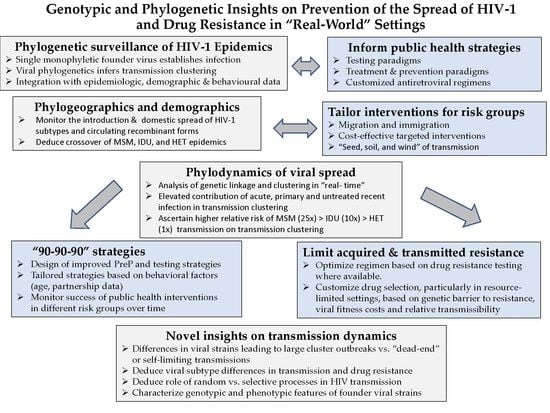
Genotypic and Phylogenetic Insights on Prevention of the Spread of HIV-1 and Drug Resistance in “Real-World” Settings
Abstract
:1. Introduction
2. Results and Discussion
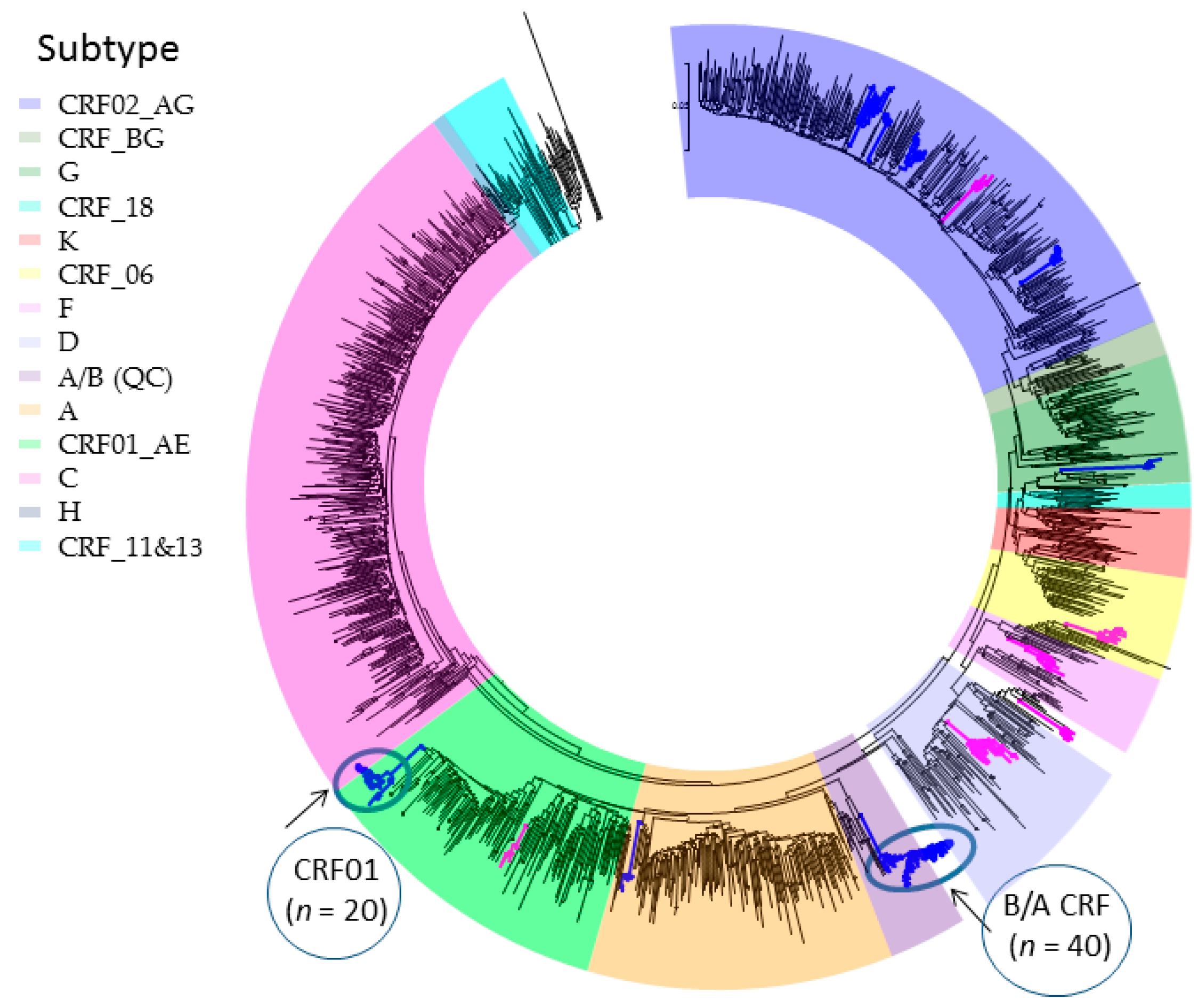
2.1. Phylogenetic Inferences on the Geographic Introduction and Spread of HIV
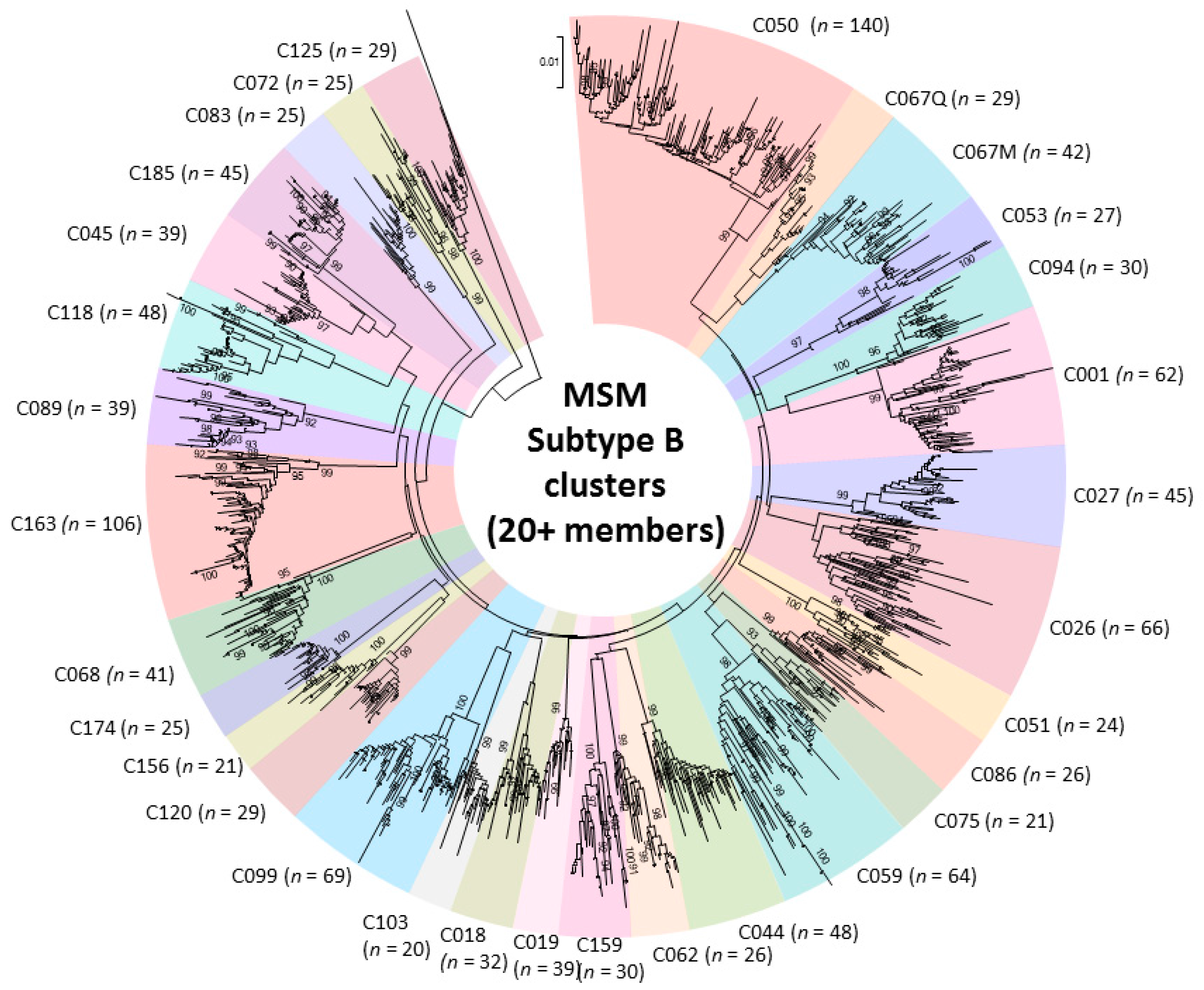
2.2. Phylogenetic Analysis of Transmission Clustering
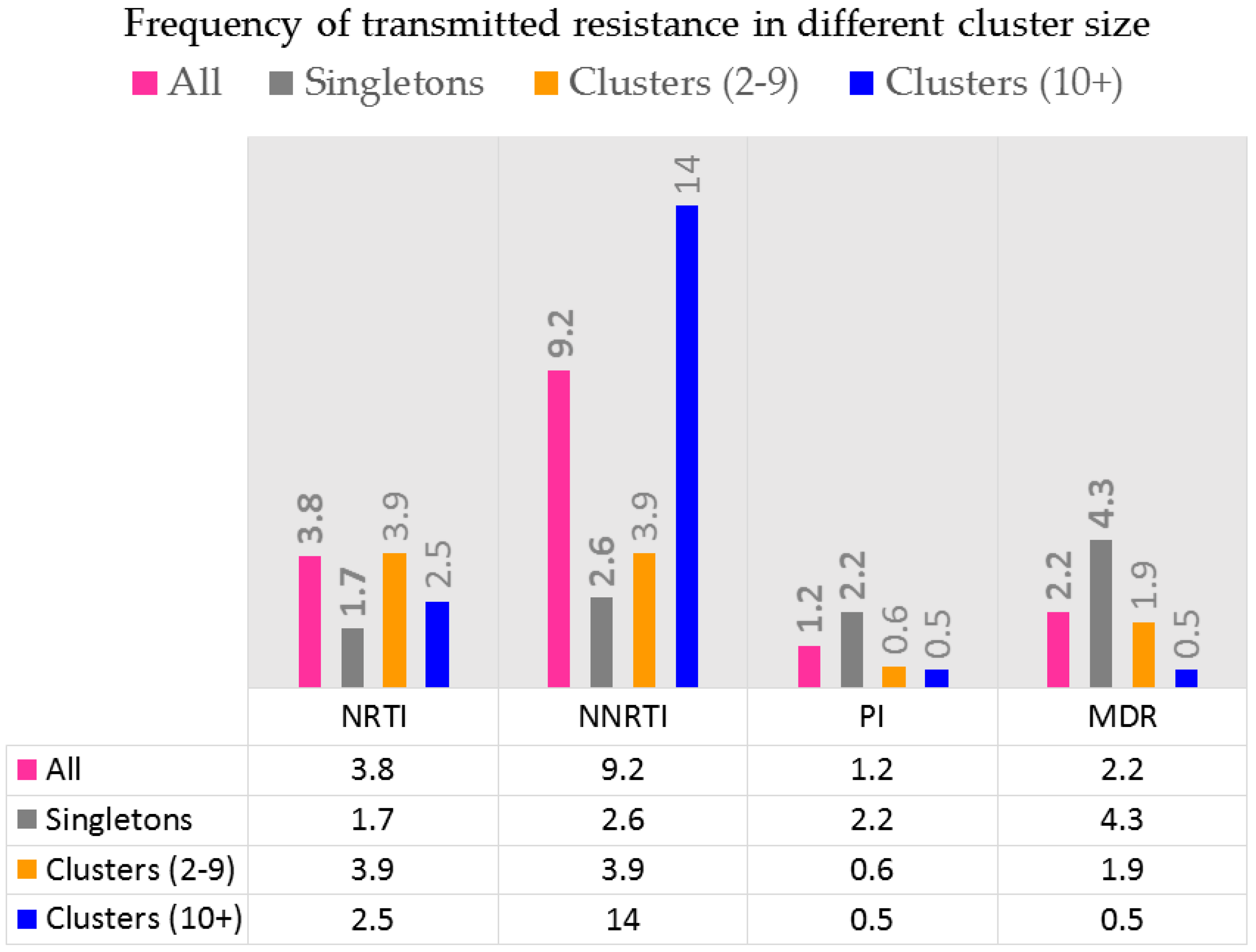
2.3. Phylogenetic Surveillance of Transmission Dynamics of the Quebec MSM Epidemic
2.4. Phylogenetic Inferences on Public Health Strategies to Control MSM and IDU Epidemics
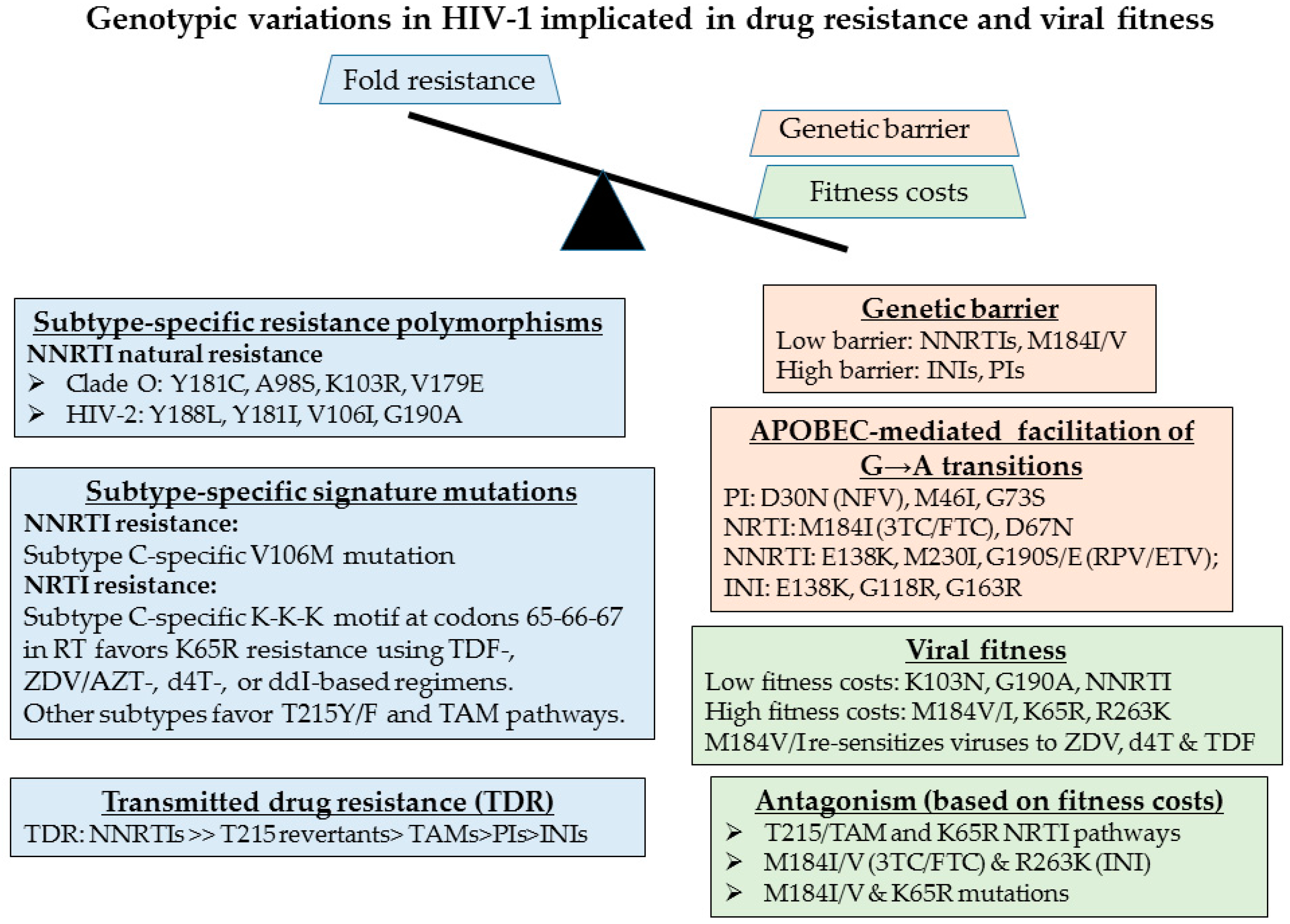
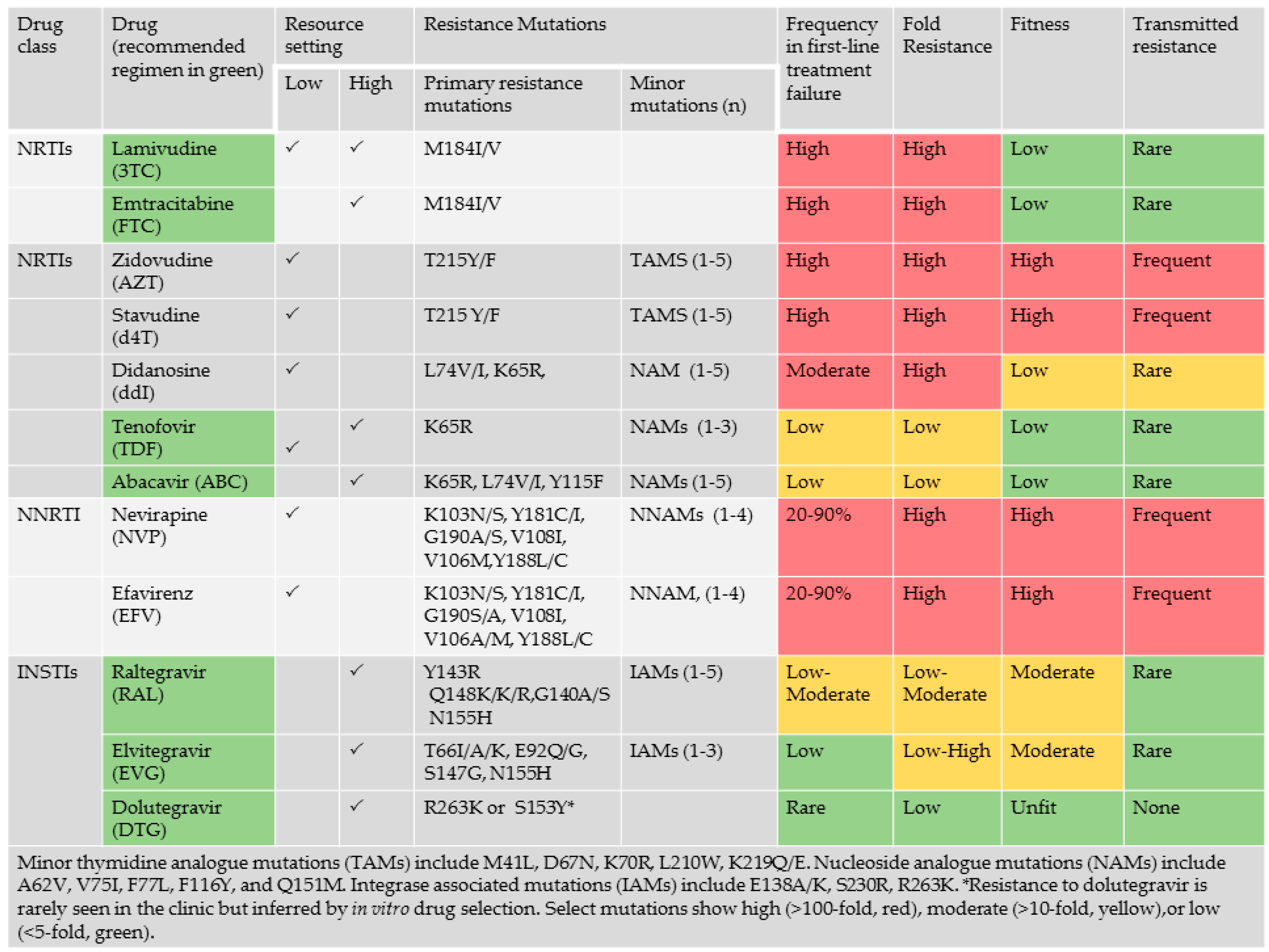
2.5. Genotypic Analysis Reveals Importance of Drug Regimen Selection in Long-Term HIV Management
2.6. The Need for Better Drugs in Africa
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Tebit, D.M.; Arts, E.J. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 2011, 11, 45–56. [Google Scholar] [CrossRef]
- Spira, S.; Wainberg, M.A.; Loemba, H.; Turner, D.; Brenner, B.G. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J. Antimicrob. Chemother. 2003, 51, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Beyrer, C.; Sullivan, P.; Sanchez, J.; Baral, S.D.; Collins, C.; Wirtz, A.L.; Altman, D.; Trapence, G.; Mayer, K. The increase in global HIV epidemics in MSM. AIDS 2013, 27, 2665–2678. [Google Scholar] [CrossRef] [PubMed]
- Wolf, E.; Herbeck, J.T.; van Rompaey, S.; Kitahata, M.; Thomas, K.; Pepper, G.; Frenkel, L. Short Communication: Phylogenetic Evidence of HIV-1 Transmission Between Adult and Adolescent Men Who Have Sex with Men. AIDS Res. Hum. Retrovir. 2017, 33, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Magiorkinis, G.; Angelis, K.; Mamais, I.; Katzourakis, A.; Hatzakis, A.; Albert, J.; Lawyer, G.; Hamouda, O.; Struck, D.; Vercauteren, J.; et al. The global spread of HIV-1 subtype B epidemic. Infect. Genet. Evol. 2016, 46, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Beloukas, A.; Psarris, A.; Giannelou, P.; Kostaki, E.; Hatzakis, A.; Paraskevis, D. Molecular epidemiology of HIV-1 infection in Europe: An overview. Infect. Genet. Evol. 2016, 46, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Montaner, J.S.; Lima, V.D.; Barrios, R.; Yip, B.; Wood, E.; Kerr, T.; Shannon, K.; Harrigan, P.R.; Hogg, R.S.; Daly, P.; et al. Association of highly active antiretroviral therapy coverage, population viral load and yearly new HIV diagnoses in British Columbia, Canada: A population-based study. Lancet 2010, 376, 532–539. [Google Scholar] [CrossRef]
- Jones, A.; Cremin, I.; Abdullah, F.; Idoko, J.; Cherutich, P.; Kilonzo, N.; Rees, H.; Hallett, T.; O’Reilly, K.; Koechlin, F.; et al. Transformation of HIV from pandemic to low-endemic levels: A public health approach to combination prevention. Lancet 2014, 384, 272–279. [Google Scholar] [CrossRef]
- Quinn, T.C.; Wawer, M.J.; Sewankambo, N.; Serwadda, D.; Li, C.; Wabwire-Mangen, F.; Meehan, M.O.; Lutalo, T.; Gray, R.H. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 2000, 342, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Powers, K.A.; Kretzschmar, M.E.; Miller, W.C.; Cohen, M.S. Impact of early-stage HIV transmission on treatment as prevention. Proc. Natl. Acad. Sci. USA 2014, 111, 15867–15868. