Biomass Modeling of Larch (Larix spp.) Plantations in China Based on the Mixed Model, Dummy Variable Model, and Bayesian Hierarchical Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Biomass Data
2.3. Statistical Analysis
2.3.1. General Model
2.3.2. Dummy Variable Model
2.3.3. Linear Mixed Effects Model
2.3.4. Bayesian Hierarchical Model
2.3.5. Model Fitting and Evaluating
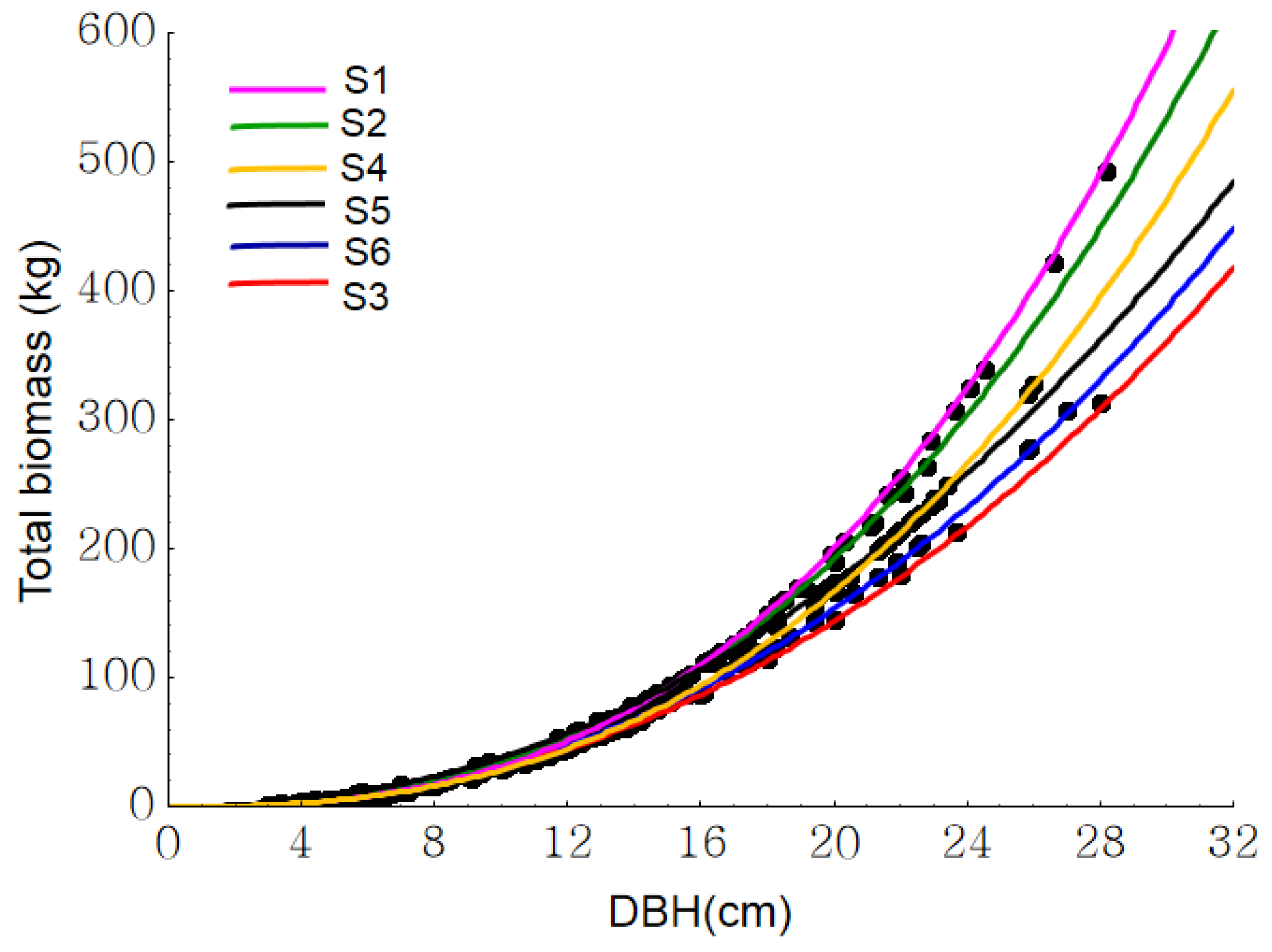
3. Results
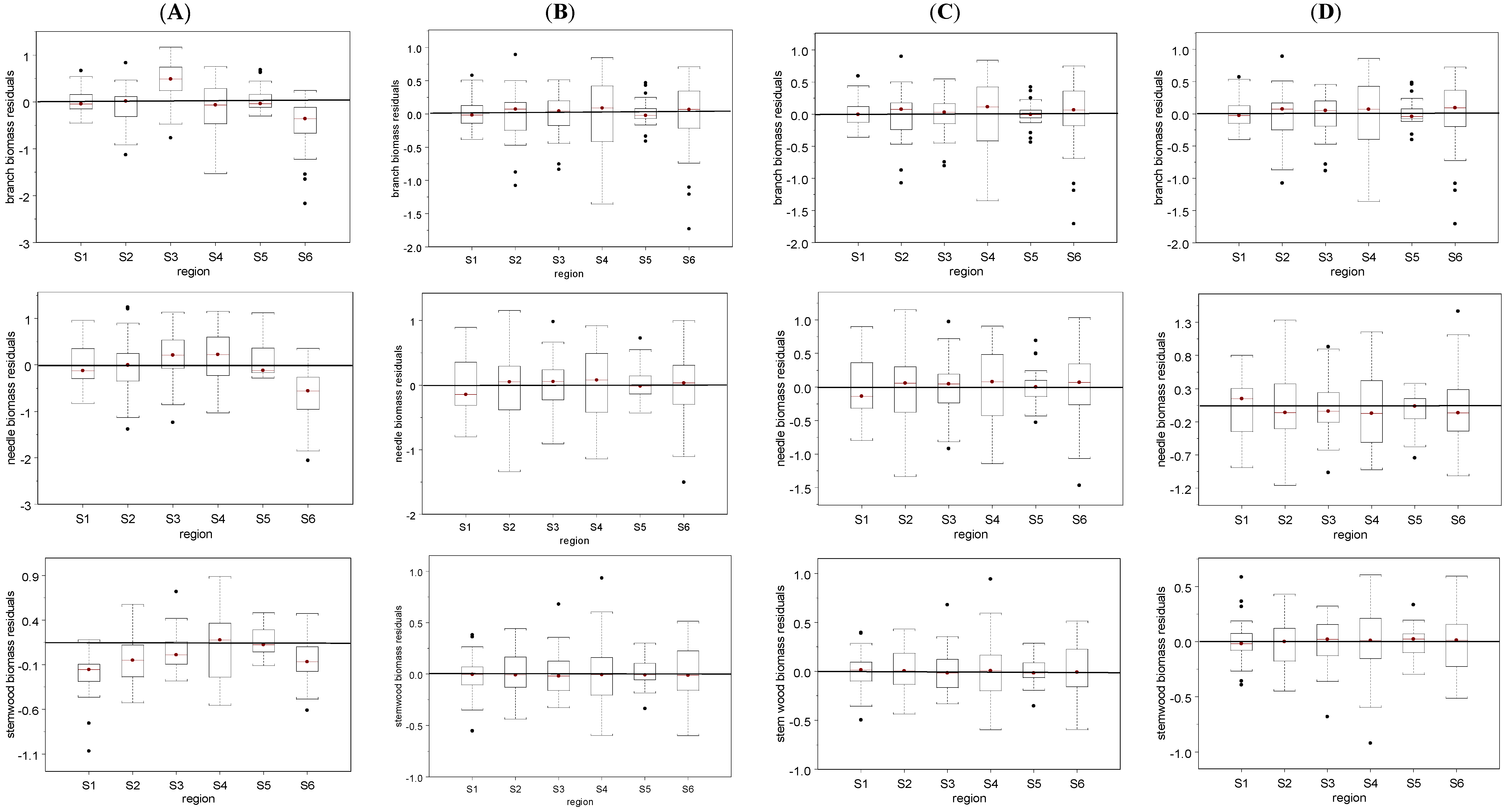
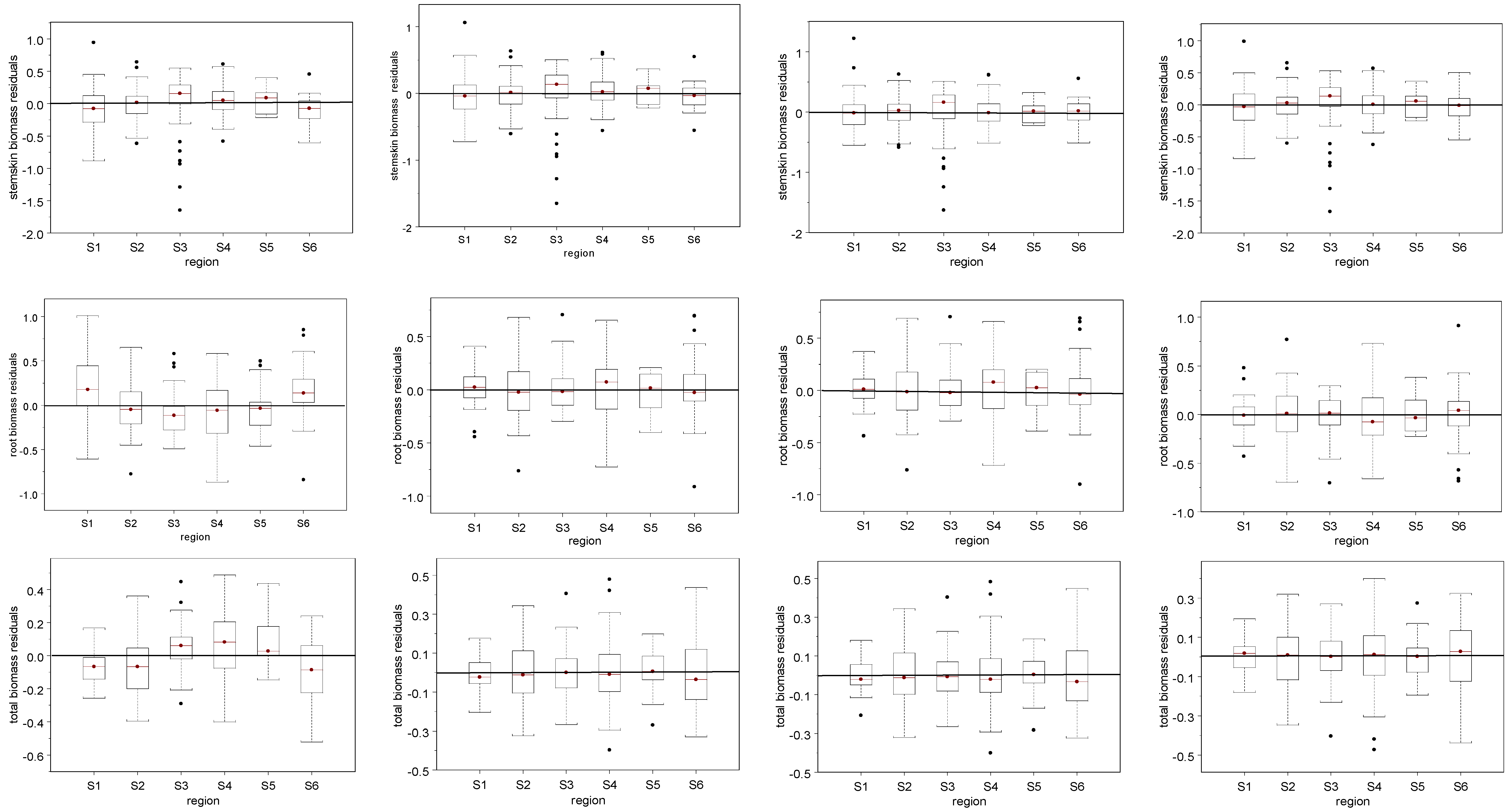
4. Discussion
5. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
Appendix
| No. | Total | Root | Stem wood | Stem Bark | Branch | Foliage | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | a | b | a | b | a | b | a | b | a | b | ||
| 1 | −1.378 | 0.760 | −0.929 | 0.368 | −2.957 | 0.911 | −4.962 | 0.913 | −1.448 | 0.307 | −4.017 | −1.378 | (Wang, 2010) [38] |
| 2 | −1.952 | 0.844 | −4.075 | 0.915 | −2.797 | 0.888 | −3.352 | 0.670 | −2.957 | 0.633 | −3.817 | −1.952 | (Shen et al, 2011) [39] |
| 3 | −2.087 | 0.892 | −4.510 | 0.981 | −2.465 | 0.849 | −3.381 | 0.684 | −4.423 | 0.990 | −5.521 | −2.087 | (Liu, 2012) [40] |
| 4 | −2.079 | 0.901 | −4.510 | 0.982 | −2.659 | 0.927 | −3.352 | 0.694 | −3.037 | 0.628 | −3.058 | −2.079 | (Zhang, 2010) [41] |
| 5 | −1.091 | 0.735 | −5.116 | 1.024 | −1.845 | 0.789 | −1.917 | 0.389 | −1.514 | 0.318 | −0.942 | −1.091 | (Min, 2010) [42] |
| 6 | −2.419 | 0.929 | −3.474 | 0.866 | −3.170 | 0.960 | −4.269 | 0.813 | −5.298 | 0.972 | −4.962 | −2.419 | (Li, 2013) [43] |
References
- Jenkins, J.C.; Chojnacky, D.C.; Heath, L.S.; Birdsey, R.A. National-scale biomass estimators for United States tree species. For. Sci. 2003, 49, 12–35. [Google Scholar]
