Soybean Thrips (Thysanoptera: Thripidae) Harbor Highly Diverse Populations of Arthropod, Fungal and Plant Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Processing of Soybean Thrips Samples
2.2. Bioinformatic Analyses
2.3. Nucleotide Sequence Accession Numbers
3. Results
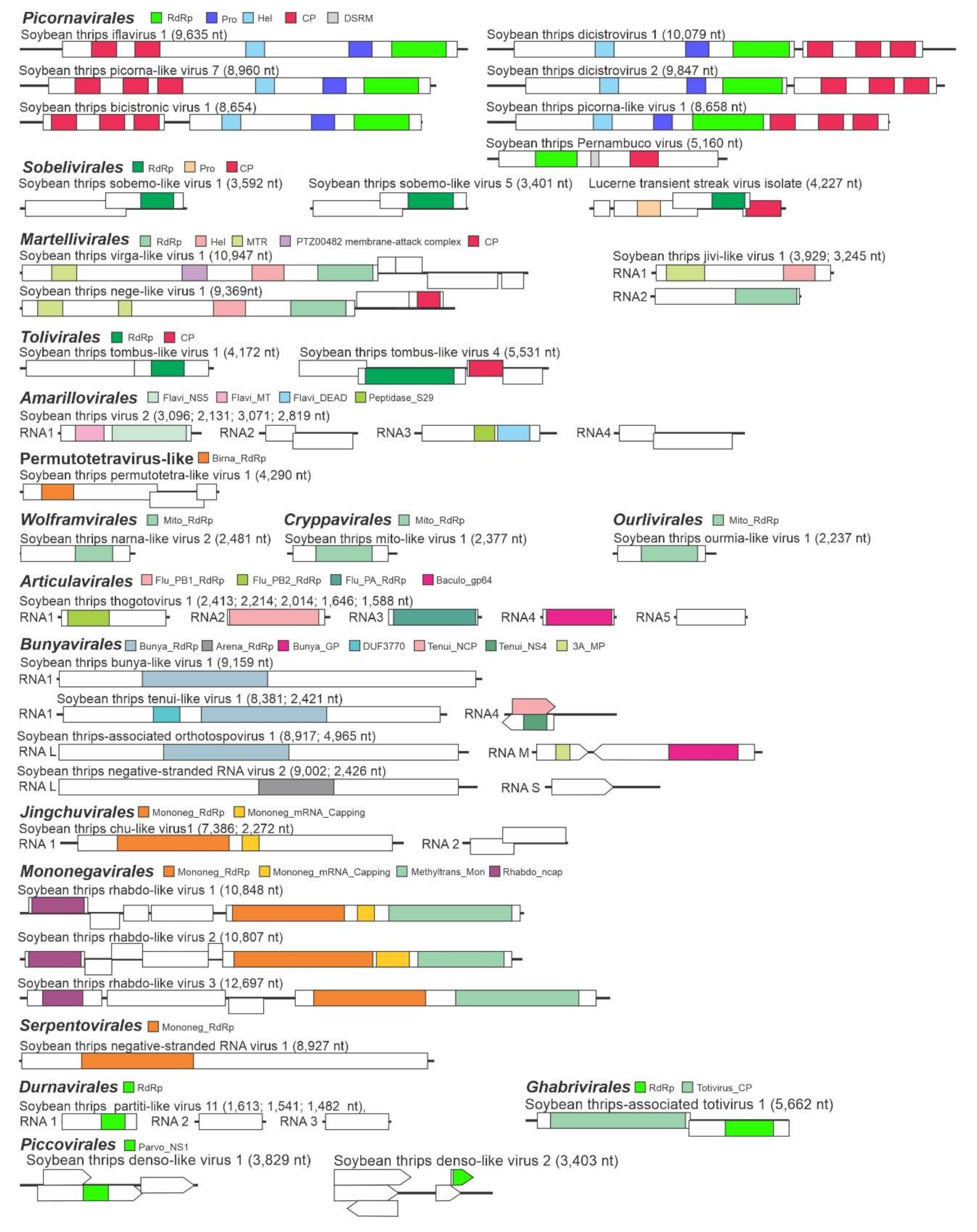
3.1. Virus-Like Sequences Associated with Soybean Thrips
3.2. Sequences Related to Single-Stranded Positive-Sense RNA Viruses
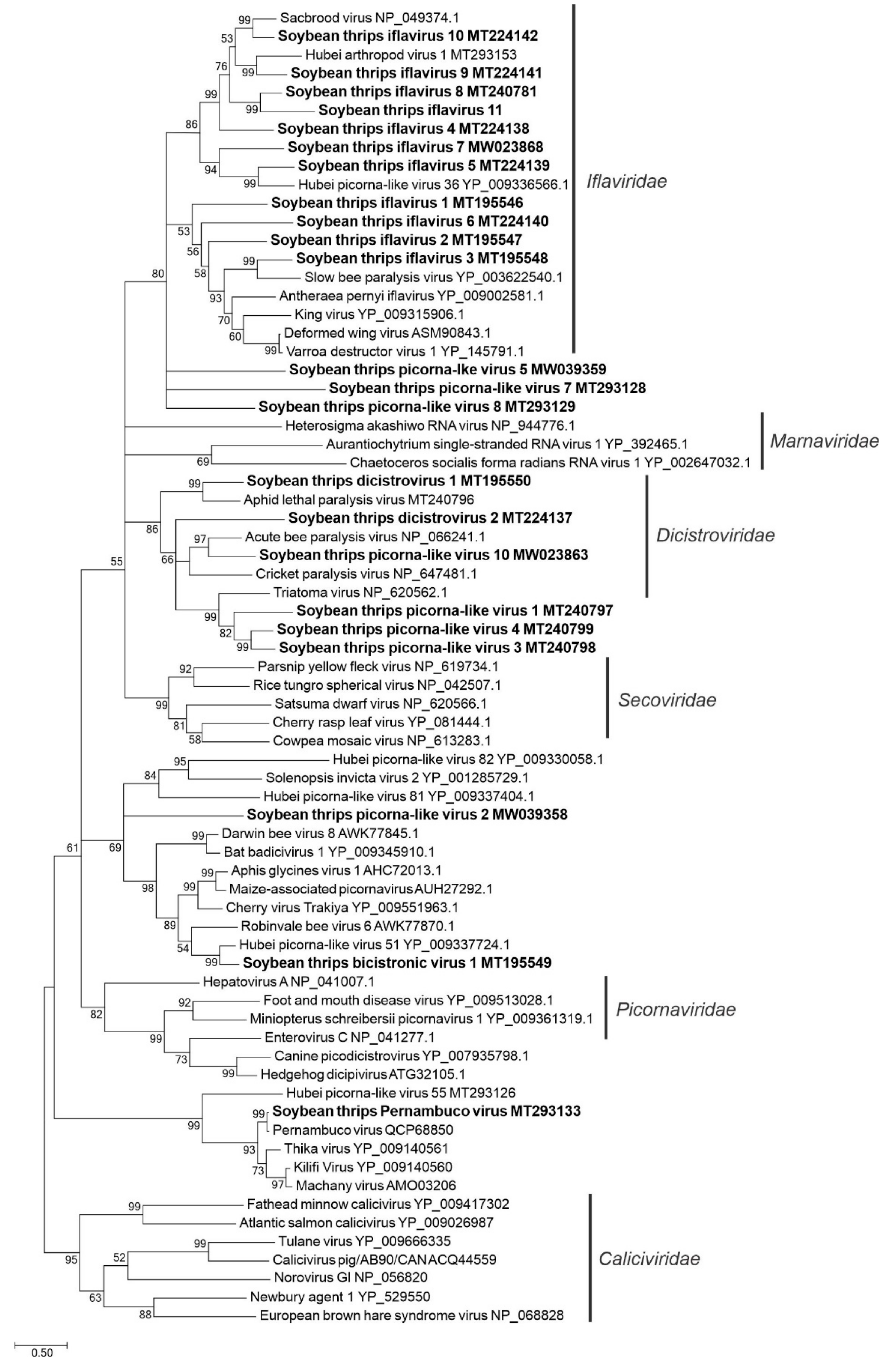
3.2.1. Sequences Related to Members of the Picornavirales
3.2.2. Sequences Related to Members of the Sobelivirales
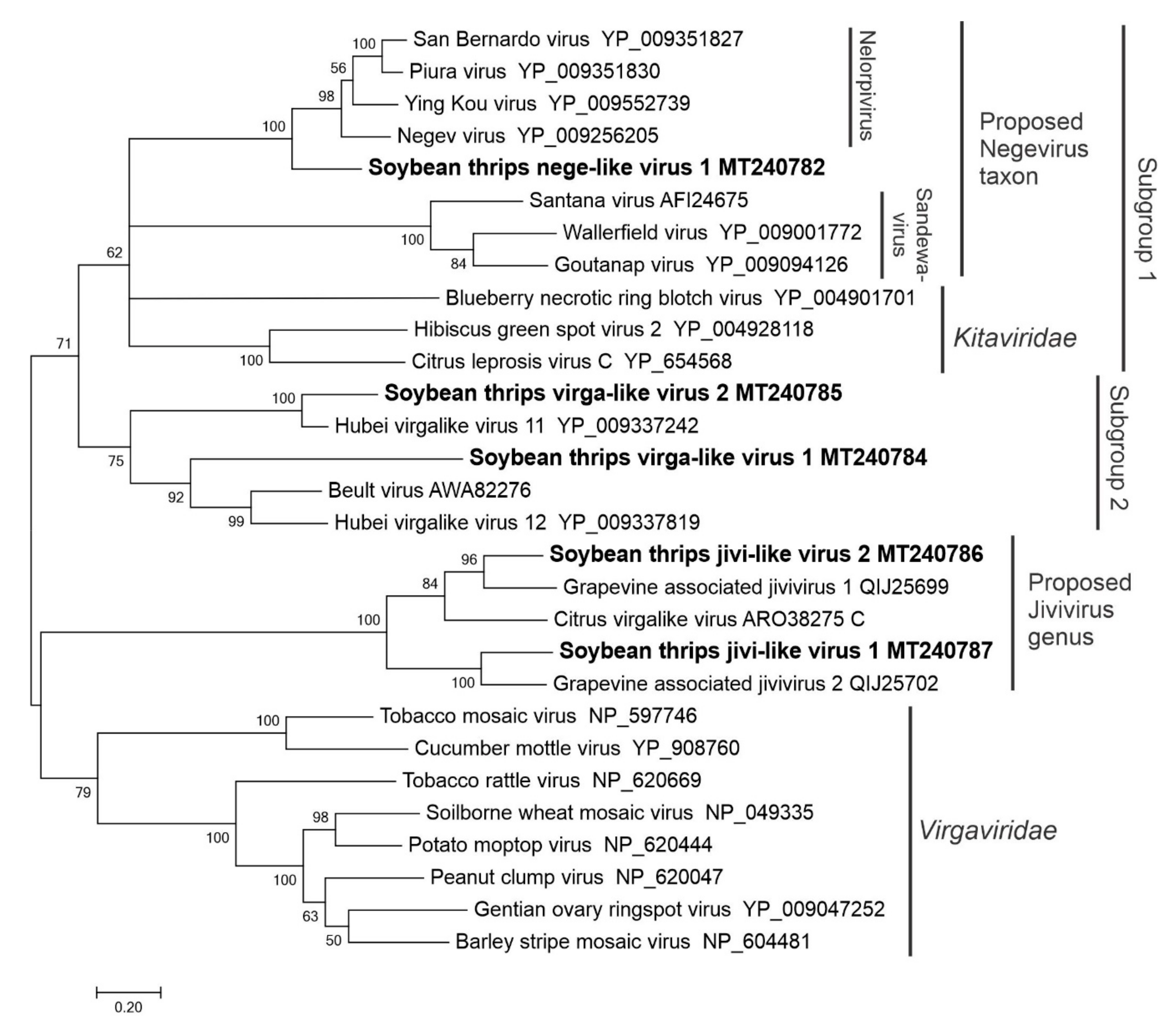
3.2.3. Sequences Related to Members of the Martellivirales
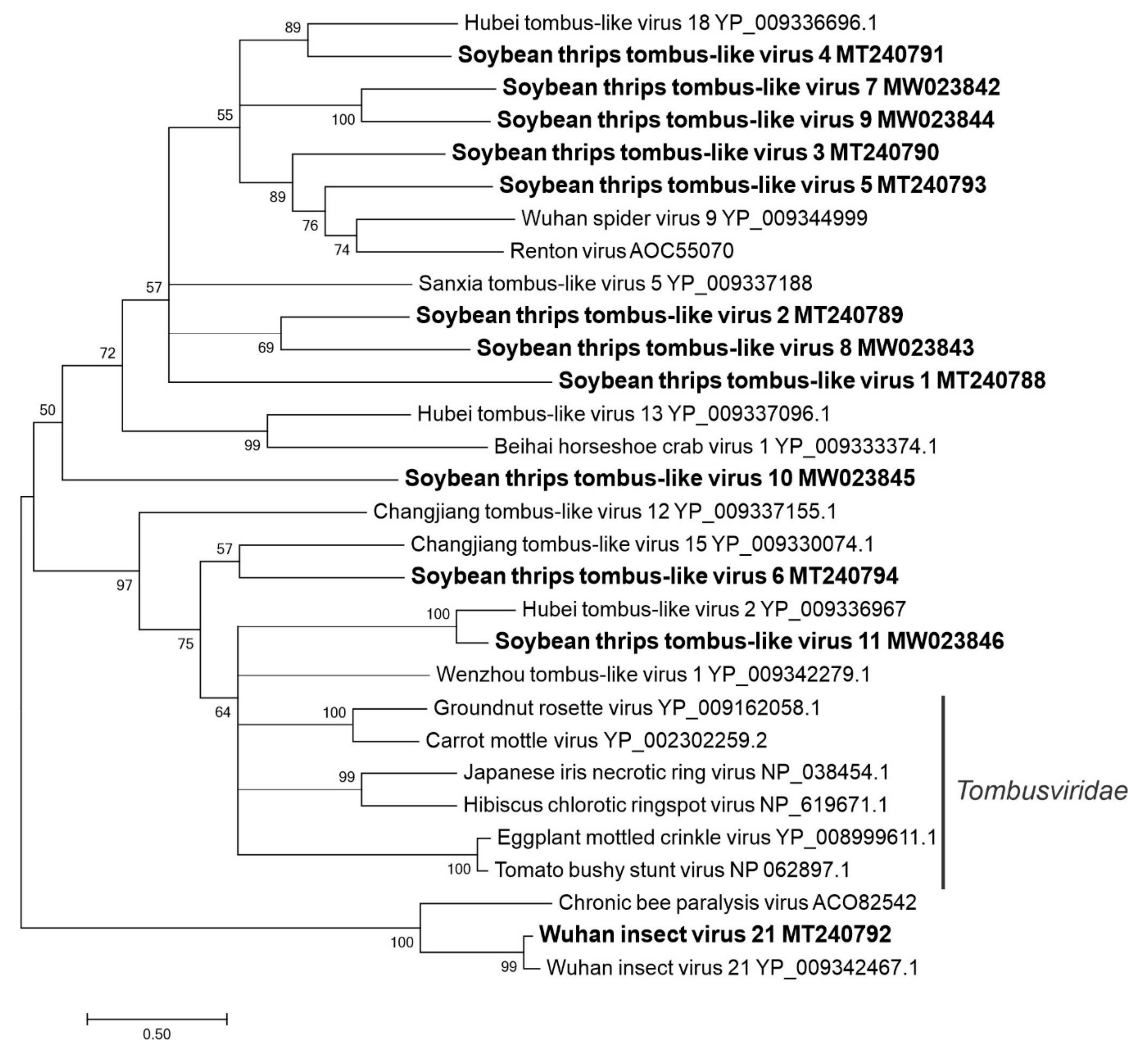
3.2.4. Sequences Related to Members of the Tolivirales
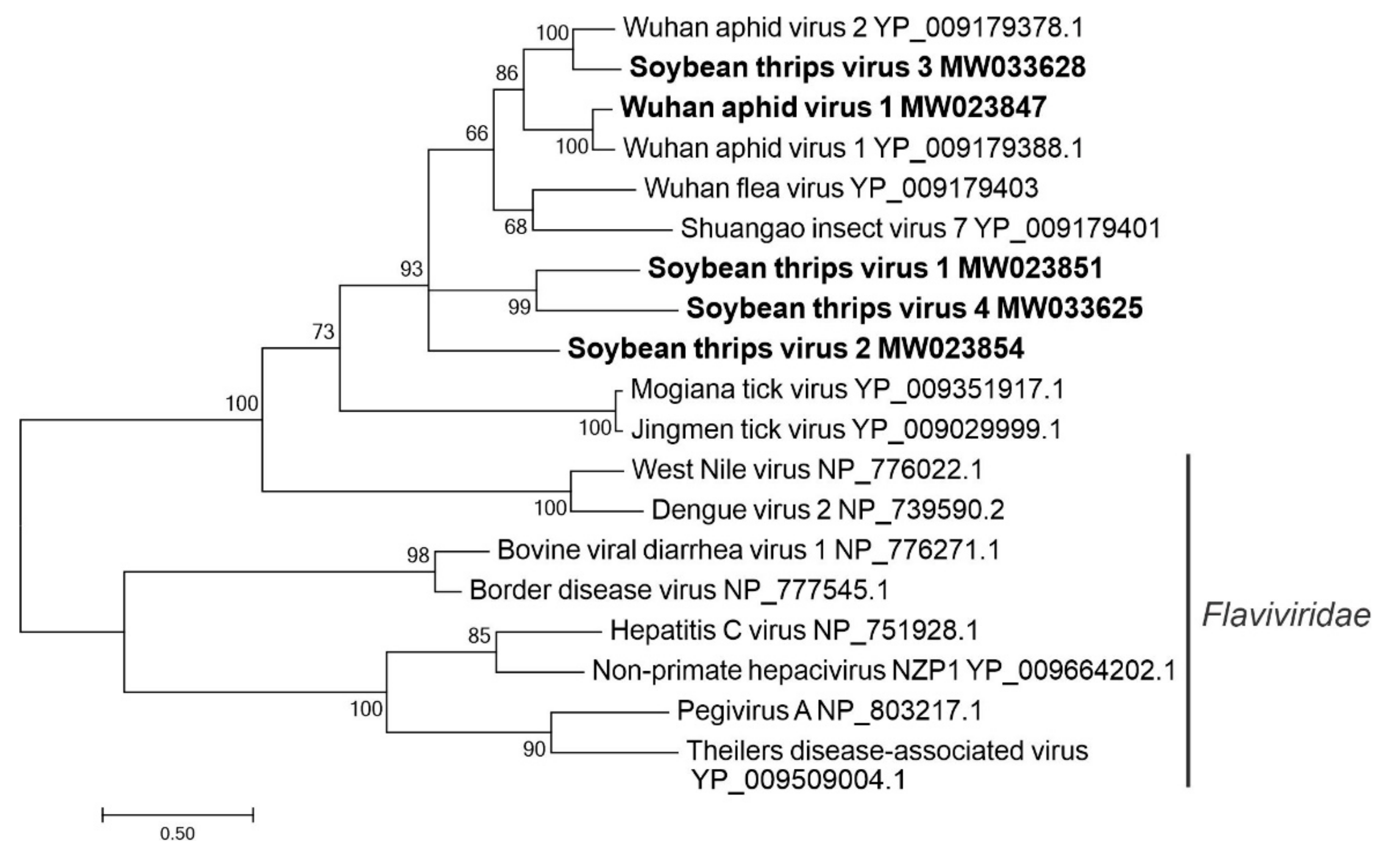
3.2.5. Sequences Related to Members of the Amarillovirales
3.2.6. Sequences Related to Members of the Tymovirales
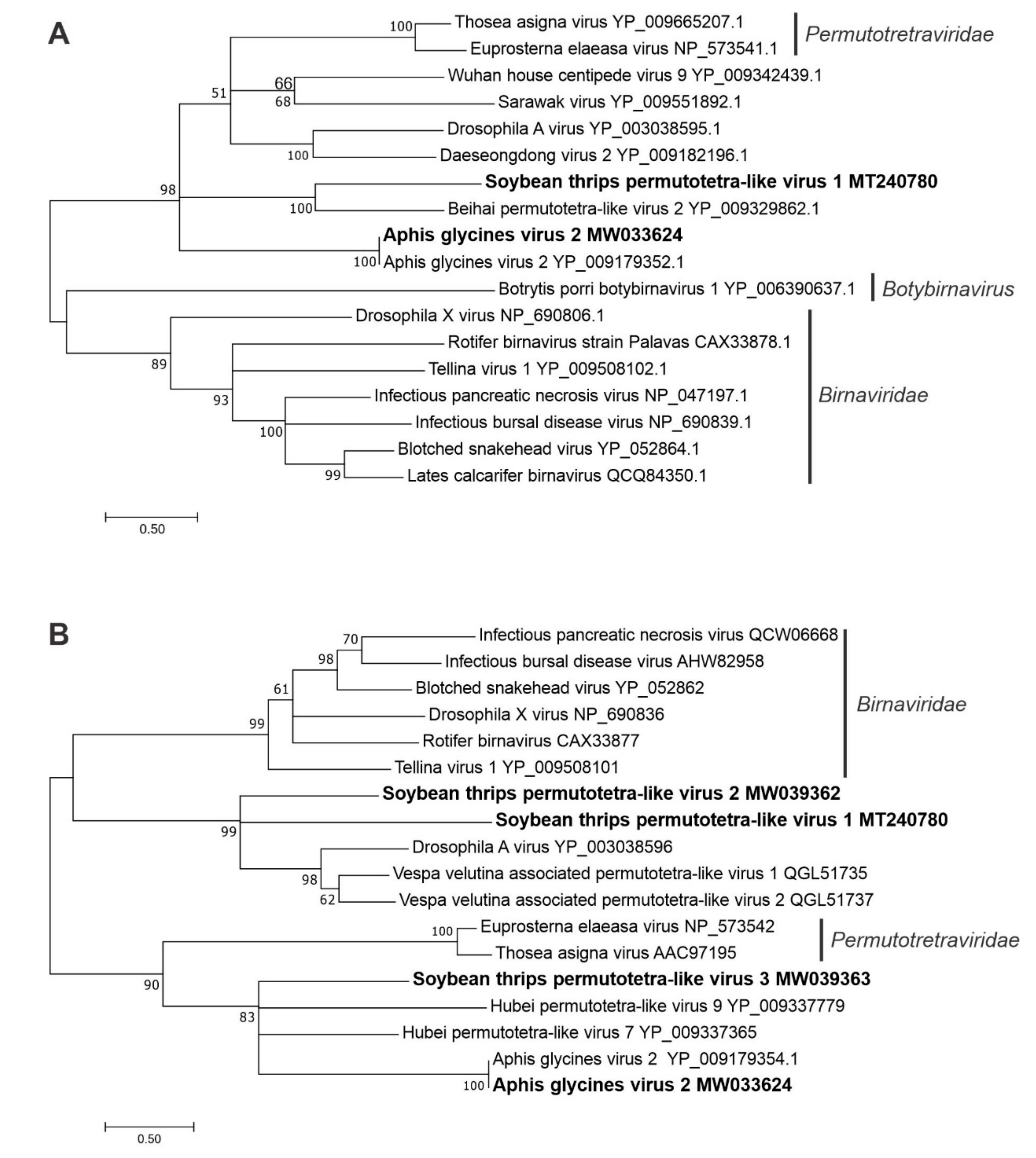
3.2.7. Sequences Related to Members of the Permutotetraviridae
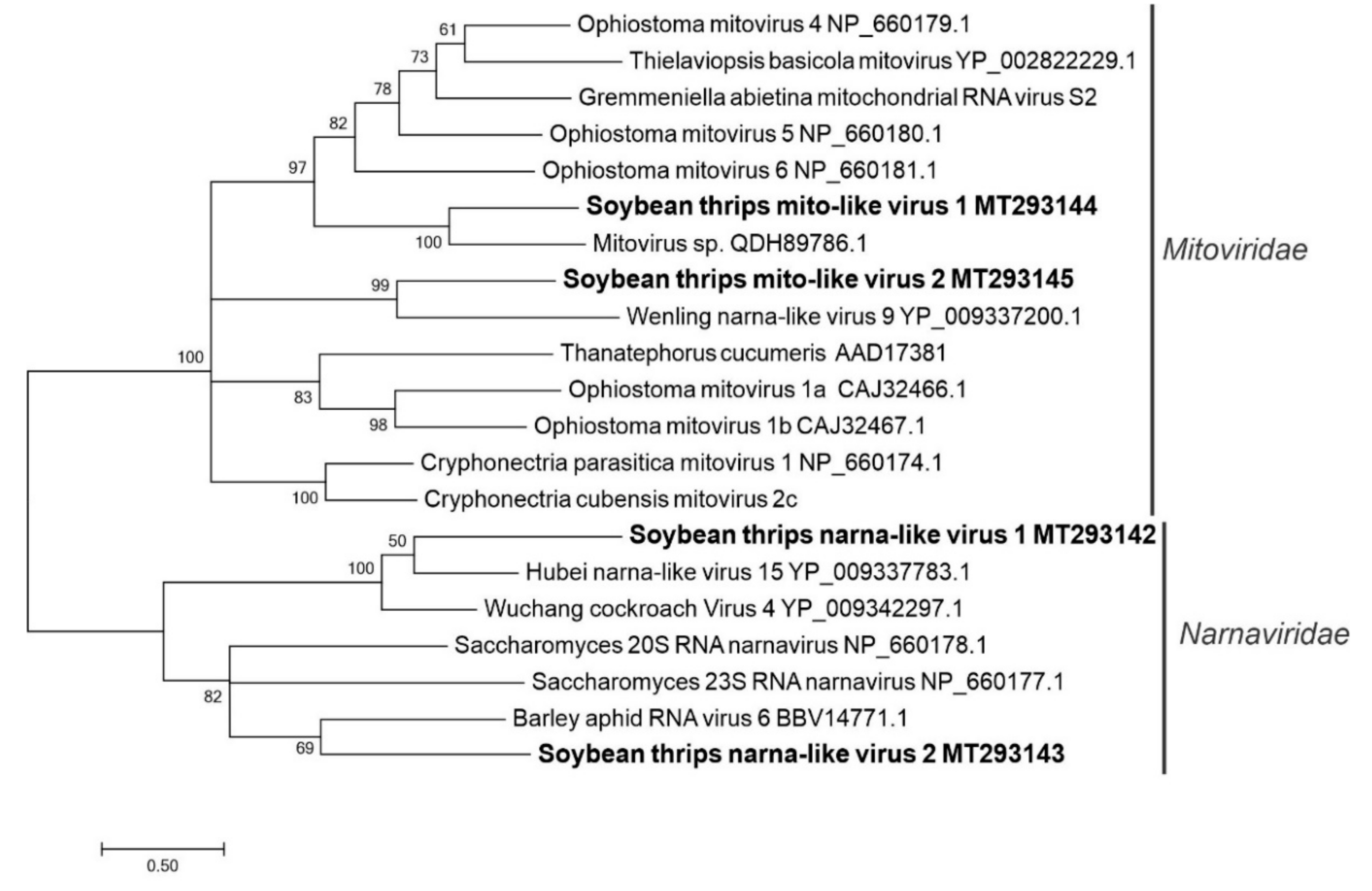
3.2.8. Sequences Related to Members of the Wolframvirales and Cryppavirales
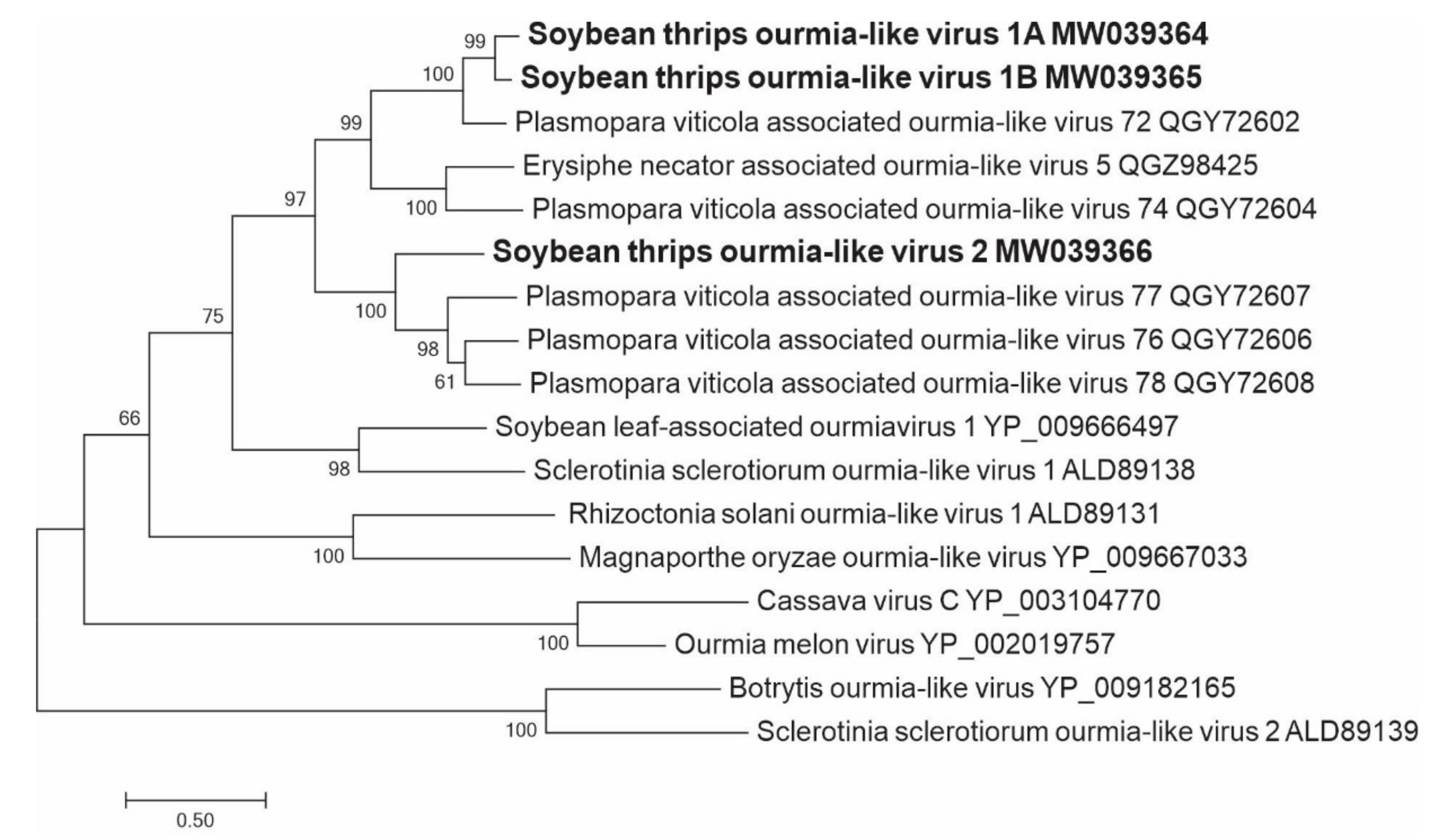
3.2.9. Sequences Related to Members of the Ourlivirales
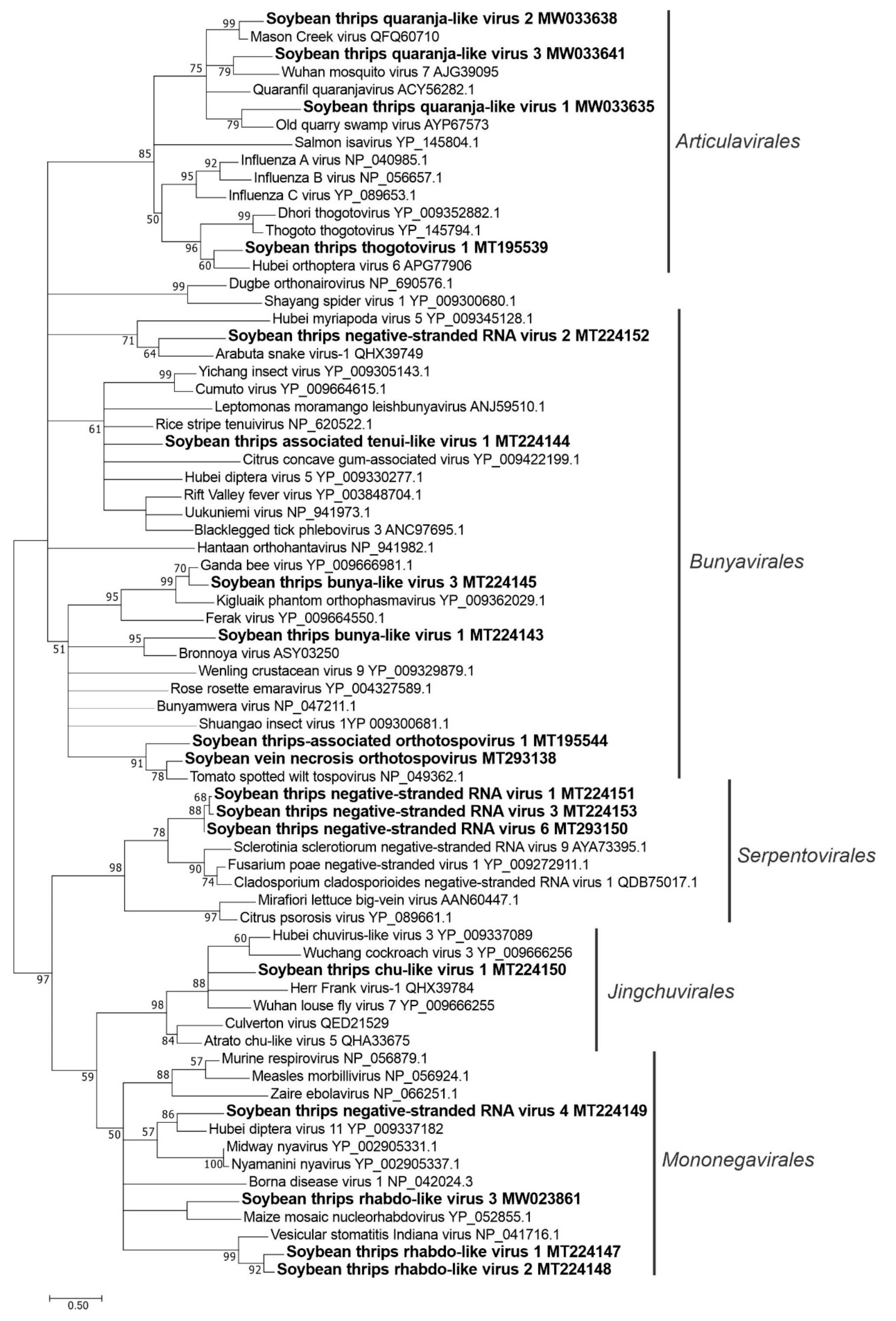
3.3. Sequences Related to Negative-Stranded RNA Viruses
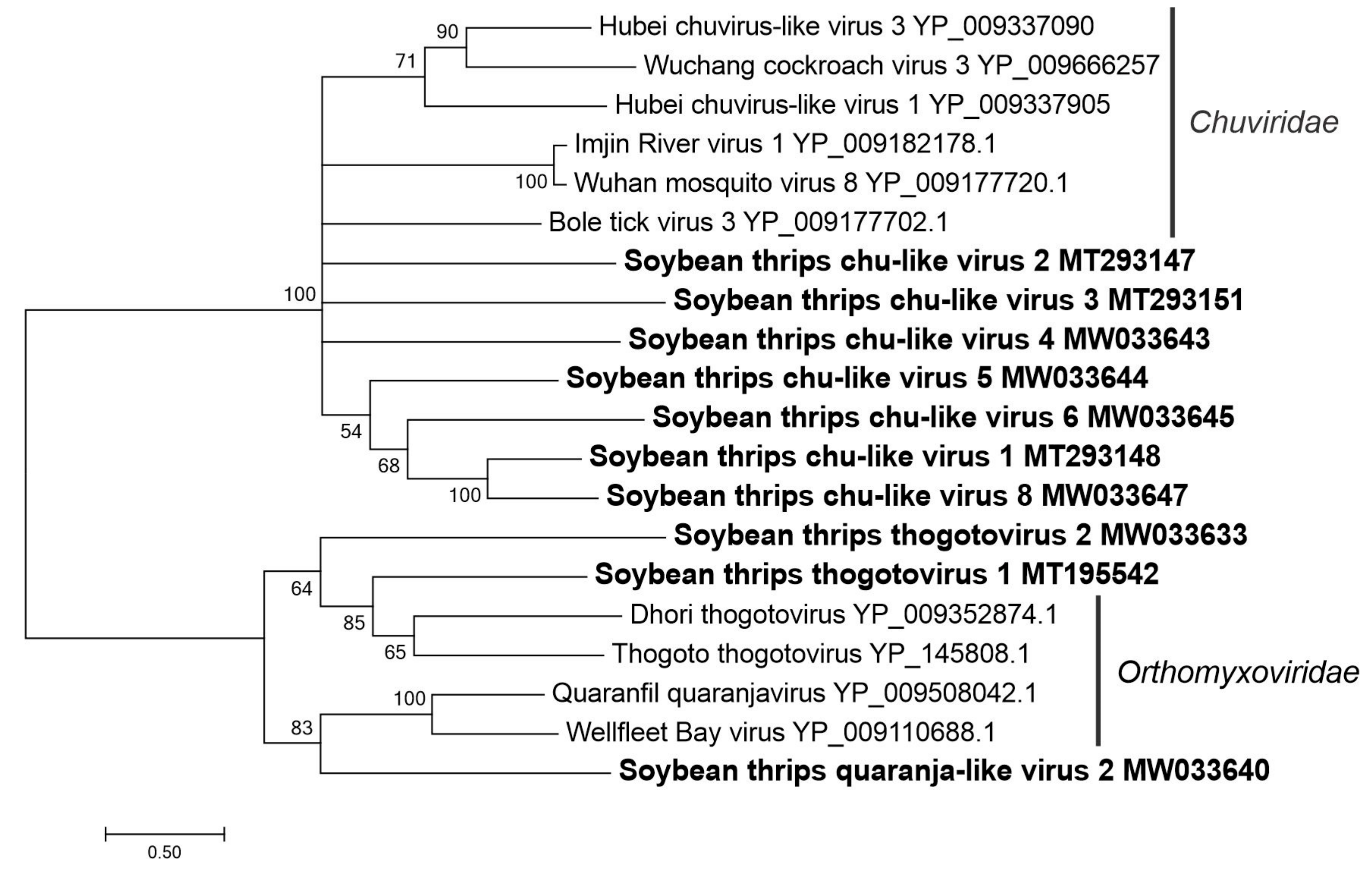
3.3.1. Sequences Related to Members of the Articulavirales
3.3.2. Sequences Related to Members of the Bunyavirales
3.3.3. Sequences Related to Members of the Jingchuvirales
3.3.4. Sequences Related to Members of the Serpentovirales
3.3.5. Sequences Related to Members of the Mononegavirales
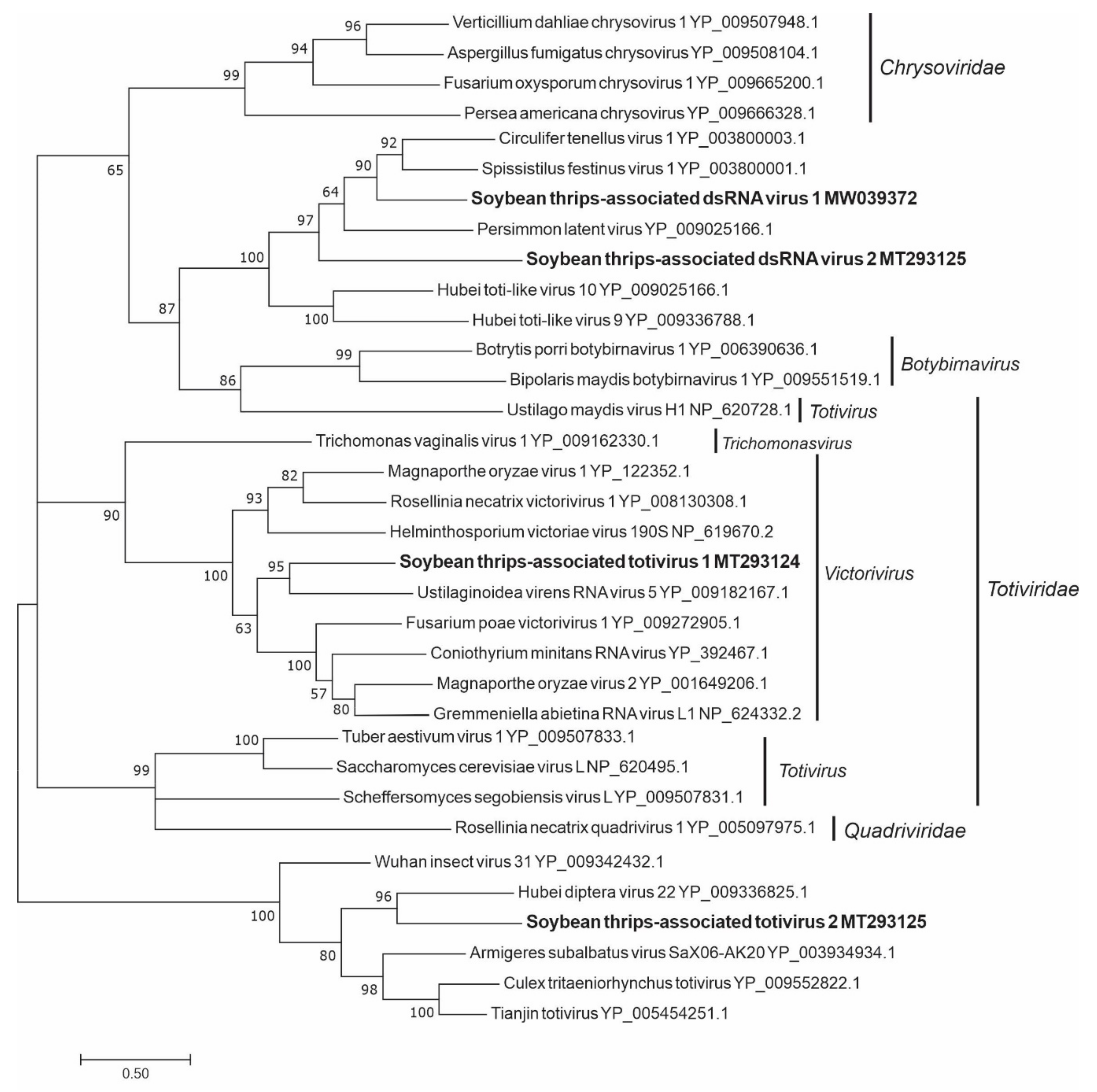
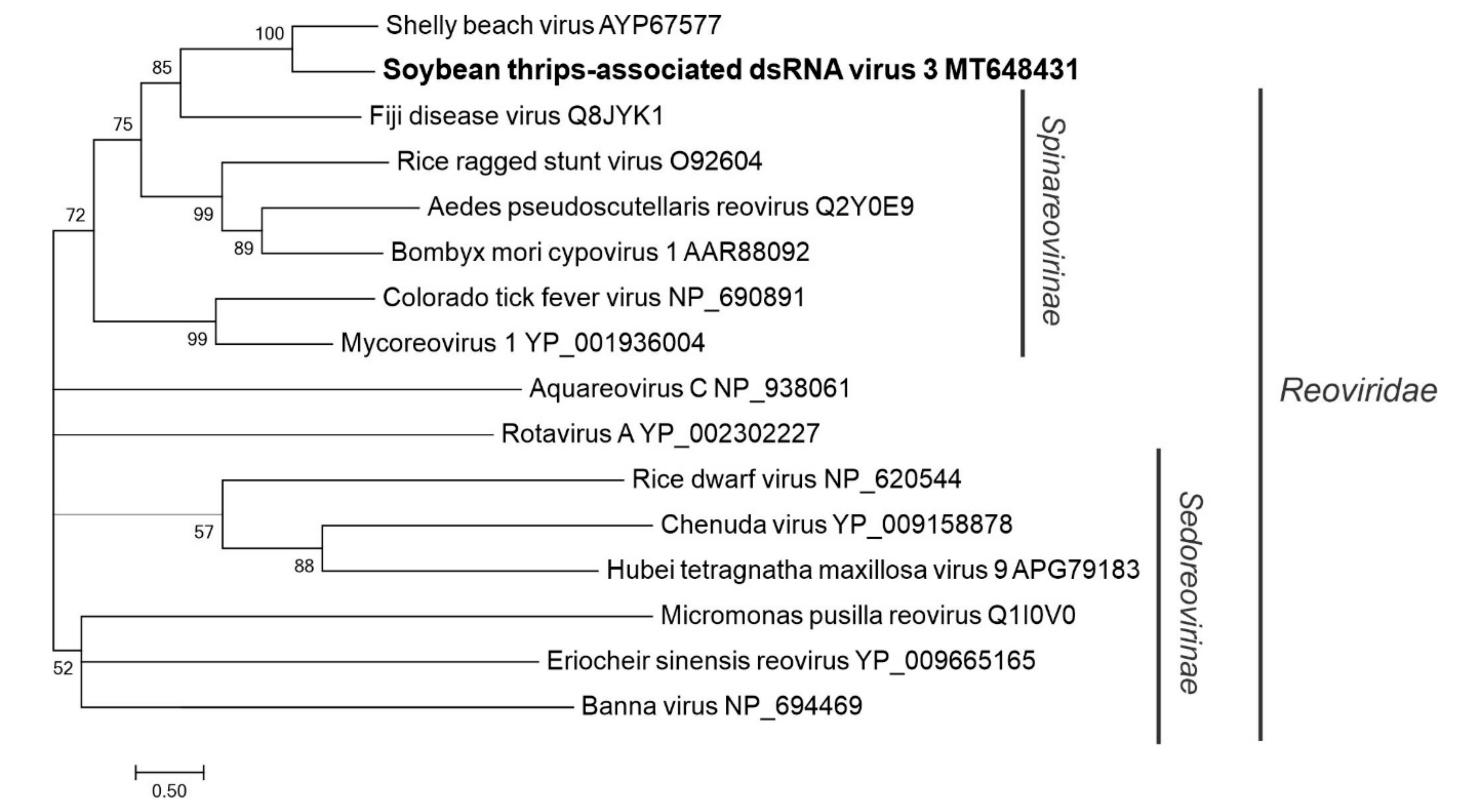
3.4. Sequences Related to dsRNA Viruses
3.4.1. Sequences Related to Members of the Durnavirales
3.4.2. Sequences Related to Members of the Ghabrivirales
3.4.3. Sequences Related to Members of the Reovirales
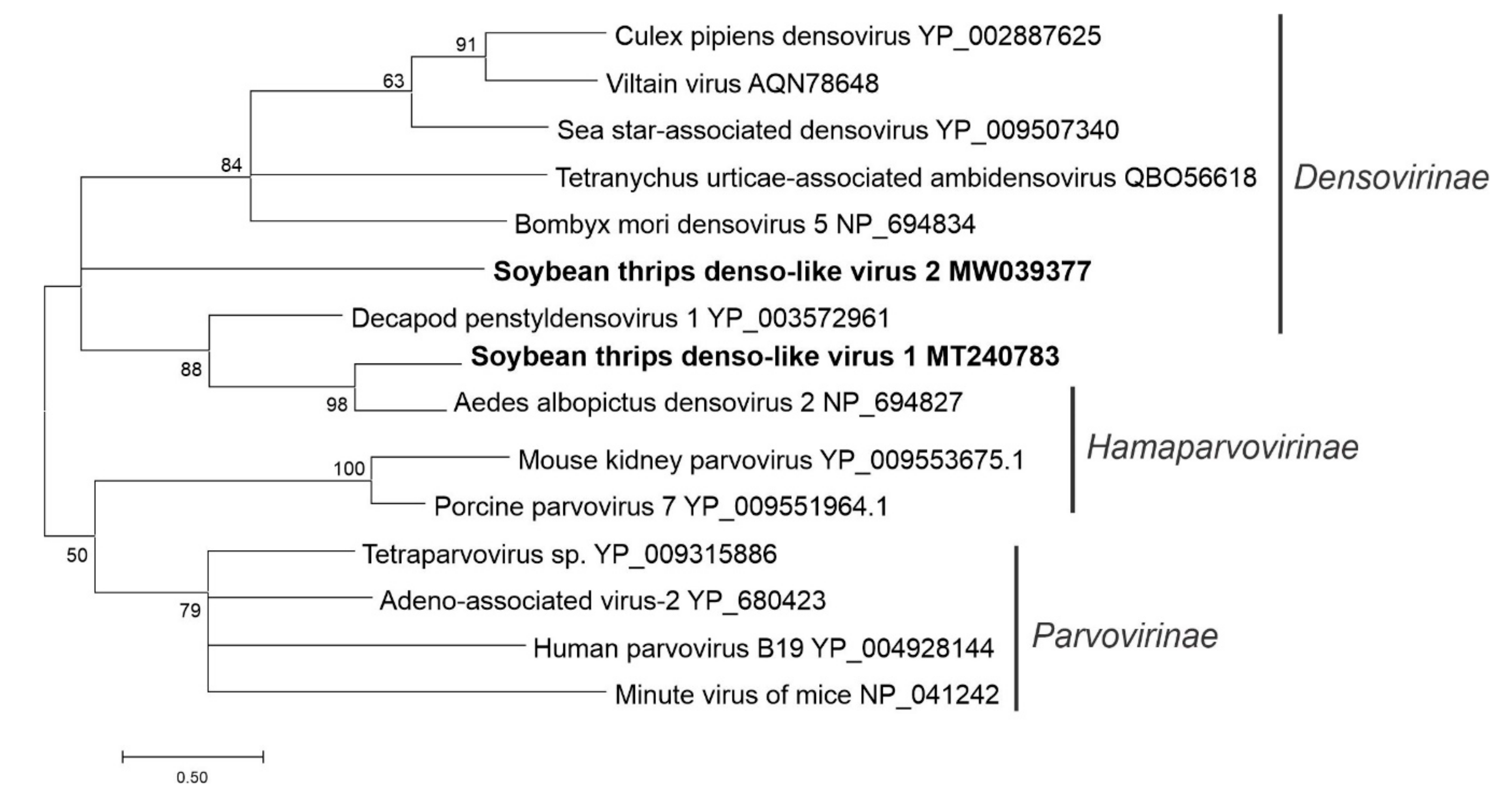
3.5. Sequences Related to Single-Stranded DNA Viruses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jones, D.R. Plant viruses transmitted by thrips. Eur. J. Plant Pathol. 2005, 113, 119–157. [Google Scholar] [CrossRef]
- Pappu, H.R.; Jones, R.A.C.; Jain, R.K. Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res. 2009, 141, 219–236. [Google Scholar] [CrossRef]
- Rotenberg, D.; Jacobson, A.L.; Schneweis, D.J.; Whitfield, A.E. Thrips transmission of tospoviruses. Curr. Opin. Virol. 2015, 15, 80–89. [Google Scholar] [CrossRef]
- Zhou, J.; Tzanetakis, I.E. Soybean vein necrosis virus: An emerging virus in North America. Virus Genes 2019, 55, 12–21. [Google Scholar] [CrossRef]