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.S.; Chen, Y.Q.; McCauley, M.; Gamble, T.; Hosseinipour, M.C.; Kumarasamy, N.; Hakim, J.G.; Kumwenda, J.; Grinsztejn, B.; Pilotto, J.H.; et al. Prevention of HIV-1 infection with early antiretroviral therapy. N. Engl. J. Med. 2011, 365, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Kityo, C.; Thompson, J.; Nankya, I.; Hoppe, A.; Ndashimye, E.; Warambwa, C.; Mambule, I.; van Oosterhout, J.J.; Wools-Kaloustian, K.; Bertagnolio, S.; et al. Europe Africa Research Network for Evaluation of Second-line Therapy Trial, T.; HIV Drug Resistance Mutations in Non-B Subtypes After Prolonged Virological Failure on NNRTI-Based First-Line Regimens in Sub-Saharan Africa. J. Acquir. Immune Defic. Syndr. 2017, 75, e45–e54. [Google Scholar] [CrossRef] [PubMed]
- Villabona-Arenas, C.J.; Vidal, N.; Guichet, E.; Serrano, L.; Delaporte, E.; Gascuel, O.; Peeters, M. In-depth analysis of HIV-1 drug resistance mutations in HIV-infected individuals failing first-line regimens in West and Central Africa. AIDS 2016, 30, 2577–2589. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.; Wainberg, M.A. We need to use the best antiretroviral drugs worldwide to prevent HIV drug resistance. AIDS 2016, 30, 2725–2727. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Wainberg, M.A. Clinical benefit of dolutegravir in HIV-1 management related to the high genetic barrier to drug resistance. Virus Res. 2017, 239, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Beyrer, C.; Pozniak, A. HIV Drug Resistance—An Emerging Threat to Epidemic Control. N. Engl. J. Med. 2017, 377, 1605–1607. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.A.; Zaharatos, G.J.; Brenner, B.G. Development of antiretroviral drug resistance. N. Engl. J. Med. 2011, 365, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Kantor, R.; Katzenstein, D.A.; Efron, B.; Carvalho, A.P.; Wynhoven, B.; Cane, P.; Clarke, J.; Sirivichayakul, S.; Soares, M.A.; Snoeck, J.; et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: Results of a global collaboration. PLoS Med. 2005, 2, e112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, S.Y.; Sankaran, K.; Varghese, V.; Winters, M.A.; Hurt, C.B.; Eron, J.J.; Parkin, N.; Holmes, S.P.; Holodniy, M.; Shafer, R.W. HIV-1 Protease, Reverse Transcriptase and Integrase Variation. J. Virol. 2016, 90, 6058–6070. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Varghese, V.; Holmes, S.P.; van Zyl, G.U.; Steegen, K.; Boyd, M.A.; Cooper, D.A.; Nsanzimana, S.; Saravanan, S.; Charpentier, C.; et al. Mutational Correlates of Virological Failure in Individuals Receiving a WHO-Recommended Tenofovir-Containing First-Line Regimen: An International Collaboration. EBioMedicine 2017, 18, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Fessel, W.J.; Liu, T.F.; Marlowe, N.M.; Rowland, C.M.; Rode, R.A.; Vandamme, A.M.; van Laethem, K.; Brun-Vezinet, F.; Calvez, V.; et al. Predictive value of HIV-1 genotypic resistance test interpretation algorithms. J. Infect. Dis. 2009, 200, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Granich, R.; Crowley, S.; Vitoria, M.; Smyth, C.; Kahn, J.G.; Bennett, R.; Lo, Y.R.; Souteyrand, Y.; Williams, B. Highly active antiretroviral treatment as prevention of HIV transmission: Review of scientific evidence and update. Curr. Opin. HIV AIDS 2010, 5, 298–304. [Google Scholar] [CrossRef] [PubMed]
- DeHovitz, J.; Uuskula, A.; El-Bassel, N. The HIV epidemic in Eastern Europe and Central Asia. Curr. HIV/AIDS Rep. 2014, 11, 168–176. [Google Scholar] [CrossRef] [PubMed]