- Muukkonen, P. Generalized allometric volume biomass equations for some tree species in Europe. Eur. J. For. Res. 2007, 126, 157–166. [Google Scholar] [CrossRef]
- Case, B.; Hall, R.J. Assessing prediction errors of generalized tree biomass and volume equations for the boreal forest region of west-central Canada. Can. J. For. Res. 2008, 38, 878–889. [Google Scholar] [CrossRef]
- Lydia, R.O.; Halloran, E.T.; Borer, E.W.; Seabloom, A.S.; MacDougall, E.E.; Cleland, R.L.; McCulley, S.H.; Hobbie, W.; StanHarploe, N.M.; DeCrappeo, J.D.; et al. Regional Contingencies in the relationship between aboveground biomass and litter in the world’s grasslands. PLoS ONE 2013, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ter-Mikaelian, M.T.; Korzukhin, M.D. Biomass equations for sixty-five North American tree species. For. Ecol. Manag. 1997, 97, 1–24. [Google Scholar] [CrossRef]
- Zianis, D.; Muukkonen, P.; Makipaa, R.; Mencuccini, M. Biomass and stem volume equations for tree species in Europe. Silva Fenn. Monogr. 2005, 4, 1–63. [Google Scholar]
- Návar, J. Allometric equations for tree species and carbon stocks for forests of northwestern Mexico. For. Ecol. Manag. 2009, 257, 427–434. [Google Scholar] [CrossRef]
- Adriano, L.; Rempei, S.; Gabriel, H.P.M.R.; Takuya, K.J.; Joaquim, D.S.; Roseana, P.S.; Cacilda, A.S.S.; Priscila, C.B.; Hideyuki, N.; Moriyoshi, I.; et al. Allometric models for estimating above- and below-ground biomass in Amazonian forests at Sao Gabriel da Cachoeira in the upper Rio Negro. Brazil. For. Ecol. Manag. 2012, 277, 163–172. [Google Scholar]
