Genetic, Molecular, and Pathogenic Characterization of the H9N2 Avian Influenza Viruses Currently Circulating in South China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Pigs
2.2. Swabs Collection and Virus Isolation
2.3. Sequencing and Phylogenetic Analysis
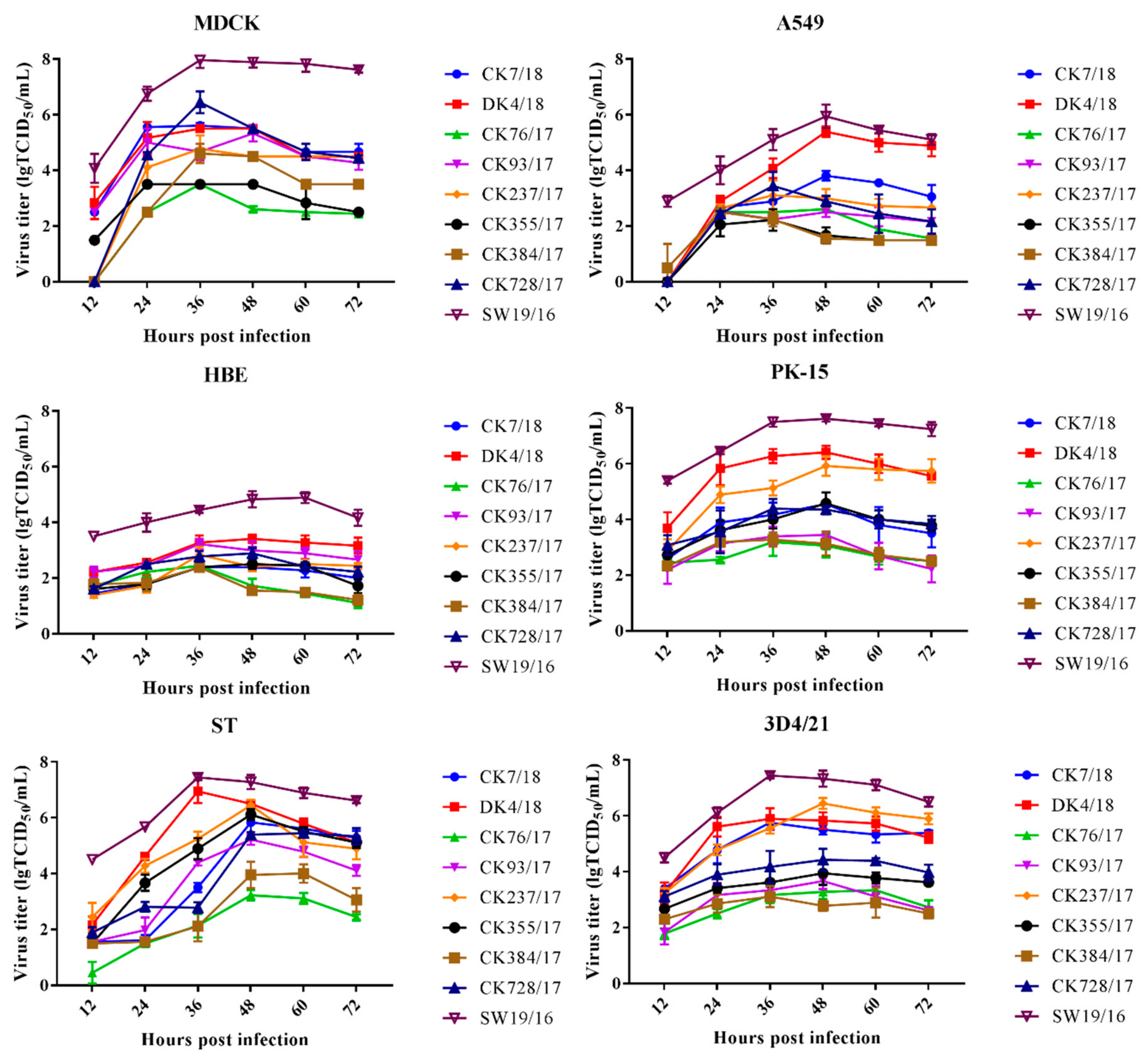
2.4. Viral Replication Kinetics in Cells
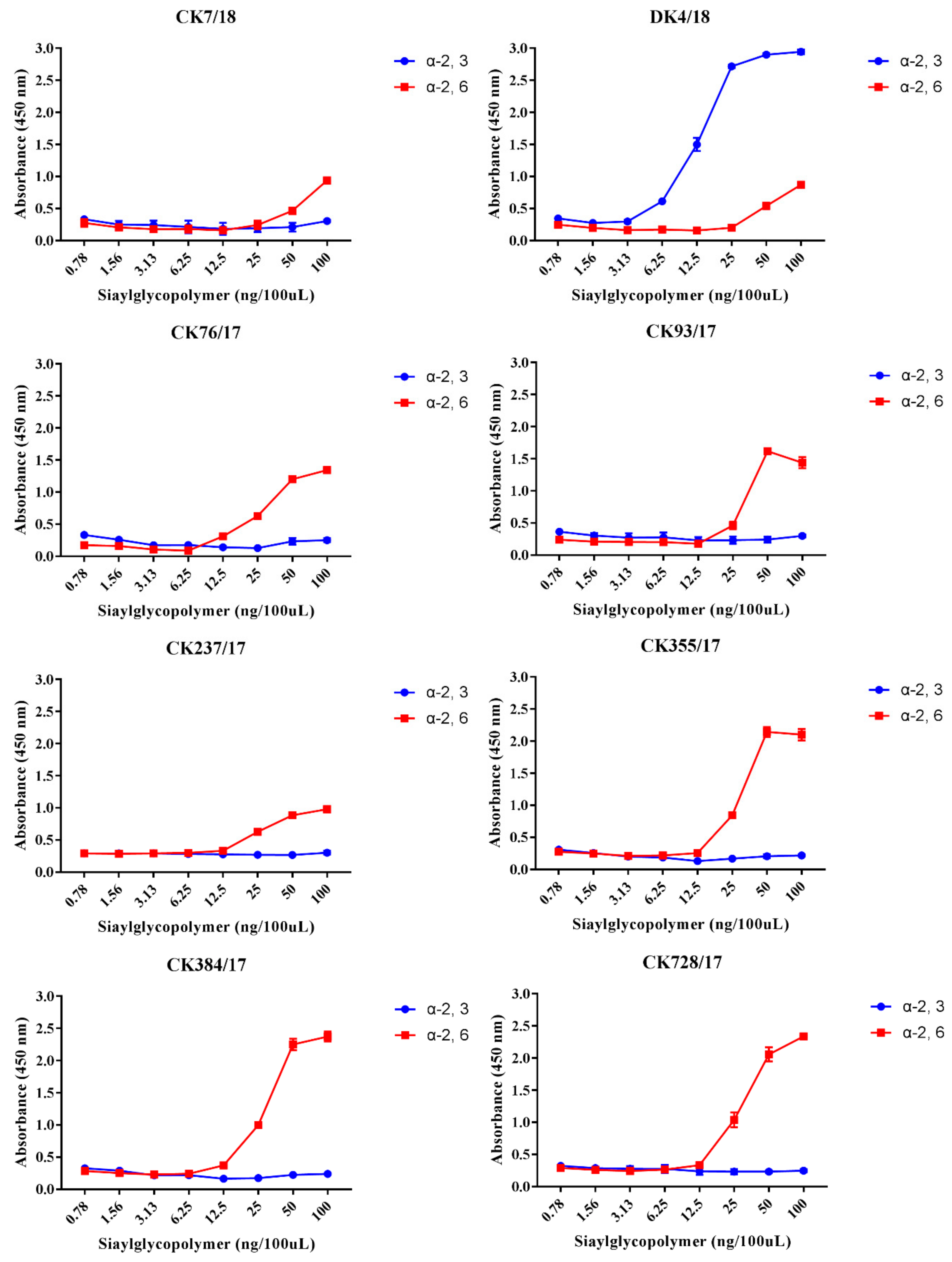
2.5. Receptor-Binding Assay
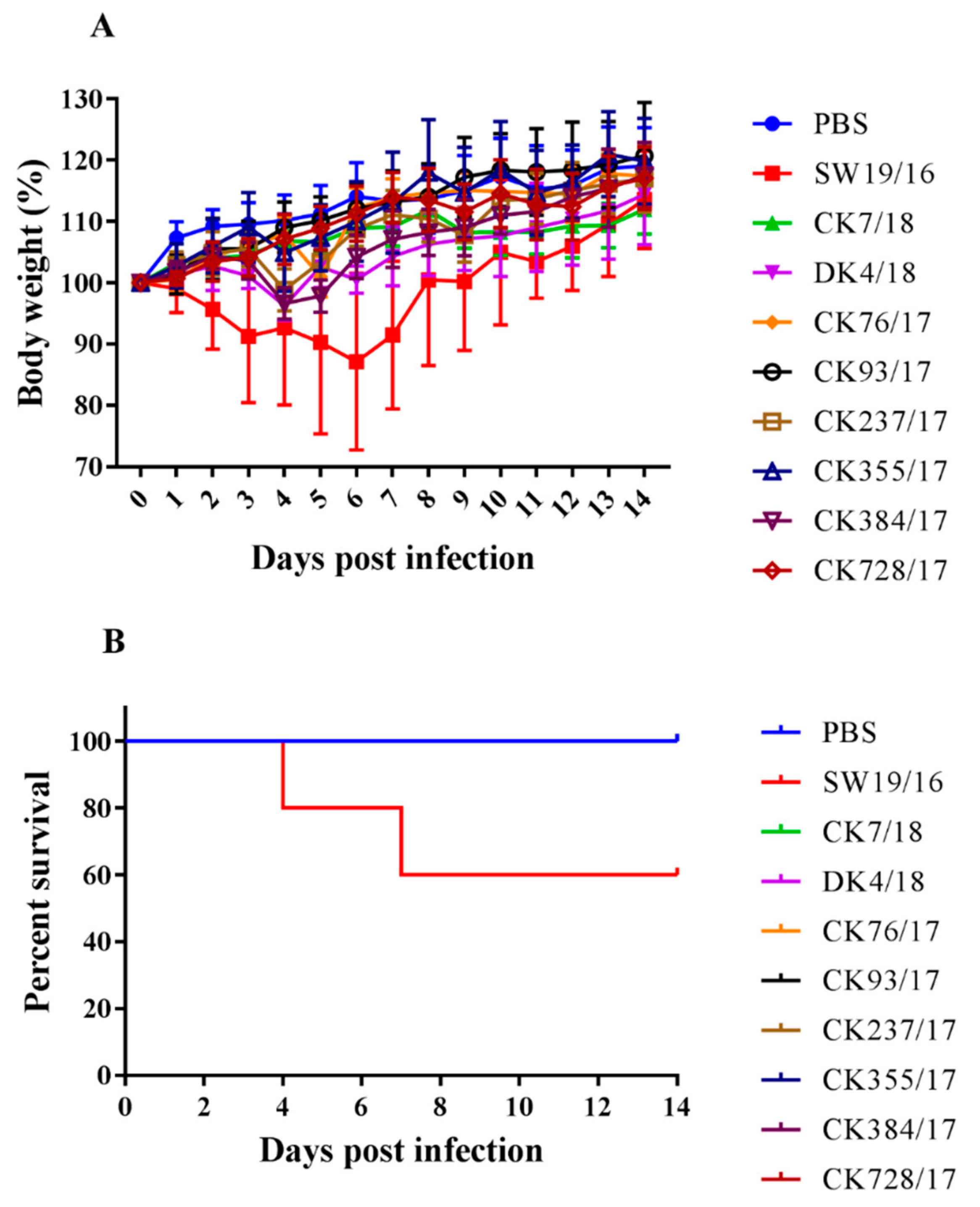
2.6. Pathogenicity in Mice
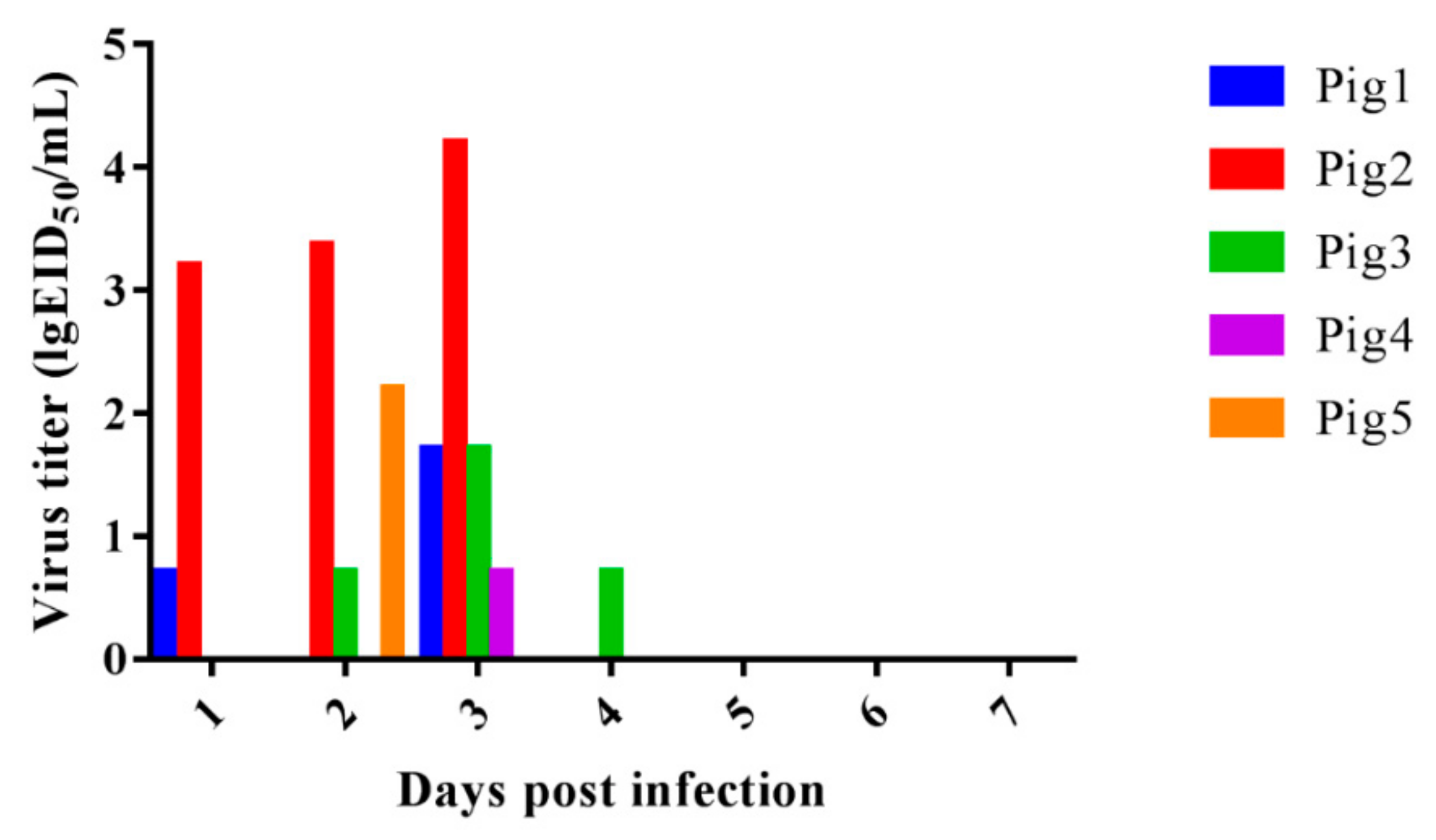
2.7. Infection of Pigs
2.8. Ethics Animal Handling
3. Results
3.1. H9N2 Viruses Form a Novel Genotype
3.2. H9N2 Viruses Effectively Replicate In Vitro
3.3. H9N2 Viruses Preferentially Bind to the α-2,6-linked SA Receptor
3.4. Virus Replication in the Lung of Mice
3.5. Virus Replication in the Turbinate of Pigs and Virus Shedding Last for Four Days
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Guo, Y.J.; Krauss, S.; Senne, D.A.; Mo, I.P.; Lo, K.S.; Xiong, X.P.; Norwood, M.; Shortridge, K.F.; Webster, R.G.; Guan, Y. Characterization of the Pathogenicity of Members of the Newly Established H9N2 Influenza Virus Lineages in Asia. Virology 2000, 267, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, D.J.; Brown, I.H. Recent Zoonoses Caused by Influenza A Viruses. Rev. Sci. Et Tech. 2000, 19, 197–225. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Chin, P.S.; Dyrting, K.C.; Ellis, T.M.; Webster, R.G.; Peiris, M. H9N2 Influenza Viruses Possessing H5N1-Like Internal Genomes Continue to Circulate in Poultry in Southeastern CHINA. J. Virol. 2000, 74, 9372–9380. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.; Shkoda, I.; Golender, N.; Perk, S.; Lapin, K.; Khinich, Y.; Panshin, A. Genetic Characterization of HA Gene of Low Pathogenic H9N2 Influenza Viruses Isolated in Israel During 2006–2012 Periods. Virus Genes 2013, 46, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.K.; Ozaki, H.; Webby, R.J.; Webster, R.G.; Peiris, J.S.; Poon, L.; Butt, C.; Leung, Y.H.; Guan, Y. Continuing Evolution of H9N2 Influenza Viruses in Southeastern China. J. Virol. 2004, 78, 8609–8614. [Google Scholar] [CrossRef] [Green Version]
- Kwon, H.J.; Cho, S.H.; Kim, M.C.; Ahn, Y.J.; Kim, S.J. Molecular Epizootiology of Recurrent Low Pathogenic Avian Influenza by H9N2 Subtype Virus in Korea. Avian Pathol. J. WVPA 2006, 35, 309–315. [Google Scholar] [CrossRef]
- Lee, D.C.W.; Mok, C.K.P.; Law, A.H.Y.; Peiris, M.; Lau, A.S.Y. Differential Replication of Avian Influenza H9N2 Viruses in Human Alveolar Epithelial A549 Cells. Virol. J. 2010, 7, 71. [Google Scholar] [CrossRef]