- Irizarry, M.D.; Elmore, M.G.; Batzer, J.C.; Whitham, S.A.; Mueller, D.S. Alternative hosts for soybean vein necrosis virus and feeding preferences of its vector soybean thrips. Plant Health Prog. 2018, 19, 176–181. [Google Scholar] [CrossRef] [Green Version]
- Anderson, N.R. Effect of Soybean Vein Necrosis on Soybean Yield and Seed Quality, and Symptom Expression on Soybean and Alternative Hosts; Purdue University: West Lafayette, IND, USA, 2017. [Google Scholar]
- Zhou, J.; Tzanetakis, I.E. Epidemiology of Soybean vein necrosis-associated virus. Phytopathology 2013, 103, 966–971. [Google Scholar] [CrossRef] [Green Version]
- Keough, S.; Han, J.; Shuman, T.; Wise, K.; Nachappa, P. Effects of soybean vein necrosis virus on life history and host preference of its vector, Neohydatothrips variabilis, and evaluation of vector status of Frankliniella tritici and Frankliniella fusca. J. Econ. Entomol. 2016, 109, 1979–1987. [Google Scholar] [CrossRef]
- Beach, A.M. Contributions to a knowledge of the Thripidae of Iowa. Proc. Iowa Acad. Sci. 1897, 3, 214–227. [Google Scholar] [CrossRef] [Green Version]
- Akin, D.S.; Lorenz, G.M., III; Studebaker, G.E.; Howard, J.E. Regional survey 2009–2010: Thrips species composition across the upland cotton belt. Res. Ser. Ark. Agric. Exp. Stn. 2011, 589, 132–137. [Google Scholar]
- Dupree, M. Control of thrips and the bean leaf beetle on lima beans with systemic insecticides. Ga. Entomol. Soc. J. 1970, 51, 48–52. [Google Scholar]
- Irwin, M.E.; Yeargan, K.V. Sampling phytophagous thrips on soybean. In Sampling Methods in Soybean Entomology; Kogan, M., Herzog, D.C., Eds.; Springer: New York, NY, YSA, 1980; pp. 283–304. [Google Scholar]
- Nault, B.A.; Speese, J., III. Major insect pests and economics of fresh-market tomato in eastern Virginia. Crop Prot. 2002, 21, 359–366. [Google Scholar] [CrossRef]
- Feng, Y.; Krueger, E.N.; Liu, S.; Dorman, K.; Bonning, B.C.; Miller, W.A. Discovery of known and novel viral genomes in soybean aphid by deep sequencing. Phytobiomes J. 2017, 1, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Wamonje, F.O.; Michuki, G.N.; Braidwood, L.A.; Njuguna, J.N.; Musembi-Mutuku, J.; Djikeng, A.; Harvey, J.J.W.; Carr, J.P. Viral metagenomics of aphids present in bean and maize plots on mixed-use farms in Kenya reveals the presence of three dicistroviruses including a novel Big Sioux River virus-like dicistrovirus. Virol. J. 2017, 14, 188. [Google Scholar] [CrossRef] [Green Version]
- Kamali, M.; Heydarnejad, J.; Pouramini, N.; Masumi, H.; Farkas, K.; Kraberger, S.; Varsani, A. Genome sequences of Beet curly top Iran virus, Oat dwarf virus, Turnip curly top virus, and Wheat dwarf virus identified in leafhoppers. Genome Announc. 2017, 5, e01674-16. [Google Scholar] [CrossRef] [Green Version]