- Clark, D.B.; Clark, D.A. Landscape-scale variation in forest structure and biomass in a tropical rain forest. For. Ecol. Manag. 2000, 137, 185–198. [Google Scholar] [CrossRef]
- Chambers, J.Q.; Santos, J.D.; Ribeiro, R.J.; Higuchi, N. Tree damage, allometric relationships, and above-ground net primary production in central Amazon forest. For. Ecol. Manag. 2001, 152, 73–84. [Google Scholar] [CrossRef]
- Li, R.; Stewart, B.; Weiskittel, A. A Bayesian approach for modeling nonlinear longitudinal/hierarchical data with random effects in forestry. Forestry 2012, 85, 17–25. [Google Scholar] [CrossRef]
- Zianis, D.; Mencuccini, M. On simplifying allometric analyses of forest biomass. For. Ecol. Manag. 2004, 187, 311–332. [Google Scholar] [CrossRef]
- Pilli, R.; Anfodillo, T.; Carrer, M. Towards a functional and simplified allometry for estimating forest biomass. For. Ecol. Manag. 2006, 237, 583–593. [Google Scholar] [CrossRef]
- Ramon, C.L.; George, A.M.; Walter, W.S.; Russell, D.W.; Oliver, S. SAS for Mixed Model, 2nd ed.; SAS Institute Inc.: Cary, NC, USA, 2006. [Google Scholar]
- Fang, Z.; Bailey, R.L. Nonlinear mixed effects modeling for Slash pine dominant height growth following intensive silvicultural treatments. For. Sci. 2001, 47, 287–300. [Google Scholar]
- Daniel, B.H.; Michael, C. Multivariate multilevel nonlinear mixed effects models for timber yield predictions. Biometrics 2004, 60, 16–24. [Google Scholar]
- Zhang, Y.J.; Borders, B.E. Using a system mixed effects modeling method to estimate tree compartment biomass for intensively managed loblolly pines-an allometric approach. For. Ecol. Manag. 2004, 194, 145–157. [Google Scholar] [CrossRef]