- Zhu, Y.B.; Yang, Y.; Liu, W.; Liu, X.; Yang, D.; Sun, Z.H.; Ju, Y.; Chen, S.J.; Peng, D.X.; Liu, X.F. Comparison of Biological Characteristics of H9N2 Avian Influenza Viruses Isolated from Different Hosts. Arch. Virol. 2015, 160, 917–927. [Google Scholar] [CrossRef]
- Li, C.J.; Yu, K.Z.; Tian, G.B.; Yu, D.D.; Liu, L.L.; Jing, B.; Ping, J.H.; Chen, H.L. Evolution of H9N2 Influenza Viruses from Domestic Poultry in Mainland China. Virology 2005, 340, 70–83. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Ji, J.; Zuo, K.J.; Xie, Q.M.; Li, H.M.; Liu, J.; Chen, F.; Xue, C.Y.; Ma, J.Y.; Bi, Y.Z. Cloning and Phylogenetic Analysis of Hemagglutinin Gene of H9N2 Subtype Avian Influenza Virus from Different Isolates in China During 2002 to 2009. Poult. Sci. 2010, 89, 1136–1143. [Google Scholar] [CrossRef]
- Pu, J.; Wang, S.; Yin, Y.; Zhang, G.; Carter, R.A.; Wang, J.; Xu, G.; Sun, H.; Wang, M.; Wen, C.; et al. Evolution of the H9N2 Influenza Genotype that Facilitated the Genesis of the Novel H7N9 Virus. Proc. Natl. Acad. Sci. USA 2015, 112, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Zhang, Y.; Li, X.; Bo, H.; Wei, H.; Dong, L.; Yang, L.; Dong, J.; Liu, J.; Shu, Y.; et al. Molecular Characterization and Receptor Binding Specificity of H9N2 Avian Influenza Viruses Based on Poultry-Related Environmental Surveillance in China Between 2013 and 2016. Virology 2019, 529, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.M.; Deng, G.C.; Dong, J.; Kong, F.L.; Li, X.Z.; Xu, Q.A.; Zhang, M.J.; Zhao, L.H.; Qiao, J.A. Phylogenetic and Molecular Characterization of H9N2 Influenza Isolates from Chickens in Northern China from 2007-2009. PLoS ONE 2010, 5, e13063. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Webster, R.G. Molecular Characterization of H9N2 Influenza Viruses: Were they the Donors of the “Internal” Genes of H5N1 Viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 1999, 96, 9363–9367. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhou, L.; Zhou, M.; Chen, Z.; Li, F.; Wu, H.; Xiang, N.; Chen, E.; Tang, F.; Wang, D.; et al. Epidemiology of Human Infections with Avian Influenza A(H7N9) Virus in China. New Engl. J. Med. 2014, 370, 520–532. [Google Scholar] [CrossRef]
- Chen, Y.; Liang, W.; Yang, S.; Wu, N.; Gao, H.; Sheng, J.; Yao, H.; Wo, J.; Fang, Q.; Cui, D.; et al. Human Infections with the Emerging Avian Influenza A H7N9 Virus from Wet Market Poultry: Clinical Analysis and Characterisation of Viral Genome. Lancet 2013, 381, 1916–1925. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, H.; Gao, R.; Zhang, J.; Wang, D.; Xiong, Y.; Fan, G.; Yang, F.; Li, X.; Zhou, J.; et al. Clinical and Epidemiological Characteristics of a Fatal Case of Avian Influenza A H10N8 Virus Infection: A Descriptive Study. Lancet 2014, 383, 714–721. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Yuan, H.; Zhou, J.; Wu, J.; Bo, H.; Xia, W.; Xiong, Y.; Yang, L.; Gao, R.; et al. Genetic Diversity of Avian Influenza A (H10N8) Virus in Live Poultry Markets and its Association with Human Infections in China. Sci. Rep. 2015, 5, 7632. [Google Scholar] [CrossRef]
- Shen, H.; Wu, B.; Chen, Y.; Bi, Y.; Xie, Q. Influenza A(H5N6) Virus Reassortant, Southern China, 2014. Emerg. Infect. Dis. 2015, 21, 1261–1262. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Guan, W.; Yuan, B.; Wang, Y.; Li, Z.; Song, Y.; Li, S.; Yang, Z.; Zhong, N.; Zhang, Y.; et al. Emergence of Triple-Subtype Reassortants of Fatal Human H5N6 Avian Influenza Virus in Yunnan, China. J. Infect. 2016, 72, 753–756. [Google Scholar] [CrossRef]
- He, J.; Duan, J. First Human Case of Avian Influenza A (H5N6) in Yunnan Province, China. SAGE Open Med. Case Rep. 2015, 3, 2050313X15596484. [Google Scholar] [CrossRef] [PubMed]
- Butt, K.M.; Smith, G.J.D.; Chen, H.L.; Zhang, L.J.; Leung, Y.H.C.; Xu, K.M.; Lim, W.; Webster, R.G.; Yuen, K.Y.; Peiris, J.S.M.; et al. Human Infection with an Avian H9N2 Influenza a Virus in Hong Kong in 2003. J. Clin. Microbiol. 2005, 43, 5760–5767. [Google Scholar] [CrossRef] [PubMed]
- Peiris, M.; Yuen, K.Y.; Leung, C.W.; Chan, K.H.; Ip, P.L.; Lai, R.W.; Orr, W.K.; Shortridge, K.F. Human Infection with Influenza H9N2. Lancet 1999, 354, 916–917. [Google Scholar] [CrossRef]
- Zhou, P.; Zhu, W.; Gu, H.; Fu, X.; Wang, L.; Zheng, Y.; He, S.; Ke, C.; Wang, H.; Yuan, Z.; et al. Avian Influenza H9N2 Seroprevalence Among Swine Farm Residents in China. J. Med Virol. 2014, 86, 597–600. [Google Scholar] [CrossRef]
- Huang, Y.; Li, X.; Zhang, H.; Chen, B.; Jiang, Y.; Yang, L.; Zhu, W.; Hu, S.; Zhou, S.; Tang, Y.; et al. Human Infection with an Avian Influenza A (H9N2) Virus in the Middle Region of China. J. Med Virol. 2015, 87, 1641–1648. [Google Scholar] [CrossRef]
- Gao, S.; Kang, Y.; Li, S.; Xiang, B.; Ma, H.; Yuan, R. Increasing Genetic Diversity of H5N6 Avian Influenza Virus in China: A Serious Threat to Persistence and Dissemination in Guangdong Province. J. Infect. 2017, 75, 585–590. [Google Scholar] [CrossRef]
- Pan, Y.; Cui, S.; Sun, Y.; Zhang, X.; Ma, C.; Shi, W.; Peng, X.; Lu, G.; Zhang, D.; Liu, Y.; et al. Human Infection with H9N2 Avian Influenza in Northern China. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2018, 24, 321–323. [Google Scholar]