- Fontenele, R.S.; Alves-Freitas, D.M.T.; Silva, P.I.T.; Foresti, J.; Silva, P.R.; Godinho, M.T.; Varsani, A.; Ribeiro, S.G. Discovery of the first maize-infecting mastrevirus in the Americas using a vector-enabled metagenomics approach. Arch. Virol. 2018, 163, 263–267. [Google Scholar] [CrossRef]
- Nouri, S.; Salem, N.; Nigg, J.C.; Falk, B.W. Diverse array of new viral sequences identified in worldwide populations of the Asian citrus psyllid (Diaphorina citri) using viral metagenomics. J. Virol. 2016, 90, 2434–2445. [Google Scholar] [CrossRef] [Green Version]
- Ng, T.F.F.; Duffy, S.; Polston, J.E.; Bixby, E.; Vallad, G.E.; Breitbart, M. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE 2011, 6, e19050. [Google Scholar] [CrossRef] [Green Version]
- Rosario, K.; Capobianco, H.; Ng, T.F.F.; Breitbart, M.; Polston, J.E. RNA viral metagenome of whiteflies leads to the discovery and characterization of a whitefly-transmitted carlavirus in North America. PLoS ONE 2014, 9, e86748. [Google Scholar] [CrossRef]
- Rosario, K.; Seah, Y.M.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Duffy, S.; Breitbart, M. Vector-enabled metagenomic (VEM) surveys using whiteflies (Aleyrodidae) reveal novel Begomovirus species in the New and Old Worlds. Viruses 2015, 7, 5553–5570. [Google Scholar] [CrossRef] [Green Version]
- Lagos-Kutz, D.; Voegtlin, D.J.; Onstad, D.; Hogg, D.; Ragsdale, D.; Tilmon, K.; Hodgson, E.; DiFonzo, C.; Groves, R.; Krupke, C.; et al. The soybean aphid suction trap network: Sampling the aerobiological “Soup”. Am. Entomol. 2020, 66, 48–55. [Google Scholar] [CrossRef]
- Hoddle, M.S.; Mound, L.A.; Paris, D.L. Thrips of California. Available online: https://keys.lucidcentral.org/keys/v3/thrips_of_california/Thrips_of_California.html (accessed on 28 September 2020).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.M.A.; Hashimoto, Y.; Herrero, S.; de Miranda, J.R.; Ryabov, E. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Iflaviridae. J. Gen. Virol. 2017, 98, 527–528. [Google Scholar] [CrossRef]
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.M.A.; Hashimoto, Y.; Herrero, S.; de Miranda, J.R.; Ryabov, E. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Dicistroviridae. J. Gen. Virol. 2017, 98, 355–356. [Google Scholar] [CrossRef]
- de Souza, W.M.; Fumagalli, M.J.; de Araujo, J.; Ometto, T.; Modha, S.; Thomazelli, L.M.; Durigon, E.L.; Murcia, P.R.; Figueiredo, L.T.M. Discovery of novel astrovirus and calicivirus identified in ruddy turnstones in Brazil. Sci. Rep. 2019, 9, 5556. [Google Scholar] [CrossRef] [Green Version]
- Adams, I.P.; Braidwood, L.A.; Stomeo, F.; Phiri, N.; Uwumukiza, B.; Feyissa, B.; Mahuku, G.; Wangi, A.; Smith, J.; Mumford, R.; et al. Characterising maize viruses associated with maize lethal necrosis symptoms in sub Saharan Africa. bioRxiv 2017, 161489. [Google Scholar] [CrossRef] [Green Version]