- El-Bassel, N.; Shaw, S.A.; Dasgupta, A.; Strathdee, S.A. Drug use as a driver of HIV risks: Re-emerging and emerging issues. Curr. Opin. HIV AIDS 2014, 9, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Lebouche, B.; Engler, K.; Levy, J.J.; Gilmore, N.; Spire, B.; Rozenbaum, W.; Lacene, T.; Routy, J.P. French HIV experts on early antiretroviral treatment for prevention: Uncertainty and heterogeneity. J. Int. Assoc. Provid. AIDS Care 2014, 13, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.Y.; Buchbinder, S.P. CROI 2017: HIV Epidemic Trends and Advances in Prevention. Top. Antivir. Med. 2017, 25, 35–50. [Google Scholar] [PubMed]
- Brenner, B.G.; Ibanescu, R.I.; Hardy, I.; Stephens, D.; Otis, J.; Moodie, E.; Grossman, Z.; Vandamme, A.M.; Roger, M.; Wainberg, M.A. Large cluster outbreaks sustain the HIV epidemic among MSM in Quebec. AIDS 2017, 31, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Poon, A.F.; Gustafson, R.; Daly, P.; Zerr, L.; Demlow, S.E.; Wong, J.; Woods, C.K.; Hogg, R.S.; Krajden, M.; Moore, D.; et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: An implementation case study. Lancet HIV 2016, 3, e231–e238. [Google Scholar] [CrossRef]
- Brenner, B.; Wainberg, M.A.; Roger, M. Phylogenetic inferences on HIV-1 transmission: Implications for the design of prevention and treatment interventions. AIDS 2013, 27, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Roger, M.; Routy, J.P.; Moisi, D.; Ntemgwa, M.; Matte, C.; Baril, J.G.; Thomas, R.; Rouleau, D.; Bruneau, J.; et al. High rates of forward transmission events after acute/early HIV-1 infection. J. Infect. Dis. 2007, 195, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Bezemer, D.; Cori, A.; Ratmann, O.; van Sighem, A.; Hermanides, H.S.; Dutilh, B.E.; Gras, L.; Rodrigues Faria, N.; van den Hengel, R.; Duits, A.J.; et al. Dispersion of the HIV-1 Epidemic in Men Who Have Sex with Men in the Netherlands: A Combined Mathematical Model and Phylogenetic Analysis. PLoS Med. 2015, 12, e1001898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezemer, D.; Faria, N.R.; Hassan, A.; Hamers, R.L.; Mutua, G.; Anzala, O.; Mandaliya, K.; Cane, P.; Berkley, J.A.; Rinke de Wit, T.F.; et al. HIV Type 1 transmission networks among men having sex with men and heterosexuals in Kenya. AIDS Res. Hum. Retrovir. 2014, 30, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Ratmann, O.; Hodcroft, E.B.; Pickles, M.; Cori, A.; Hall, M.; Lycett, S.; Colijn, C.; Dearlove, B.; Didelot, X.; Frost, S.; et al. Phylogenetic Tools for Generalized HIV-1 Epidemics: Findings from the PANGEA-HIV Methods Comparison. Mol. Biol. Evol. 2017, 34, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Ratmann, O.; van Sighem, A.; Bezemer, D.; Gavryushkina, A.; Jurriaans, S.; Wensing, A.; de Wolf, F.; Reiss, P.; Fraser, C. Sources of HIV infection among men having sex with men and implications for prevention. Sci. Transl. Med. 2016, 8, 320ra2. [Google Scholar] [CrossRef] [PubMed]
- Kouyos, R.D.; von Wyl, V.; Yerly, S.; Boni, J.; Taffe, P.; Shah, C.; Burgisser, P.; Klimkait, T.; Weber, R.; Hirschel, B.; et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J. Infect. Dis. 2010, 201, 1488–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzel, A.; Shilaih, M.; Yang, W.L.; Boni, J.; Yerly, S.; Klimkait, T.; Aubert, V.; Braun, D.L.; Calmy, A.; Furrer, H.; et al. HIV-1 Transmission During Recent Infection and During Treatment Interruptions as Major Drivers of New Infections in the Swiss HIV Cohort Study. Clin. Infect. Dis. 2016, 62, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Shilaih, M.; Marzel, A.; Yang, W.L.; Scherrer, A.U.; Schupbach, J.; Boni, J.; Yerly, S.; Hirsch, H.H.; Aubert, V.; Cavassini, M.; et al. Genotypic Resistance Tests Sequences Reveal the Role of Marginalized Populations in HIV-1 Transmission in Switzerland. Sci. Rep. 2016, 6, 27580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villandre, L.; Stephens, D.A.; Labbe, A.; Gunthard, H.F.; Kouyos, R.; Stadler, T.; Swiss HIV Cohort Study. Assessment of Overlap of Phylogenetic Transmission Clusters and Communities in Simple Sexual Contact Networks: Applications to HIV-1. PLoS ONE 2016, 11, e0148459. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Roger, M.; Stephens, D.; Moisi, D.; Hardy, I.; Weinberg, J.; Turgel, R.; Charest, H.; Koopman, J.; Wainberg, M.A. Transmission clustering drives the onward spread of the HIV epidemic among men who have sex with men in Quebec. J. Infect. Dis. 2011, 204, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Roger, M.; Moisi, D.D.; Oliveira, M.; Hardy, I.; Turgel, R.; Charest, H.; Routy, J.P.; Wainberg, M.A. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS 2008, 22, 2509–2515. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Wainberg, M.A. Future of phylogeny in HIV prevention. J. Acquir. Immune Defic. Syndr. 2013, 63 (Suppl. 2), S248–S254. [Google Scholar] [CrossRef] [PubMed]
- Bezemer, D.; van Sighem, A.; Lukashov, V.V.; van der Hoek, L.; Back, N.; Schuurman, R.; Boucher, C.A.; Claas, E.C.; Boerlijst, M.C.; Coutinho, R.A.; et al. Transmission networks of HIV-1 among men having sex with men in the Netherlands. AIDS 2010, 24, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Leigh Brown, A.J.; Lycett, S.J.; Weinert, L.; Hughes, G.J.; Fearnhill, E.; Dunn, D.T. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J. Infect. Dis. 2011, 204, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.A.; Mesplede, T.; Raffi, F. What if HIV were unable to develop resistance against a new therapeutic agent? BMC Med. 2013, 11, 249. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.A.; Friedland, G. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA 1998, 279, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- German, D.; Grabowski, M.K.; Beyrer, C. Enhanced use of phylogenetic data to inform public health approaches to HIV among men who have sex with men. Sex. Health 2016, 14, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Brenner BG, I.R.; Roger, M.; Oliveira, M.; Hardy, I.; Wainberg, M.A. Phylogenetic, Epidemiological and Virological Insights on the Rise of Large Cluster Outbreaks Fueling the HIV-1 Epidemic among Men Having Sex with Men within Quebec. In Proceedings of the 12th International Workshop on HIV Transmission-Principles of Intervention; Paris, France, 21–22 July 2017; p. 3. [Google Scholar]
- Esbjornsson, J.; Mild, M.; Audelin, A.; Fonager, J.; Skar, H.; Bruun Jorgensen, L.; Liitsola, K.; Bjorkman, P.; Bratt, G.; Gisslen, M.; et al. HIV-1 transmission between MSM and heterosexuals and increasing proportions of circulating recombinant forms in the Nordic Countries. Virus Evol. 2016, 2, vew010. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.S.; Pybus, O.G.; Sanders, E.J.; Albert, J.; Esbjornsson, J. Defining HIV-1 transmission clusters based on sequence data. AIDS 2017, 31, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Paraskevis, D.; Nikolopoulos, G.; Tsiara, C.; Paraskeva, D.; Antoniadou, A.; Lazanas, M.; Gargalianos, P.; Psychogiou, M.; Malliori, M.; Kremastinou, J.; et al. HIV-1 outbreak among injecting drug users in Greece, 2011: A preliminary report. Euro Surveill. 2011, 16, 19962. [Google Scholar] [CrossRef] [PubMed]