- Wang, Q.; Ju, L.; Liu, P.; Zhou, J.; Lv, X.; Li, L.; Shen, H.; Su, H.; Jiang, L.; Jiang, Q. Serological and virological Surveillance of Avian influenza A Virus H9N2 Subtype in Humans and Poultry in Shanghai, China, between 2008 and 2010. Zoonoses Public Health 2015, 62, 131–140. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, L.Q.; Pu, J.; Zhao, J.Y.; Sun, Y.P.; Shen, G.N.; Wei, H.T.; Zhu, J.J.; Zheng, R.F.; Xiong, D.Y.; et al. Risk Perceptions for Avian Influenza Virus Infection among Poultry Workers, China. Emerg. Infect. Dis. 2013, 19, 313–316. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zheng, Q.; Yang, K.; Zeng, F.; Lau, S.Y.; Wu, W.L.; Huang, S.; Zhang, J.; Chen, H.; Xia, N. Serological Survey of Antibodies to Influenza A Viruses in a Group of People without a History of Influenza Vaccination. Clin. Microbiol. Infec. 2011, 17, 1347–1349. [Google Scholar] [CrossRef]
- Huang, R.; Wang, A.R.; Liu, Z.H.; Liang, W.; Li, X.X.; Tang, Y.J.; Miao, Z.M.; Chai, T.J. Seroprevalence of Avian Influenza H9N2 Among Poultry Workers in Shandong Province, China. Eur. J. Clin. Microbiol. 2013, 32, 1347–1351. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fan, W.; Wei, R.; Zhao, H. Isolation and Identification of Swine Influenza Recombinant A/Swine/Shandong/1/2003(H9N2) Virus. Microbes Infect. 2004, 6, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.L.; Wang, C.F.; Yan, C.M.; Peng, J.S.; Jiang, Z.L.; Liu, J.H. Swine Infection with H9N2 Influenza Viruses in China in 2004. Virus Genes 2008, 36, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.L.; Pu, J.; Liu, Q.F.; Wang, S.; Zhang, G.Z.; Zhang, X.L.; Fan, W.X.; Brown, E.G.; Liu, J.H. Antigenic and Genetic Characterization of H9N2 Swine Influenza Viruses in China. J. Gen. Virol. 2007, 88, 2035–2041. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.; Guan, Y.; Markwell, D.; Ghose, P.; Webster, R.G.; Shortridge, K.F. Cocirculation of Avian H9N2 and Contemporary "Human" H3N2 Influenza A Viruses in Pigs in Southeastern China: Potential for Genetic Reassortment? J. Virol. 2001, 75, 9679–9686. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, A.; Takada, A.; Okazaki, K.; Shortridge, K.F.; Kida, H. Seroepidemiological Evidence of Avian H4, H5, and H9 Influenza A Virus Transmission to Pigs in Southeastern China. Vet. Microbiol. 2002, 88, 107–114. [Google Scholar] [CrossRef]
- Li, S.; Zhou, Y.; Zhao, Y.; Li, W.; Song, W.; Miao, Z. Avian Influenza H9N2 Seroprevalence Among Pig Population and Pig Farm Staff in Shandong, China. Virol. J. 2015, 12, 34. [Google Scholar] [CrossRef]