- Yinda, C.K.; Zell, R.; Deboutte, W.; Zeller, M.; Conceição-Neto, N.; Heylen, E.; Maes, P.; Knowles, N.J.; Ghogomu, S.M.; van Ranst, M.; et al. Highly diverse population of Picornaviridae and other members of the Picornavirales, in Cameroonian fruit bats. BMC Genom. 2017, 18, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- François, S.; Mutuel, D.; Duncan, A.B.; Rodrigues, L.R.; Danzelle, C.; Lefevre, S.; Santos, I.; Frayssinet, M.; Fernandez, E.; Filloux, D.; et al. A new prevalent densovirus discovered in Acari. insight from metagenomics in viral communities associated with two-spotted mite (Tetranychus urticae) populations. Viruses 2019, 11, 233. [Google Scholar]
- Shi, M.; Lin, X.-D.; Vasilakis, N.; Tian, J.-H.; Li, C.-X.; Chen, L.-J.; Eastwood, G.; Diao, X.-N.; Chen, M.-H.; Chen, X.; et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 2016, 90, 659–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasmin, T.; Thekke-Veetil, T.; Hobbs, H.A.; Nelson, B.D.; McCoppin, N.K.; Lagos-Kutz, D.; Hartman, G.L.; Lambert, K.N.; Walker, D.R.; Domier, L.L. Aphis glycines virus 1, a new bicistronic virus with two functional internal ribosome entry sites, is related to a group of unclassified viruses in the Picornavirales. J Gen Virol 2020, 101, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, Y. Virus diseases of alfalfa and biology of alfalfa mosaic virus in Ontario and western Quebec. Can. J. Plant Pathol. 1982, 4, 175–179. [Google Scholar] [CrossRef]
- Paliwal, Y. Identification and distribution in eastern Canada of lucerne transient streak, a virus newly recognized in North America. Can. J. Plant Pathol. 1983, 5, 75–80. [Google Scholar] [CrossRef]
- Medd, N.C.; Fellous, S.; Waldron, F.M.; Xuéreb, A.; Nakai, M.; Cross, J.V.; Obbard, D.J. The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus Evol. 2018, 4, vey009. [Google Scholar] [CrossRef]
- Nunes, M.R.T.; Contreras-Gutierrez, M.A.; Guzman, H.; Martins, L.C.; Barbirato, M.F.; Savit, C.; Balta, V.; Uribe, S.; Vivero, R.; Suaza, J.D.; et al. Genetic characterization, molecular epidemiology, and phylogenetic relationships of insect-specific viruses in the taxon Negevirus. Virology 2017, 504, 152–167. [Google Scholar] [CrossRef]
- Vasilakis, N.; Forrester, N.L.; Palacios, G.; Nasar, F.; Savji, N.; Rossi, S.L.; Guzman, H.; Wood, T.G.; Popov, V.; Gorchakov, R.; et al. Negevirus: A proposed new taxon of insect-specific viruses with wide geographic distribution. J. Virol. 2013, 87, 2475–2488. [Google Scholar] [CrossRef] [Green Version]