- Trincado, G.; Burkhart, H.E. A generalized approach for modeling and localizing stem profile curves. For. Sci. 2006, 52, 670–682. [Google Scholar]
- Fehrmann, L.; Lehtonen, A.; Kleinn, C.; Tomppo, R. Comparison of linear and mixed-effect regression models and a k-nearest neighbor approach for estimation of singletree biomass. Can. J. For. Res. 2008, 38, 1–9. [Google Scholar] [CrossRef]
- Derek, F.S.; Philip, G.C.; Alexis, A. Branch models for white spruce (Piceaglauca. (Moench.) Voss) in naturally regenerated stands. For. Ecol. Manag. 2014, 325, 74–89. [Google Scholar]
- Anholt, B.R.; Werner, E.; Skelly, D.K. Effect of food and predators on the activity of four larval ranid frogs. Ecology 2000, 81, 3509–3521. [Google Scholar] [CrossRef]
- Berger, J.; Berry, D.A. Statistical analysis and the illusion of objectivity. Am. Sci. 1988, 76, 159–165. [Google Scholar]
- Jaynes, E.T. Probability Theory: The Logic of Science; Cambridge University Press: New York, NY, USA, 2003; pp. 19–55. [Google Scholar]
- Ellison, A.M. Bayesian inference in ecology. Ecol. Lett. 2004, 7, 509–520. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Duan, A.G.; Zhang, J.G. Tree Biomass Estimation of Chinese fir (Cunninghamialanceolata.) based on Bayesian method. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef]
- Bullock, B.P.; Boone, E.L. Deriving tree diameter distributions using Bayesian model averaging. For. Ecol. Manag. 2007, 242, 127–132. [Google Scholar] [CrossRef]