- Yebra, G.; Ragonnet-Cronin, M.; Ssemwanga, D.; Parry, C.M.; Logue, C.H.; Cane, P.A.; Kaleebu, P.; Brown, A.J. Analysis of the history and spread of HIV-1 in Uganda using phylodynamics. J. Gen. Virol. 2015, 96 Pt 7, 1890–1898. [Google Scholar] [CrossRef] [PubMed]
- Hue, S.; Brown, A.E.; Ragonnet-Cronin, M.; Lycett, S.J.; Dunn, D.T.; Fearnhill, E.; Dolling, D.I.; Pozniak, A.; Pillay, D.; Delpech, V.C.; et al. Phylogenetic analyses reveal HIV-1 infections between men misclassified as heterosexual transmissions. AIDS 2014, 28, 1967–1975. [Google Scholar] [CrossRef] [PubMed]
- Ragonnet-Cronin, M.; Lycett, S.J.; Hodcroft, E.B.; Hue, S.; Fearnhill, E.; Brown, A.E.; Delpech, V.; Dunn, D.; Leigh Brown, A.J.; United Kingdom HIV Drug Resistance Database. Transmission of Non-B HIV Subtypes in the United Kingdom Is Increasingly Driven by Large Non-Heterosexual Transmission Clusters. J. Infect. Dis. 2016, 213, 1410–1418. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Borkowf, C.B.; Brooks, J.T.; Lasry, A.; Lansky, A.; Mermin, J. Estimating per-act HIV transmission risk: A systematic review. AIDS 2014, 28, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Baeten, J.M.; Overbaugh, J. Measuring the infectiousness of persons with HIV-1: Opportunities for preventing sexual HIV-1 transmission. Curr. HIV Res. 2003, 1, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Keele, B.F.; Giorgi, E.E.; Salazar-Gonzalez, J.F.; Decker, J.M.; Pham, K.T.; Salazar, M.G.; Sun, C.; Grayson, T.; Wang, S.; Li, H.; et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 2008, 105, 7552–7557. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Gonzalez, J.F.; Bailes, E.; Pham, K.T.; Salazar, M.G.; Guffey, M.B.; Keele, B.F.; Derdeyn, C.A.; Farmer, P.; Hunter, E.; Allen, S.; et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 2008, 82, 3952–39570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, M.S.; Shaw, G.M.; McMichael, A.J.; Haynes, B.F. Acute HIV-1 Infection. N. Engl. J. Med. 2011, 364, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, G.S.; Heath, L.; Nickle, D.C.; Wong, K.G.; Leach, S.E.; Jacobs, B.; Gezahegne, S.; van’t Wout, A.B.; Jacobson, L.P.; Margolick, J.B.; et al. HIV-1 variation before seroconversion in men who have sex with men: Analysis of acute/early HIV infection in the multicenter AIDS cohort study. J. Infect. Dis. 2008, 197, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Vrbik, I.; Stephens, D.A.; Roger, M.; Brenner, B.G. The Gap Procedure: For the identification of phylogenetic clusters in HIV-1 sequence data. BMC Bioinform. 2015, 16, 355. [Google Scholar] [CrossRef] [PubMed]
- Lessells, R.J.; Stott, K.E.; Manasa, J.; Naidu, K.K.; Skingsley, A.; Rossouw, T.; de Oliveira, T. Implementing antiretroviral resistance testing in a primary health care HIV treatment programme in rural KwaZulu-Natal, South Africa: Early experiences, achievements and challenges. BMC Health Serv. Res. 2014, 14, 116. [Google Scholar] [CrossRef] [PubMed]
- Lessells, R.J.; Avalos, A.; de Oliveira, T. Implementing HIV-1 genotypic resistance testing in antiretroviral therapy programs in Africa: Needs, opportunities and challenges. AIDS Rev. 2013, 15, 221. [Google Scholar] [PubMed]