- Chiapello, M.; Rodríguez-Romero, J.; Nerva, L.; Forgia, M.; Chitarra, W.; Ayllón, M.A.; Turina, M. Putative new plant viruses associated with Plasmopara viticola-infected grapevine samples. Ann. Appl. Biol. 2020, 176, 180–191. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Qin, X.-C.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Gao, D.-Y.; He, J.-R.; Wang, J.-B.; Li, C.-X.; Kang, Y.-J.; Yu, B.; et al. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. USA 2014, 111, 6744–6749. [Google Scholar] [CrossRef] [Green Version]
- Delmas, B.; Attoui, H.; Ghosh, S.; Malik, Y.S.; Mundt, E.; Vakharia, V.N. ICTV Report Consortium, ICTV virus taxonomy profile: Birnaviridae. J. Gen. Virol. 2019, 100, 5–6. [Google Scholar] [CrossRef]
- Liu, S.; Vijayendran, D.; Chen, Y.; Bonning, B.C. Aphis glycines virus 2, a novel insect virus with a unique genome structure. Viruses 2016, 8, 315. [Google Scholar] [CrossRef] [Green Version]
- de Risi, J.L.; Huber, G.; Kistler, A.; Retallack, H.; Wilkinson, M.; Yllanes, D. An exploration of ambigrammatic sequences in narnaviruses. Sci. Rep. 2019, 9, 17982. [Google Scholar] [CrossRef] [Green Version]
- Lye, L.-F.; Akopyants, N.S.; Dobson, D.E.; Beverley, S.M. A narnavirus-like element from the trypanosomatid protozoan parasite Leptomonas seymouri. Genome Announc. 2016, 4, e00713-16. [Google Scholar] [CrossRef] [Green Version]
- Ayllón, M.A.; Turina, M.; Xie, J.; Nerva, L.; Marzano, S.-Y.L.; Donaire, L.; Jiang, D. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Botourmiaviridae. J. Gen. Virol. 2020, 101, 454–455. [Google Scholar]
- Wang, Q.; Mu, F.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D. A single ssRNA segment encoding RdRp is sufficient for replication, infection, and transmission of ourmia-like virus in fungi. Front. Microbiol. 2020, 11, 379. [Google Scholar] [CrossRef] [Green Version]
- Lambert, A.J.; Velez, J.O.; Brault, A.C.; Calvert, A.E.; Bell-Sakyi, L.; Bosco-Lauth, A.M.; Staples, J.E.; Kosoy, O.I. Molecular, serological and in vitro culture-based characterization of Bourbon virus, a newly described human pathogen of the genus Thogotovirus. J. Clin. Virol. 2015, 73, 127–132. [Google Scholar] [CrossRef] [Green Version]
- Temmam, S.; Chrétien, D.; Bigot, T.; Dufour, E.; Petres, S.; Desquesnes, M.; Devillers, E.; Dumarest, M.; Yousfi, L.; Jittapalapong, S.; et al. Monitoring silent spillovers before emergence: A pilot study at the tick/human interface in Thailand. Front. Microbiol. 2019, 10, 2315. [Google Scholar] [CrossRef]
- Plyusnin, A.; Beaty, B.J.; Elliott, R.M.; Goldbach, R.; Kormelink, R.; Lundkvist, Å.; Schmaljohn, C.S.; Tesh, R.B. Family Bunyaviridae. In Virus Taxonomy -Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: New York, NY, USA, 2012; pp. 729–741. [Google Scholar]