- Green, E.J.; Roesch, F.A.; Smith, F.M.; Strawderman, W.E. Bayesian estimation for the three-parameter Weibull distribution with tree diameter data. Biometrics 1994, 50, 254–269. [Google Scholar] [CrossRef]
- Clark, J.S.; Wolosin, M.; Dietze, M.; Ibanez, I.; Ladeau, S.; Welsh, M.; Kloeppel, B. Tree growth inference and prediction from diameter censuses and ring widths. Ecol. Appl. 2007, 17, 1942–1953. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, C.J.E.; McMahon, S.M.; Clark, J.S. Overcoming data sparseness and parametric constraints in modeling of tree mortality: A new nonparametric Bayesian model. Can. J. For. Res. 2009, 39, 1677–1687. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Zhang, J.G.; Duan, A.G. A hierarchical Bayesian model to predict self-thinning line for Chinese Fir in Southern China. PLoS ONE 2015, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zapata-Cuartas, M.; Sierra, C.A.; Alleman, L. Probability distribution of allometric coefficients and Bayesian estimation of aboveground tree biomass. For. Ecol. Manag. 2012, 277, 173–179. [Google Scholar] [CrossRef]
- Tang, S.Z.; Li, Y. Statistical Foundation for Biomathematical Models; Science Press: Beijing, China, 2002; pp. 168–194. (In Chinese) [Google Scholar]
- Tang, S.Z.; Lang, K.J.; Li, H.K. Statistics and Computation of Biomathematical Models (For Stat Course); Science Press: Beijing, China, 2008; pp. 115–261. (In Chinese) [Google Scholar]
- Fu, L.Y.; Zeng, W.S.; Tang, S.Z.; Sharma, R.P.; Li, H.K. Using linear mixed model and dummy variable model approaches to construct compatible single tree biomass equations at different scales—A case study for Masson pine in Southern China. J. For. Sci. 2012, 58, 101–115. [Google Scholar]
