruses. Antivir. Res. 2021, 191, 105087. [Google Scholar] [CrossRef] [PubMed]
- Karpenko, I.; Deev, S.; Kiselev, O.; Charushin, V.; Rusinov, V.; Ulomsky, E.; Deeva, E.; Yanvarev, D.; Ivanov, A.; Smirnova, O.; et al. Antiviral properties, metabolism, and pharmacokinetics of a novel azolo-1,2,4-triazine-derived inhibitor of influenza A and B virus replication. Antimicrob. Agents Chemother. 2010, 54, 2017–2022. [Google Scholar] [CrossRef]
- Sapozhnikova, I.M.; Ulomsky, E.N.; Rusinov, V.L.; Chupakhin, O.N.; Stepanov, A.V.; Savateeva-Lyubimova, T.N.; Sivak, K.V. 3-Cyanoazolo [5,1-c][1,2,4]triazines: Synthesis and antiviral activity. Chem. Heterocycl. Comp. 2021, 57, 467–472. [Google Scholar] [CrossRef]
- Sindac, J.A.; Barraza, S.J.; Dobry, C.J.; Xiang, J.; Blakely, P.K.; Irani, D.N.; Keep, R.F.; Miller, D.J.; Larsen, S.D. Optimization of Novel Indole-2-carboxamide Inhibitors of Neurotropic Alphavirus Replication. J. Med. Chem. 2013, 56, 9222–9241. [Google Scholar] [CrossRef]
- Barraza, S.J.; Sindac, J.A.; Dobry, C.J.; Delekta, P.C.; Lee, P.H.; Miller, D.J.; Larsen, S.D. Synthesis and biological activity of conformationally restricted indole-based inhibitors of neurotropic alphavirus replication: Generation of a three-dimensional pharmacophore. Bioorg. Med. Chem. Lett. 2021, 46, 128171. [Google Scholar] [CrossRef]
- Saul, S.; Huang, P.T.; Einav, S.; Asquith, C.R.M. Identification and evaluation of 4-anilinoquin(az)olines as potent inhibitors of both dengue virus (DENV) and Venezuelan equine encephalitis virus (VEEV). Bioorg. Med. Chem. Lett. 2021, 52, 128407. [Google Scholar] [CrossRef]
- Huang, P.T.; Saul, S.; Einav, S.; Asquith, C.R.M. Optimization of 4-Anilinoquinolines as Dengue Virus Inhibitors. Molecules 2021, 26, 7338. [Google Scholar] [CrossRef]
- NIAID. NIAID Emerging Infectious Diseases/Pathogens | Category B Pathogens. Available online: https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens (accessed on 31 January 2023).
- Villari, P.; Spielman, A.; Komar, N.; McDowell, M.; Timperi, R.J. The economic burden imposed by a residual case of eastern encephalitis. Am. J. Trop. Med. Hyg. 1995, 52, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Earnest, M.P.; Goolishian, H.A.; Calverley, J.R.; Hayes, R.O.; Hill, H.R. Neurologic, intellectual, and psychologic sequelae following western encephalitis. A follow-up study of 35 cases. Neurology 1971, 21, 969–974. [Google Scholar] [CrossRef]
- Allio, T. The FDA Animal Rule and its role in protecting human safety. Expert Opin. Drug Saf. 2018, 17, 971–973. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, P. FDA Experience with Medical Countermeasures under the Animal Rule. Adv. Prev. Med. 2012, 2012, 507571. [Google Scholar] [CrossRef] [PubMed]
- Painter, G.R.; Natchus, M.G.; Cohen, O.; Holman, W.; Painter, W.P. Developing a direct acting, orally available antiviral agent in a pandemic: The evolution of molnupiravir as a potential treatment for COVID-19. Curr. Opin. Virol. 2021, 50, 17–22. [Google Scholar] [CrossRef]
- Jonsson, C.B.; Golden, J.E.; Meibohm, B. Time to ‘Mind the Gap’ in novel small molecule drug discovery for direct-acting antivirals for SARS-CoV-2. Curr. Opin. Virol. 2021, 50, 1–7. [Google Scholar] [CrossRef]
- Morens, D.M.; Folkers, G.K.; Fauci, A.S. Eastern Equine Encephalitis Virus—Another Emergent Arbovirus in the United States. N. Engl. J. Med. 2019, 381, 1989–1992. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).