- Li, C.-X.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Kang, Y.-J.; Chen, L.-J.; Qin, X.-C.; Xu, J.; Holmes, E.C.; Zhang, Y.-Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 2015, 4, e05378. [Google Scholar] [CrossRef]
- Gondard, M.; Temmam, S.; Devillers, E.; Pinarello, V.; Bigot, T.; Chrétien, D.; Aprelon, R.; Vayssier-Taussat, M.; Albina, E.; Eloit, M.; et al. RNA viruses of Amblyomma variegatum and Rhipicephalus microplus and cattle susceptibility in the French Antilles. Viruses 2020, 12, 144. [Google Scholar] [CrossRef] [Green Version]
- García, M.L.; Bó, E.D.; da Graça, J.V.; Gago-Zachert, S.; Hammond, J.; Moreno, P.; Natsuaki, T.; Pallás, V.; Navarro, J.A.; Reyes, C.A.; et al. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Ophioviridae. J. Gen. Virol. 2017, 98, 1161–1162. [Google Scholar] [CrossRef] [Green Version]
- Osaki, H.; Sasaki, A.; Nomiyama, K.; Tomioka, K. Multiple virus infection in a single strain of Fusarium poae shown by deep sequencing. Virus Genes 2016, 52, 835–847. [Google Scholar] [CrossRef]
- Nerva, L.; Turina, M.; Zanzotto, A.; Gardiman, M.; Gaiotti, F.; Gambino, G.; Chitarra, W. Isolation, molecular characterization and virome analysis of culturable wood fungal endophytes in esca symptomatic and asymptomatic grapevine plants. Environ. Microbiol. 2019, 21, 2886–2904. [Google Scholar] [CrossRef]
- Walker, P.J.; Blasdell, K.R.; Calisher, C.H.; Dietzgen, R.G.; Kondo, H.; Kurath, G.; Longdon, B.; Stone, D.M.; Tesh, R.B.; Tordo, N.; et al. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Rhabdoviridae. J. Gen. Virol. 2018, 99, 447–448. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Huot, O.B.; Martin, K.M.; Kondo, H.; Dietzgen, R.G. Plant rhabdoviruses—Their origins and vector interactions. Curr. Opin. Virol. 2018, 33, 198–207. [Google Scholar] [CrossRef]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef]
- Wickner, R.B.; Ghabrial, S.A.; Nibert, M.L.; Patterson, J.L.; Wang, C.C. Family Totiviridae. In Virus Taxonomy -Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: New York, NY, USA, 2012; pp. 639–650. [Google Scholar]
- Li, F.; Du, J.; Wu, Z.Q.; Zhang, W.J.; Fu, S.H.; Song, J.D.; Wang, Q.Y.; He, Y.; Lei, W.W.; Xu, S.T.; et al. Identification and genetic analysis of a totivirus isolated from the Culex tritaeniorhynchus in northern China. Arch. Microbiol. 2020, 202, 807–813. [Google Scholar] [CrossRef]
- Wu, Q.F.; Luo, Y.J.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.X.; Ding, S.W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef] [Green Version]
- Harvey, E.; Rose, K.; Eden, J.S.; Lo, N.; Abeyasuriya, T.; Shi, M.; Doggett, S.L.; Holmes, E.C. Extensive diversity of RNA viruses in Australian ticks. J. Virol. 2019, 93, e01358-18. [Google Scholar] [CrossRef] [Green Version]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Penzes, J.J.; et al. ICTV Report Consortium, ICTV Virus Taxonomy Profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef]