- Brenner, B.G.; Ibanescu, R.I.; Oliveira, M.; Roger, M.; Hardy, I.; Routy, J.P.; Kyeyune, F.; Quinones-Mateu, M.E.; Wainberg, M.A.; Montreal PHI Cohort Study Group. HIV-1 strains belonging to large phylogenetic clusters show accelerated escape from integrase inhibitors in cell culture compared with viral isolates from singleton/small clusters. J. Antimicrob. Chemother. 2017, 72, 2171–2183. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.B.; Swanstrom, R.; Kashuba, A.D.; Cohen, M.S. Bottlenecks in HIV-1 transmission: Insights from the study of founder viruses. Nat. Rev. Microbiol. 2015, 13, 414–425. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.S.; Bibollet-Ruche, F.; Sherrill-Mix, S.; Learn, G.H.; Plenderleith, L.; Smith, A.G.; Barbian, H.J.; Russell, R.M.; Gondim, M.V.; Bahari, C.Y.; et al. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc. Natl. Acad. Sci. USA 2017, 114, E590–E599. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, S.P.; Liu, A.Y. CROI 2016: Hot Spots in HIV Infection and Advances in HIV Prevention. Top. Antivir. Med. 2016, 24, 10–28. [Google Scholar] [PubMed]
- Engler, K.; Rollet, K.; Lessard, D.; Thomas, R.; Lebouche, B. Ability of a rapid HIV testing site to attract and test vulnerable populations: A cross-sectional study on Actuel sur Rue. Int. J. STD AIDS 2016, 27, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Thomas, R.; Blanco, J.L.; Ibanescu, R.I.; Oliveira, M.; Mesplede, T.; Golubkov, O.; Roger, M.; Garcia, F.; Martinez, E.; et al. Development of a G118R mutation in HIV-1 integrase following a switch to dolutegravir monotherapy leading to cross-resistance to integrase inhibitors. J. Antimicrob. Chemother. 2016, 71, 1948–1953. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, N.; Osman, N.; Ohnona, F.; Xu, H.T.; Brenner, B.; Mesplede, T.; Wainberg, M.A. Does antiretroviral treatment change HIV-1 codon usage patterns in its genes: A preliminary bioinformatics study. AIDS Res. Ther. 2017, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Lowe, M.; Moisi, D.; Hardy, I.; Gagnon, S.; Charest, H.; Baril, J.G.; Wainberg, M.A.; Roger, M. Subtype diversity associated with the development of HIV-1 resistance to integrase inhibitors. J. Med. Virol. 2011, 83, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Blanco, J.L.; Jordan, M.R.; Taylor, J.; Lemey, P.; Varghese, V.; Hamers, R.L.; Bertagnolio, S.; Rinke de Wit, T.F.; Aghokeng, A.F.; et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: An individual-patient- and sequence-level meta-analysis. PLoS Med. 2015, 12, e1001810. [Google Scholar]
- Wainberg, M.A.; Drosopoulos, W.C.; Salomon, H.; Hsu, M.; Borkow, G.; Parniak, M.; Gu, Z.; Song, Q.; Manne, J.; Islam, S.; et al. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 1996, 271, 1282–1285. [Google Scholar] [CrossRef] [PubMed]
- Petrella, M.; Wainberg, M.A. Might the M184V substitution in HIV-1 RT confer clinical benefit? AIDS Rev. 2002, 4, 224–232. [Google Scholar] [PubMed]
- Turner, D.; Brenner, B.G.; Routy, J.P.; Petrella, M.; Wainberg, M.A. Rationale for maintenance of the M184v resistance mutation in human immunodeficiency virus type 1 reverse transcriptase in treatment experienced patients. New Microbiol. 2004, 27 (Suppl. 1), 31–39. [Google Scholar] [PubMed]