- Yuan, Z.X.; Zhu, W.J.; Chen, Y.; Zhou, P.; Cao, Z.P.; Xie, J.X.; Zhang, C.H.; Ke, C.W.; Qi, W.B.; Su, S.; et al. Serological Surveillance of H5 and H9 Avian Influenza A Viral Infections Among Pigs in Southern China. Microb. Pathog. 2013, 64, 39–42. [Google Scholar] [CrossRef]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal Primer Set for the Full-Length Amplification of All Influenza A Viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef]
- Pan, W.Q.; Xie, H.J.; Li, X.B.; Guan, W.D.; Chen, P.H.; Zhang, B.W.; Zhang, M.C.; Dong, J.; Wang, Q.; Li, Z.X.; et al. Patient-Derived Avian Influenza A (H5N6) Virus is Highly Pathogenic in Mice but can be Effectively Treated by Anti-Influenza Polyclonal Antibodies. Emerg. Microbes Infec. 2018, 7, 1–10. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, W.; Qi, J.; Wang, F.; Zhou, J.; Bi, Y.; Wu, Y.; Sun, H.; Liu, J.; Huang, C.; et al. Structural Basis for Preferential Avian Receptor Binding by the Human-Infecting H10N8 Avian Influenza Virus. Nat. Commun. 2015, 6, 5600. [Google Scholar] [CrossRef] [PubMed]
- Parvin, R.; Shehata, A.A.; Heenemann, K.; Gac, M.; Rueckner, A.; Halami, M.Y.; Vahlenkamp, T.W. Differential Replication Properties Among H9N2 Avian Influenza Viruses of Eurasian Origin. Vet. Res. 2015, 46, 75. [Google Scholar] [CrossRef] [PubMed]
- Moatasim, Y.; Kandeil, A.; Mostafa, A.; Elghaffar, S.K.A.; El Shesheny, R.; Elwahy, A.H.M.; Ali, M.A. Single Gene Reassortment of Highly Pathogenic Avian Influenza A H5N1 in the Low Pathogenic H9N2 Backbone and its Impact on Pathogenicity and Infectivity of Novel Reassortant Viruses. Arch. Virol. 2017, 162, 2959–2969. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.I.; Cordeiro, M.; Sevilla, E.; Liu, J. Comparison of Egg and High Yielding MDCK Cell-Derived Live Attenuated Influenza Virus for Commercial Production of Trivalent Influenza Vaccine: In Vitro Cell Susceptibility and Influenza Virus Replication Kinetics in Permissive and Semi-Permissive Cells. Vaccine 2010, 28, 3848–3855. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.P.; Liu, Q.F.; Su, X.; Teng, Q.Y.; Bao, D.Q.; Che, G.S.; Chen, H.J.; Cui, H.R.; Ruan, T.; Li, X.S.; et al. Pathogenicity of Reassortant H9 Influenza Viruses with Different NA Genes in Mice and Chickens. Vet. Res. 2016, 47, 67. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhang, X.; Gao, W.; Wang, C.; Wang, J.; Sun, H.; Sun, Y.; Guo, L.; Zhang, R.; Chang, K.C.; et al. Prevailing PA Mutation K356R in Avian Influenza H9N2 Virus Increases Mammalian Replication and Pathogenicity. J. Virol. 2016, 90, 8105–8114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Sun, W.; Shi, Y.; Xing, Z.; Su, X. Altered Viral Replication and Cell Responses by Inserting microRNA Recognition Element into PB1 in Pandemic Influenza A Virus (H1N1) 2009. Mediat. Inflamm. 2015, 2015, 976575. [Google Scholar] [CrossRef]