- Bekal, S.; Domier, L.L.; Niblack, T.L.; Lambert, K.N. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J. Gen. Virol. 2011, 92, 1870–1879. [Google Scholar] [CrossRef]
- Ruark, C.L.; Gardner, M.; Mitchum, M.G.; Davis, E.L.; Sit, T.L. Novel RNA viruses within plant parasitic cyst nematodes. PLoS ONE 2018, 13, e0193881. [Google Scholar] [CrossRef] [Green Version]
- Chu, D.; Qu, W.-M.; Guo, L. Invasion genetics of alien insect pests in China: Research progress and future prospects. J. Integr. Agric. 2019, 18, 748–757. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, T.; Morimoto, N.; Nishida, G.M.; Kiritani, K.; Moriya, S.; Liebhold, A.M. Comparison of insect invasions in North America, Japan and their Islands. Biol. Invasions 2015, 17, 3049–3061. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Hajano, J.-U.-D.; Wang, X. New insights on the transmission mechanism of tenuiviruses by their vector insects. Curr. Opin. Virol. 2018, 33, 13–17. [Google Scholar] [CrossRef]
- Gildow, F.E. D’Arcy, Barley and oats as reservoirs for an aphid virus and the influence on barley yellow dwarf virus transmission. Phytopathology 1988, 78, 811. [Google Scholar] [CrossRef]
- Dezordi, F.Z.; Vasconcelos, C.R.d.S.; Rezende, A.M.; Wallau, G.L. In and outs of Chuviridae endogenous viral elements: Origin of a potentially new retrovirus and signature of ancient and ongoing arms race in mosquito genomes. Front. Genet. 2020, 11, 542437. [Google Scholar] [CrossRef]
- Suzuki, Y.; Frangeul, L.; Dickson, L.B.; Blanc, H.; Verdier, Y.; Vinh, J.; Lambrechts, L.; Saleh, M.C. Uncovering the repertoire of endogenous flaviviral elements in Aedes mosquito genomes. J. Virol. 2017, 91, e00571-17. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.G.; Kelly, A.G.; Enosi-Tuipulotu, D.; Tanaka, M.M.; White, P.A. Novel insights into endogenous RNA viral elements in Ixodes scapularis and other arbovirus vector genomes. Virus Evol. 2019, 5, vez010. [Google Scholar] [CrossRef]
- Ter-Horst, A.M.; Nigg, J.C.; Dekker, F.M.; Falk, B.W. Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWI-interacting RNAs. J. Virol. 2019, 93, e02124-18. [Google Scholar] [CrossRef] [Green Version]
- Houé, V.; Bonizzoni, M.; Failloux, A.B. Endogenous non-retroviral elements in genomes of Aedes mosquitoes and vector competence. Emerg. Microbes Infect. 2019, 8, 542–555. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, A.; Simmonds, P.; Lipkin, W.I. Discovery and characterization of mammalian endogenous parvoviruses. J. Virol. 2010, 84, 12628–12635. [Google Scholar] [CrossRef] [Green Version]