- Brenner, B.; Turner, D.; Oliveira, M.; Moisi, D.; Detorio, M.; Carobene, M.; Marlink, R.G.; Schapiro, J.; Roger, M.; Wainberg, M.A. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 2003, 17, F1–F5. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Coutsinos, D. The K65R mutation in HIV-1 reverse transcriptase: Genetic barriers, resistance profile and clinical implications. HIV Ther. 2009, 3, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Brenner, B.G.; Oliveira, M.; Doualla-Bell, F.; Moisi, D.D.; Ntemgwa, M.; Frankel, F.; Essex, M.; Wainberg, M.A. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS 2006, 20, F9–F13. [Google Scholar] [CrossRef] [PubMed]
- Yerly, S.; Junier, T.; Gayet-Ageron, A.; Amari, E.B.; von Wyl, V.; Gunthard, H.F.; Hirschel, B.; Zdobnov, E.; Kaiser, L. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS 2009, 23, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Charest, H.; Doualla-Bell, F.; Cantin, R.; Murphy, D.G.; Lemieux, L.; Brenner, B.; Hardy, I.; Moisi, D.; Lo, E.; Baril, J.G.; et al. A Significant Reduction in the Frequency of HIV-1 Drug Resistance in Quebec from 2001 to 2011 Is Associated with a Decrease in the Monitored Viral Load. PLoS ONE 2014, 9, e109420. [Google Scholar] [CrossRef] [PubMed]
- Raffi, F.; Jaeger, H.; Quiros-Roldan, E.; Albrecht, H.; Belonosova, E.; Gatell, J.M.; Baril, J.G.; Domingo, P.; Brennan, C.; Almond, S.; et al. Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 Week results from a randomised, double-blind, non-inferiority trial. Lancet Infect. Dis. 2013, 13, 927–935. [Google Scholar] [CrossRef]
- Walmsley, S.L.; Antela, A.; Clumeck, N.; Duiculescu, D.; Eberhard, A.; Gutierrez, F.; Hocqueloux, L.; Maggiolo, F.; Sandkovsky, U.; Granier, C.; et al. Dolutegravir plus abacavir-lamivudine for the treatment of HIV-1 infection. N. Engl. J. Med. 2013, 369, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Walmsley, S.; Baumgarten, A.; Berenguer, J.; Felizarta, F.; Florence, E.; Khuong-Josses, M.A.; Kilby, J.M.; Lutz, T.; Podzamczer, D.; Portilla, J.; et al. Brief Report: Dolutegravir Plus Abacavir/Lamivudine for the Treatment of HIV-1 Infection in Antiretroviral Therapy-Naive Patients: Week 96 and Week 144 Results from the SINGLE Randomized Clinical Trial. J. Acquir. Immune Defic. Syndr. 2015, 70, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Clotet, B.; Feinberg, J.; van Lunzen, J.; Khuong-Josses, M.A.; Antinori, A.; Dumitru, I.; Pokrovskiy, V.; Fehr, J.; Ortiz, R.; Saag, M.; et al. Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 Week results from the randomised open-label phase 3b study. Lancet 2014, 383, 2222–2231. [Google Scholar] [CrossRef]
- Miller, M.M.; Liedtke, M.D.; Lockhart, S.M.; Rathbun, R.C. The role of dolutegravir in the management of HIV infection. Infect. Drug Resist. 2015, 8, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Squires, K.; Kityo, C.; Hodder, S.; Johnson, M.; Voronin, E.; Hagins, D.; Avihingsanon, A.; Koenig, E.; Jiang, S.; White, K.; et al. Integrase inhibitor versus protease inhibitor based regimen for HIV-1 infected women (WAVES): A randomised, controlled, double-blind, phase 3 study. Lancet HIV 2016, 3, e410–e420. [Google Scholar] [CrossRef]
- Kaplan, R.; Wood, R. Resistance to first-line ART and a role for dolutegravir. Lancet HIV 2017. [Google Scholar] [CrossRef]
- Phillips, A.N.; Cambiano, V.; Nakagawa, F.; Revill, P.; Jordan, M.R.; Hallett, T.B.; Doherty, M.; De Luca, A.; Lundgren, J.D.; Mhangara, M.; et al. Cost-effectiveness of public-health policy options in the presence of pretreatment NNRTI drug resistance in sub-Saharan Africa: A modelling study. Lancet HIV 2017. [Google Scholar] [CrossRef]