- Zou, S.; Gao, R.; Zhang, Y.; Li, X.; Chen, W.; Bai, T.; Dong, L.; Wang, D.; Shu, Y. Molecular Characterization of H6 Subtype Influenza Viruses in Southern China from 2009 to 2011. Emerg. Microbes Infect. 2016, 5, e73. [Google Scholar] [CrossRef]
- Xiao, C.; Ma, W.; Sun, N.; Huang, L.; Li, Y.; Zeng, Z.; Wen, Y.; Zhang, Z.; Li, H.; Li, Q.; et al. PB2-588 V Promotes the Mammalian Adaptation of H10N8, H7N9 and H9N2 Avian Influenza Viruses. Sci. Rep. 2016, 6, 19474. [Google Scholar] [CrossRef]
- Gao, W.; Zu, Z.; Liu, J.; Song, J.; Wang, X.; Wang, C.; Liu, L.; Tong, Q.; Wang, M.; Sun, H.; et al. Prevailing I292V PB2 Mutation in Avian Influenza H9N2 Virus Increases Viral Polymerase Function and Attenuates IFN-Beta Induction in Human Cells. J. Gen. Virol. 2019. [Google Scholar] [CrossRef]
- Li, X.; Shi, J.; Guo, J.; Deng, G.; Zhang, Q.; Wang, J.; He, X.; Wang, K.; Chen, J.; Li, Y.; et al. Genetics, Receptor Binding Property, and Transmissibility in Mammals of Naturally Isolated H9N2 Avian Influenza Viruses. PLoS Pathogens 2014, 10, e1004508. [Google Scholar] [CrossRef] [PubMed]
- Li, S.C.; Li, X.H.; Zhong, S.G.; Sun, H.L.; Pan, J.J.; Chen, S.J.; Peng, D.X.; Liu, X.F. Genome Sequencing and Phylogenetic Analysis of Avian Influenza Viruses suBtype H9N2. Bing Du Xue Bao Chin. J. Virol. 2012, 28, 7–14. [Google Scholar]
- Matrosovich, M.N.; Krauss, S.; Webster, R.G. H9N2 Influenza A Viruses from Poultry in Asia Have Human Virus-Like Receptor Specificity. Virology 2001, 281, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Perez, D.R. Amino Acid 226 in the Hemagglutinin of H9N2 Influenza Viruses Determines Cell Tropism and Replication in Human Airway Epithelial Cells. J. Virol. 2007, 81, 5181–5191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, Y.; Ibrahim, M.S.; Ellakany, H.F.; Kawashita, N.; Mizuike, R.; Hiramatsu, H.; Sriwilaijaroen, N.; Takagi, T.; Suzuki, Y.; Ikuta, K. Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages During Their Emergence in Birds in Egypt. PLoS Pathogens 2011, 7, e1002068. [Google Scholar] [CrossRef] [PubMed]
- Teng, Q.; Xu, D.; Shen, W.; Liu, Q.; Rong, G.; Li, X.; Yan, L.; Yang, J.; Chen, H.; Yu, H.; et al. A Single Mutation at Position 190 in Hemagglutinin Enhances Binding Affinity for Human Type Sialic Acid Receptor and Replication of H9N2 Avian Influenza Virus in Mice. J. Virol. 2016, 90, 9806–9825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.T.; Liu, Y.Z.; Yang, J.; Huang, X.M.; Han, K.K.; Zhao, D.M.; Bi, K.R.; Li, Y. Two Genetically Similar H9N2 Influenza A Viruses Show Different Pathogenicity in Mice. Front. Microbiol. 2016, 7, 1737. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.T.; Chen, H.Z.; Huang, J.Q.; Chen, Y.X.; Gu, M.; Wang, X.Q.; Hu, S.L.; Liu, X.W.; Liu, X.F. A Nonpathogenic Duck-Origin H9N2 Influenza A Virus Adapts to High Pathogenicity in Mice. Arch. Virol. 2014, 159, 2243–2252. [Google Scholar] [CrossRef]