- Crochu, S.; Cook, S.; Attoui, H.; Charrel, R.N.; de Chesse, R.; Belhouchet, M.; Lemasson, J.-J.; de Micco, P.; de Lamballerie, X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J. Gen. Virol. 2004, 85, 1971–1980. [Google Scholar] [CrossRef]
- Shalileh, S.; Ogada, P.A.; Moualeu, D.P.; Poehling, H.M. Manipulation of Frankliniella occidentalis (Thysanoptera: Thripidae) by Tomato spotted wilt virus (Tospovirus) via the host plant nutrients to enhance its transmission and spread. Environ. Entomol. 2016, 45, 1235–1242. [Google Scholar] [CrossRef] [Green Version]
- Stafford-Banks, C.A.; Yang, L.H.; McMunn, M.S.; Ullman, D.E. Virus infection alters the predatory behavior of an omnivorous vector. Oikos 2014, 123, 1384–1390. [Google Scholar] [CrossRef]
- Stafford, C.A.; Walker, G.P.; Ullman, D.E. Infection with a plant virus modifies vector feeding behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 9350–9355. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Hussain, S.; Merchant, A.; Xu, B.; Xie, W.; Wang, S.; Zhang, Y.; Zhou, X.; Wu, Q. Tomato spotted wilt orthotospovirus influences the reproduction of its insect vector, western flower thrips, Frankliniella occidentalis, to facilitate transmission. Pest Manag. Sci. 2020, 76, 2406–2414. [Google Scholar] [CrossRef]
- Jiménez-Martínez, E.S.; Bosque-Pérez, N.A.; Berger, P.H.; Zemetra, R.S. Life history of the bird cherry-oat aphid, Rhopalosiphum padi (Homoptera: Aphididae), on transgenic and untransformed wheat challenged with barley yellow dwarf virus. J. Econ. Entomol. 2004, 97, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Käfer, S.; Paraskevopoulou, S.; Zirkel, F.; Wieseke, N.; Donath, A.; Petersen, M.; Jones, T.C.; Liu, S.; Zhou, X.; Middendorf, M.; et al. Re-assessing the diversity of negative-strand RNA viruses in insects. PLoS Pathog. 2019, 15, e1008224. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thekke-Veetil, T.; Lagos-Kutz, D.; McCoppin, N.K.; Hartman, G.L.; Ju, H.-K.; Lim, H.-S.; Domier, L.L. Soybean Thrips (Thysanoptera: Thripidae) Harbor Highly Diverse Populations of Arthropod, Fungal and Plant Viruses. Viruses 2020, 12, 1376. https://doi.org/10.3390/v12121376
Thekke-Veetil T, Lagos-Kutz D, McCoppin NK, Hartman GL, Ju H-K, Lim H-S, Domier LL. Soybean Thrips (Thysanoptera: Thripidae) Harbor Highly Diverse Populations of Arthropod, Fungal and Plant Viruses. Viruses. 2020; 12(12):1376. https://doi.org/10.3390/v12121376
Chicago/Turabian StyleThekke-Veetil, Thanuja, Doris Lagos-Kutz, Nancy K. McCoppin, Glen L. Hartman, Hye-Kyoung Ju, Hyoun-Sub Lim, and Leslie. L. Domier. 2020. "Soybean Thrips (Thysanoptera: Thripidae) Harbor Highly Diverse Populations of Arthropod, Fungal and Plant Viruses" Viruses 12, no. 12: 1376. https://doi.org/10.3390/v12121376