- Venter, W.D.F.; Clayden, P.; Serenata, C.; Consortium, O. The ADVANCE study: A groundbreaking trial to evaluate a candidate universal antiretroviral regimen. Curr. Opin. HIV AIDS 2017, 12, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Anstett, K.; Brenner, B.; Mesplede, T.; Wainberg, M.A. HIV drug resistance against strand transfer integrase inhibitors. Retrovirology 2017, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Raffi, F.; Rachlis, A.; Brinson, C.; Arasteh, K.; Gorgolas, M.; Brennan, C.; Pappa, K.; Almond, S.; Granier, C.; Nichols, W.G.; et al. Dolutegravir efficacy at 48 weeks in key subgroups of treatment-naive HIV-infected individuals in three randomized trials. AIDS 2015, 29, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Cohn, J.; Bekker, L.G.; Bygrave, H.; Calmy, A. Hit me with your best shot: Dolutegravir—A space in the next WHO guidelines? AIDS 2015, 29, 2067–2070. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.A.; Mesplede, T. Implications for the future of the HIV epidemic if drug resistance against dolutegravir cannot occur in first-line therapy. J. Int. AIDS Soc. 2015, 18, 20824. [Google Scholar] [CrossRef] [PubMed]
- Pinnetti, C.; Tintoni, M.; Ammassari, A.; Tamburrini, E.; Bernardi, S.; Liuzzi, G.; Scambia, G.; Perno, C.F.; Floridia, M.; Antinori, A.; et al. Successful prevention of HIV mother-to-child transmission with dolutegravir-based combination antiretroviral therapy in a vertically infected pregnant woman with multiclass highly drug-resistant HIV-1. AIDS 2015, 29, 2534–2537. [Google Scholar] [CrossRef] [PubMed]
- Raffi, F.; Pozniak, A.L.; Wainberg, M.A. Has the time come to abandon efavirenz for first-line antiretroviral therapy? J. Antimicrob. Chemother. 2014, 69, 1742–1747. [Google Scholar] [CrossRef] [PubMed]
- Hofstra, L.M.; Sauvageot, N.; Albert, J.; Alexiev, I.; Garcia, F.; Struck, D.; van de Vijver, D.A.; Asjo, B.; Beshkov, D.; Coughlan, S.; et al. Transmission of HIV Drug Resistance and the Predicted Effect on Current First-line Regimens in Europe. Clin. Infect. Dis. 2016, 62, 655–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamers, R.L.; Wallis, C.L.; Kityo, C.; Siwale, M.; Mandaliya, K.; Conradie, F.; Botes, M.E.; Wellington, M.; Osibogun, A.; Sigaloff, K.C.; et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: A multicentre observational study. Lancet Infect. Dis. 2011, 11, 750–759. [Google Scholar] [CrossRef]
- Ceccherini-Silberstein, F.; van Baelen, K.; Armenia, D.; Trignetti, M.; Rondelez, E.; Fabeni, L.; Scopelliti, F.; Pollicita, M.; van Wesenbeeck, L.; van Eygen, V.; et al. Secondary integrase resistance mutations found in HIV-1 minority quasispecies in integrase therapy-naive patients have little or no effect on susceptibility to integrase inhibitors. Antimicrob. Agents Chemother. 2010, 54, 3938–3948. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brenner, B.G.; Ibanescu, R.-I.; Hardy, I.; Roger, M. Genotypic and Phylogenetic Insights on Prevention of the Spread of HIV-1 and Drug Resistance in “Real-World” Settings. Viruses 2018, 10, 10. https://doi.org/10.3390/v10010010
Brenner BG, Ibanescu R-I, Hardy I, Roger M. Genotypic and Phylogenetic Insights on Prevention of the Spread of HIV-1 and Drug Resistance in “Real-World” Settings. Viruses. 2018; 10(1):10. https://doi.org/10.3390/v10010010
Chicago/Turabian StyleBrenner, Bluma G., Ruxandra-Ilinca Ibanescu, Isabelle Hardy, and Michel Roger. 2018. "Genotypic and Phylogenetic Insights on Prevention of the Spread of HIV-1 and Drug Resistance in “Real-World” Settings" Viruses 10, no. 1: 10. https://doi.org/10.3390/v10010010