- Ma, W.; Kahn, R.E.; Richt, J.A. The Pig as A Mixing Vessel for Influenza Viruses: Human and Veterinary Implications. J. Mol. Genet. Med. An Int. J. Biomed. Res. 2008, 3, 158–166. [Google Scholar] [CrossRef]
- Scholtissek, C. Source for Influenza Pandemics. Eur. J. Epidemiol. 1994, 10, 455–458. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and Ecology of Influenza A Viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [PubMed]
- Wang, J.; Wu, M.C.; Hong, W.S.; Fan, X.H.; Chen, R.R.; Zheng, Z.Y.; Zeng, Y.; Huang, R.; Zhang, Y.; Lam, T.T.Y.; et al. Infectivity and Transmissibility of Avian H9N2 Influenza Viruses in Pigs. J. Virol. 2016, 90, 3506–3514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, C.L.; Liu, Q.F.; Bawa, B.; Shen, H.G.; Qi, W.B.; Chen, Y.; Mok, C.K.P.; Garcia-Sastre, A.; Richt, J.A.; Ma, W.J. Pathogenicity and Transmissibility of Reassortant H9 Influenza Viruses with Genes from Pandemic H1N1 Virus. J. Gen. Virol. 2012, 93, 2337–2345. [Google Scholar] [CrossRef] [PubMed]
- Nomura, N.; Sakoda, Y.; Endo, M.; Yoshida, H.; Yamamoto, N.; Okamatsu, M.; Sakurai, K.; Hoang, N.V.; Nguyen, L.V.; Chu, H.D.; et al. Characterization of Avian Influenza Viruses Isolated from Domestic Ducks in Vietnam in 2009 and 2010. Arch. Virol. 2012, 157, 247–257. [Google Scholar] [CrossRef]
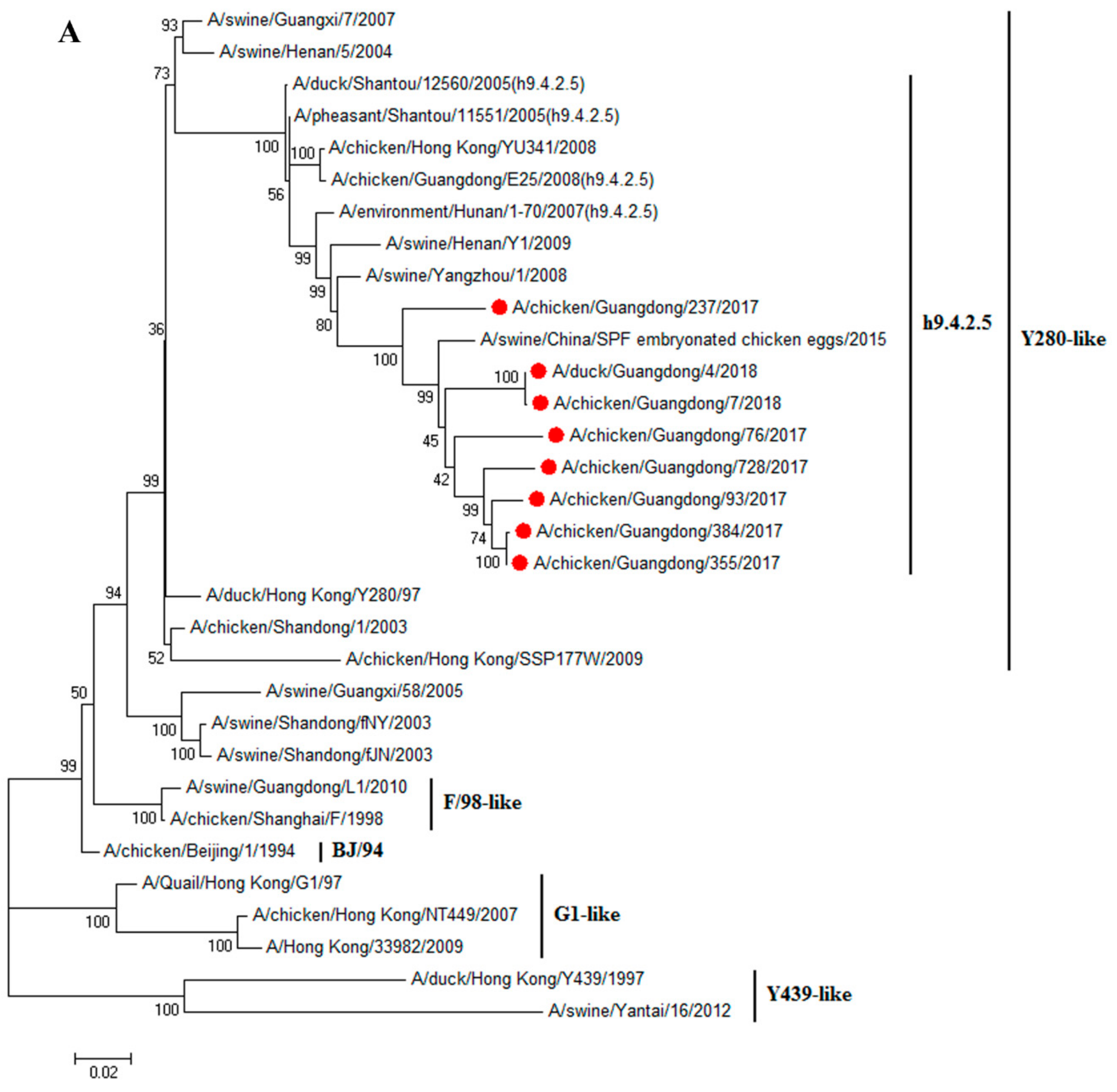
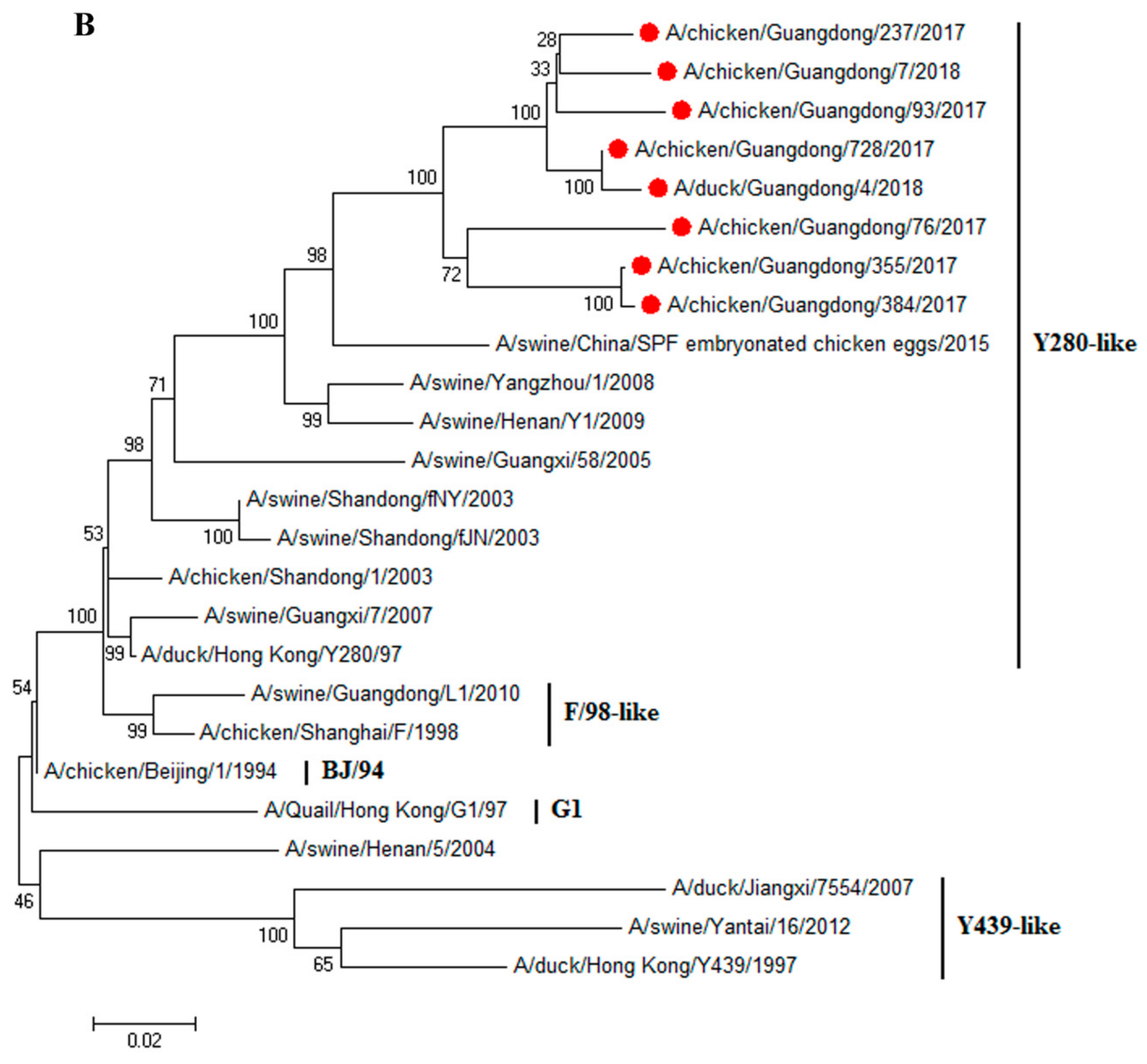
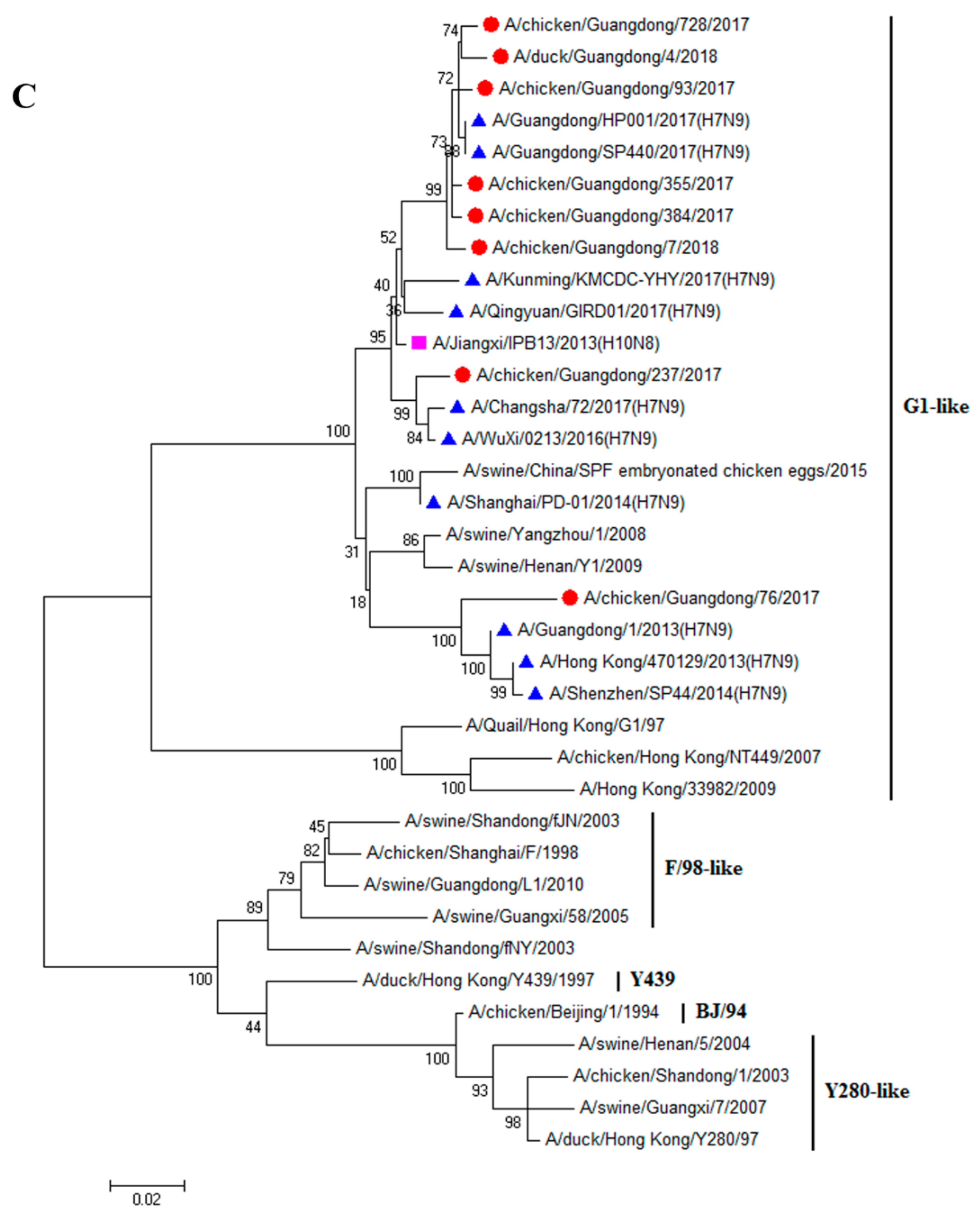
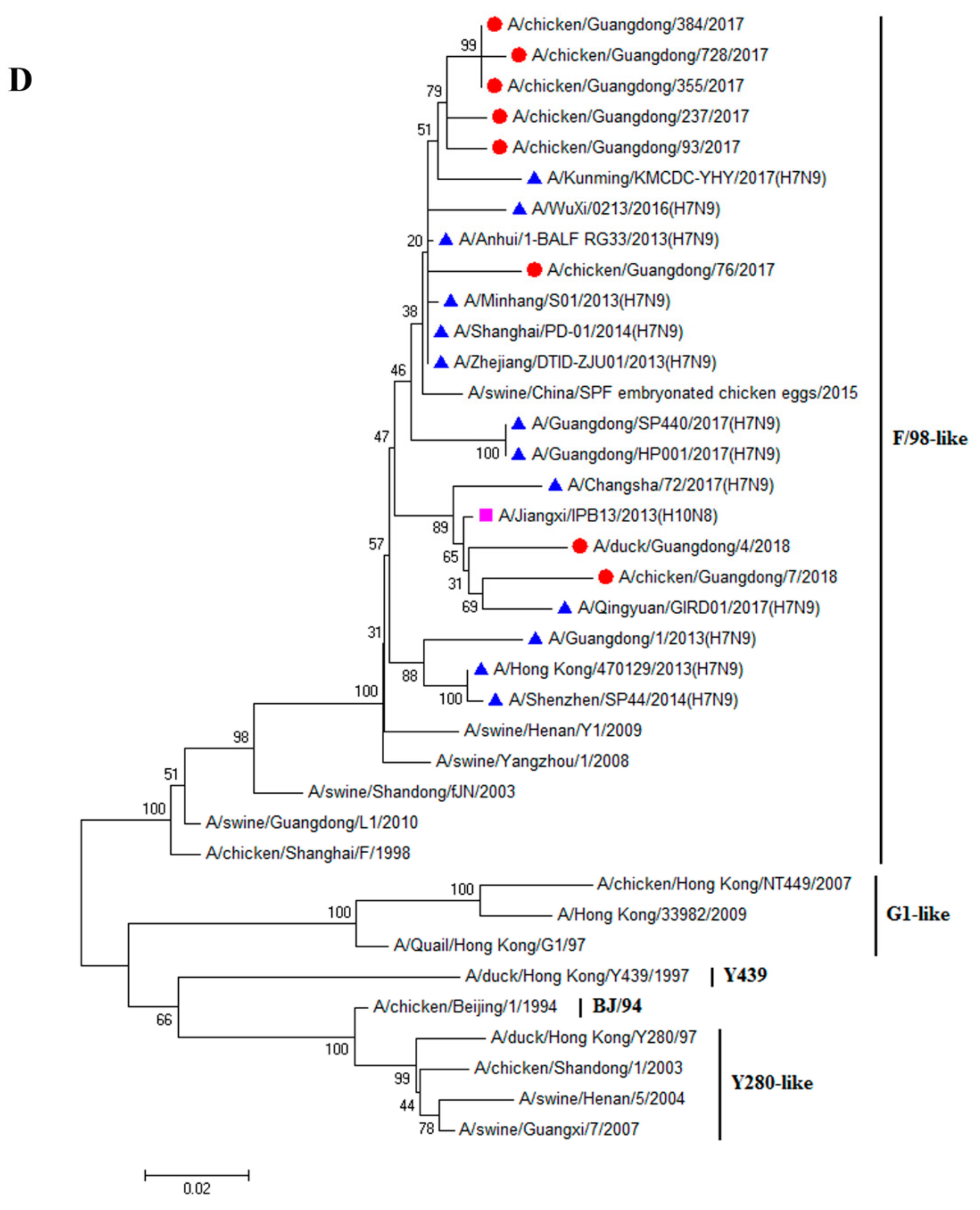
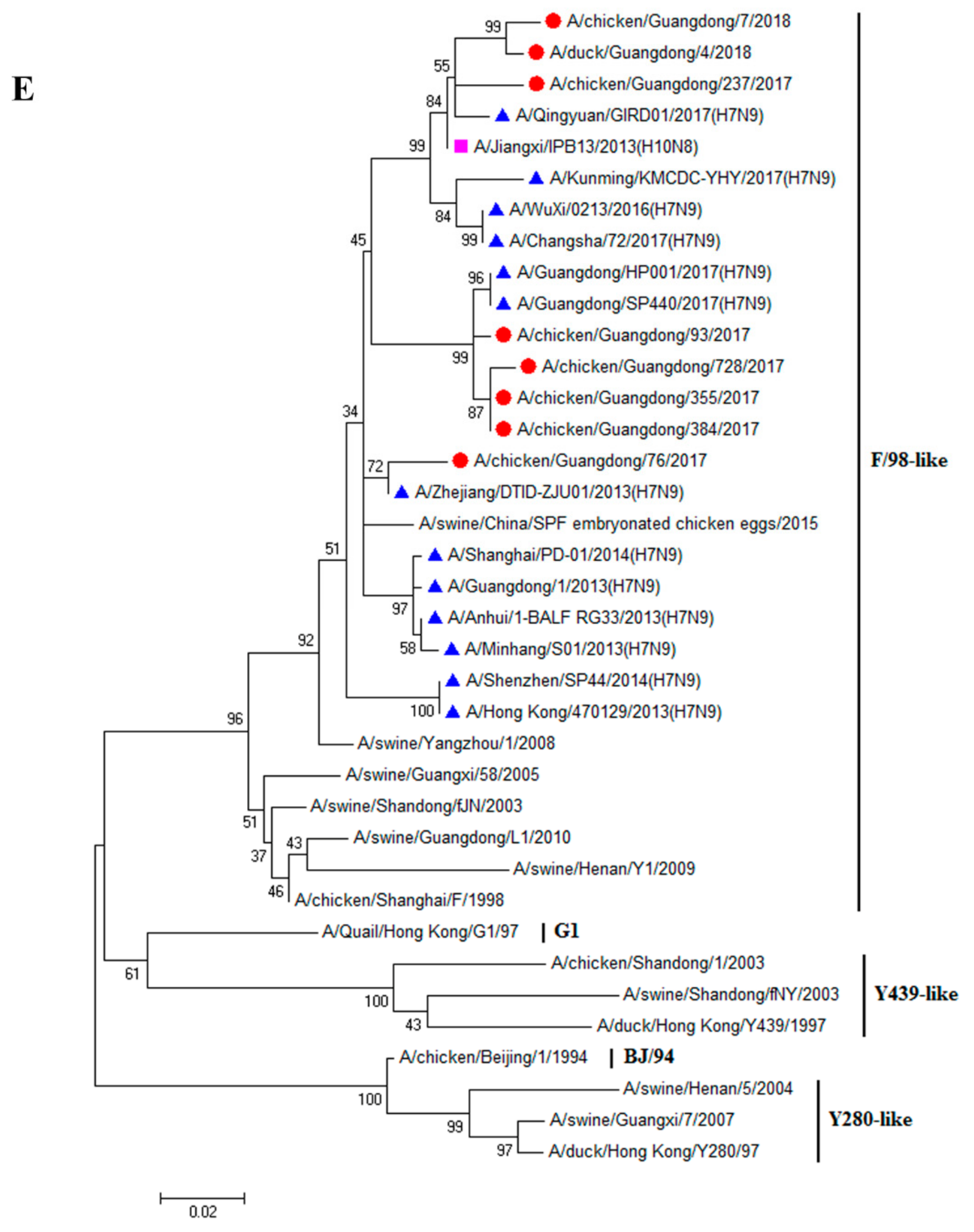
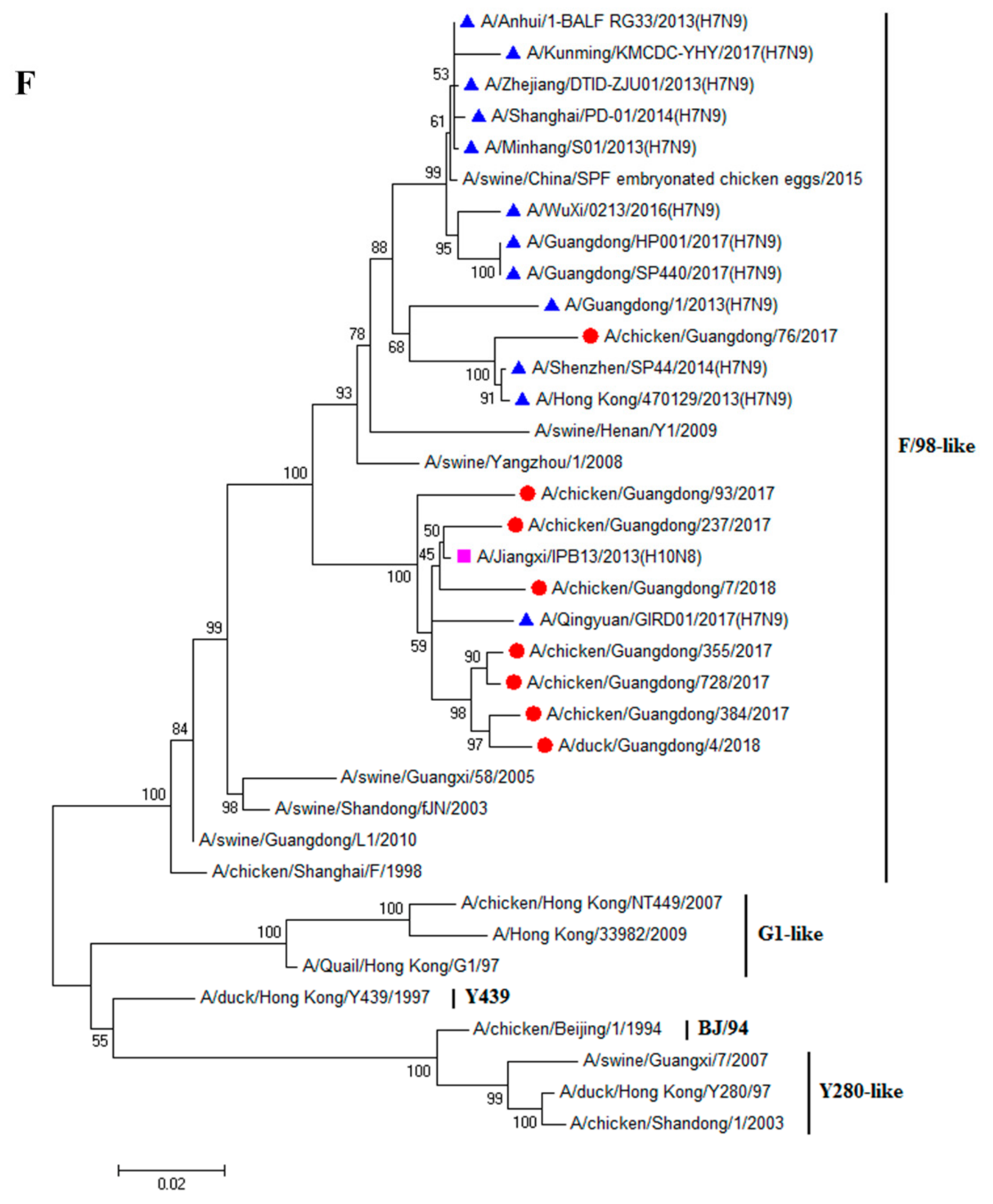
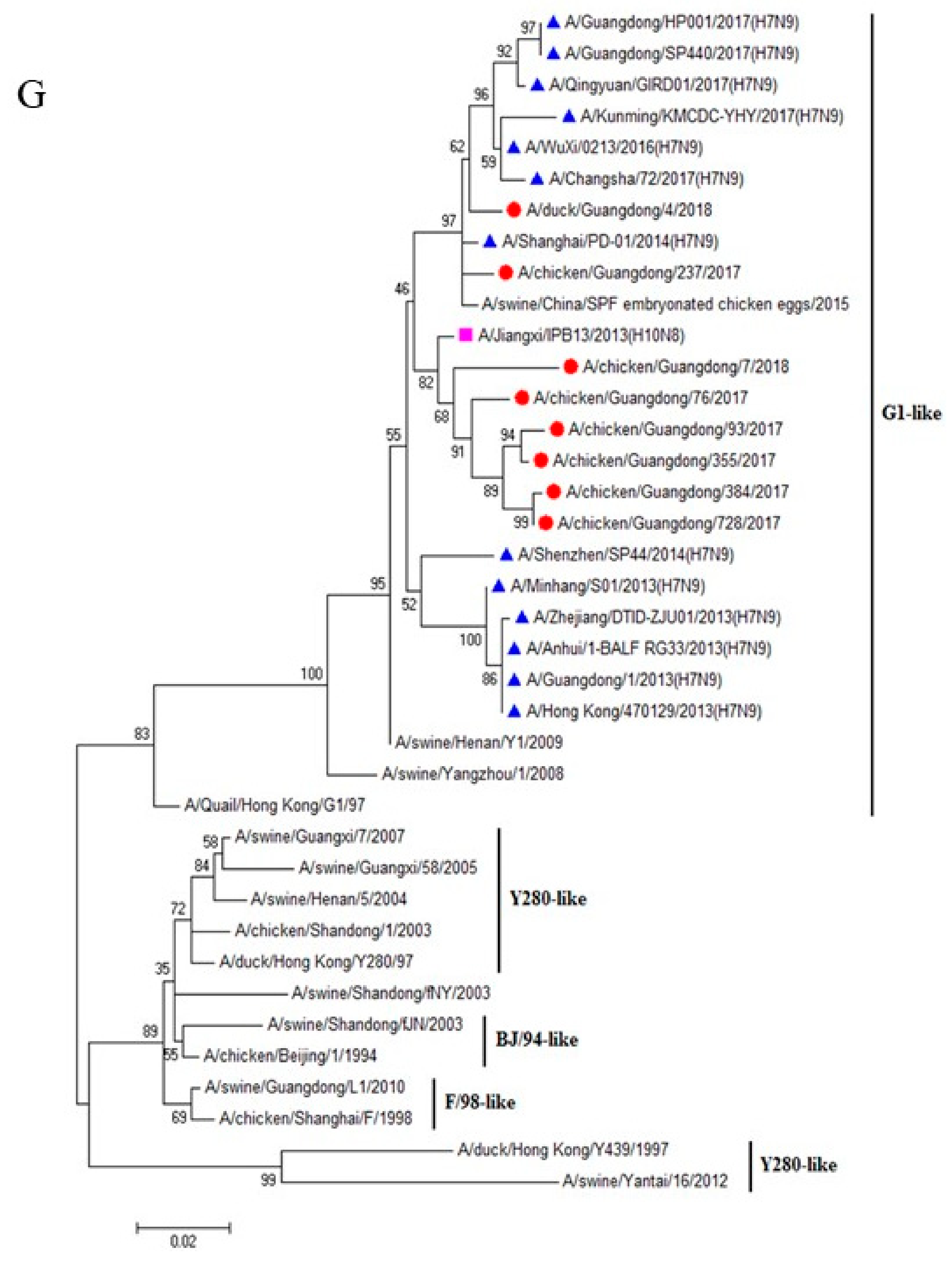
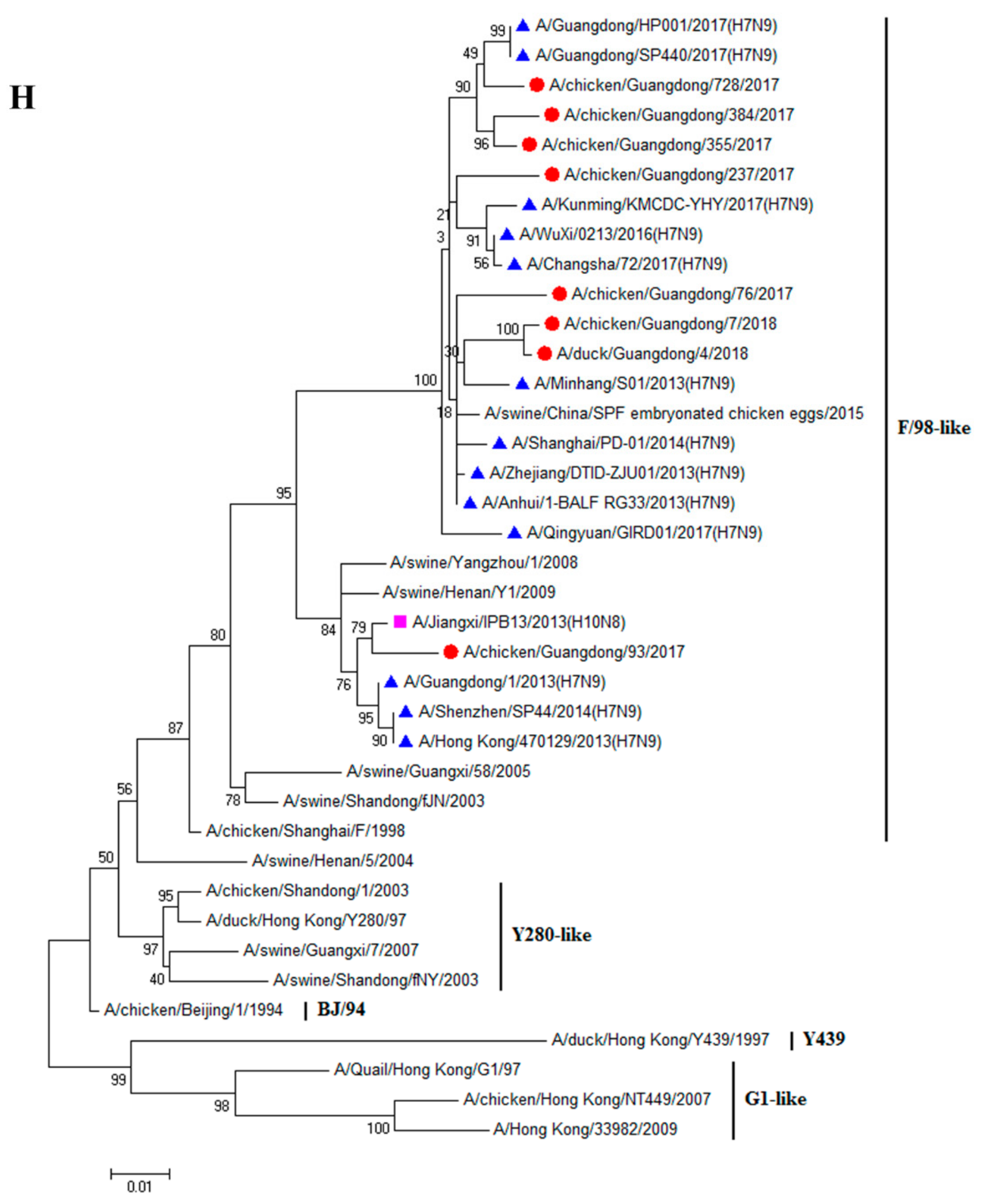
| Viruses | Cleavage Site in HA | Residues in HA (H3 Numbering) | Residues in PB2 | Residues in PB1 | Residues in PA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I155T | H183N | A190T/V | T212I | Q226L | D253N | I292V | 588V | 591K | E627K | G685R | D701N | I368V | K356R | ||
| A/chicken/Guangdong/7/2018 | PSRSSR | T | N | T | I | L | D | V | V | Q | E | G | D | V | R |
| A/duck/Guangdong/4/2018 | PSRSSR | T | N | T | I | L | D | V | I | Q | E | G | D | V | R |
| A/chicken/Guangdong/76/2017 | PSRSSR | I | N | T | V | L | D | V | A | Q | E | G | D | V | R |
| A/chicken/Guangdong/93/2017 | PSRSSR | T | N | T | I | L | D | V | V | Q | E | G | D | V | R |
| A/chicken/Guangdong/237/2017 | PSRSSR | T | N | A | I | L | D | V | T | Q | E | G | D | V | R |
| A/chicken/Guangdong/355/2017 | PSRSSR | T | N | T | I | L | D | V | V | Q | E | G | D | V | R |
| A/chicken/Guangdong/384/2017 | PSRSSR | T | N | T | I | L | D | V | V | Q | E | G | D | V | R |
| A/chicken/Guangdong/728/2017 | PSRSSR | T | N | T | I | L | D | V | I | Q | E | G | D | V | R |
| Virus | Lung | Spleen | Kidney | Brain |
|---|---|---|---|---|
| SW19/16 | 5.83 ± 0.42 | ND | ND | ND |
| CK7/18 | ND | ND | ND | ND |
| DK4/18 | 5.50 ± 0.20 | ND | ND | ND |
| CK76/17 | ND | ND | ND | ND |
| CK93/17 | ND | ND | ND | ND |
| CK237/17 | 1.08 ± 1.53 | ND | ND | ND |
| CK355/17 | ND | ND | ND | ND |
| CK384/17 | ND | ND | ND | ND |
| CK728/17 | 0.58 ± 0.82 | ND | ND | ND |
| PBS | ND | ND | ND | ND |
| Virus | Mouse 1 | Mouse 2 | Mouse 3 | Mouse 4 | Mouse 5 |
|---|---|---|---|---|---|
| SW19/16 | 80 | 160 | 80 | 80 | 80 |
| CK7/18 | 160 | 160 | 160 | 80 | 80 |
| DK4/18 | 320 | 320 | 320 | 640 | 320 |
| CK76/17 | 80 | 80 | 80 | 80 | 80 |
| CK93/17 | 80 | 40 | 40 | 40 | 20 |
| CK237/17 | 80 | 160 | 160 | 320 | 160 |
| CK355/17 | 80 | 80 | 80 | 80 | 40 |
| CK384/17 | 160 | 20 | 40 | 160 | 160 |
| CK728/17 | 160 | 320 | 160 | 160 | 160 |
| PBS | <10 | <10 | <10 | <10 | <10 |
| dpi. | Inoculated Pigs | Physical Contact Pigs | Control Pigs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pig 1 | Pig 2 | Pig 3 | Pig 4 | Pig 5 | Pig 6 | Pig 7 | Pig 8 | Pig 9 | Pig 10 | Pig 11 | Pig 12 | Pig 13 | |
| 7 | - | - | 40 | - | <10 | <10 | <10 | - | - | - | - | <10 | - |
| 14 | - | - | 160 | - | <10 | <10 | <10 | - | - | - | - | <10 | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Lin, J.; Liu, Z.; Yu, Y.; Wu, M.; Li, S.; Liu, Y.; Feng, Y.; Wu, Y.; Li, M.; et al. Genetic, Molecular, and Pathogenic Characterization of the H9N2 Avian Influenza Viruses Currently Circulating in South China. Viruses 2019, 11, 1040. https://doi.org/10.3390/v11111040
Sun H, Lin J, Liu Z, Yu Y, Wu M, Li S, Liu Y, Feng Y, Wu Y, Li M, et al. Genetic, Molecular, and Pathogenic Characterization of the H9N2 Avian Influenza Viruses Currently Circulating in South China. Viruses. 2019; 11(11):1040. https://doi.org/10.3390/v11111040
Chicago/Turabian StyleSun, Hailiang, Jiate Lin, Zhiting Liu, Yanan Yu, Meihua Wu, Shuo Li, Yang Liu, Yaling Feng, Yuqian Wu, Mingliang Li, and et al. 2019. "Genetic, Molecular, and Pathogenic Characterization of the H9N2 Avian Influenza Viruses Currently Circulating in South China" Viruses 11, no. 11: 1040. https://doi.org/10.3390/v11111040