- Wang, M.; Borders, B.E.; Zhao, D. An empirical comparison of two subject-specific approaches to dominant heights modeling, the dummy variable method and the mixed model method. For. Ecol. Manag. 2008, 255, 2659–2669. [Google Scholar] [CrossRef]
- China Forestry Bureau. The Eighth Forest Resource Survey Report; Chinese Forestry Press: Beijing, China, 2014; p. 18. (In Chinese)
- Wagner, H.; Tüchler, R. Bayesian estimation of random effects models for multivariate responses of mixed data. Comput. Stat. Data Anal. 2010, 54, 1206–1218. [Google Scholar] [CrossRef]
- Wang, H.Q. Research on the Biomass Model of Larix kaemferi Plantation in Northern Sub-Tropical Alpine area Based on Sample Plots and Remote Sensing. Master of Thesis, Chinese Academy of Forestry, Beijing, China, 2010; Science Press, Beijing, China, 2008. [Google Scholar]
- Shen, Y.Z.; Sun, X.M.; Zhang, J.T.; Du, Y.C.; Ma, J.W. Study on the individual tree biomass of Larix kaempferi plantation in Xiaolong mountain Gansu province. For. Res. 2011, 24, 517–522. (In Chinese) [Google Scholar]
- Liu, Y.Q. The Study of Single Biomass, Carbon Storage and Distribution of Larixprincipis-rupprechtii and Populus in Hebei Province. Master of Thesis, Agricultural University of Hebei, Baoding, China, 2012. (In Chinese). [Google Scholar]
- Zhang, H. Study on the Dominant Tree Biomass and Carbon Storage in Daqing Mountain. Master of Thesis, Inner Mongolia Agricultural University, Huhehaote, China, 2010. (In Chinese). [Google Scholar]
- Min, Z.Q. Study on Biomass Estimation Model of Larix olgensis. Master of Thesis, Beijing Forestry University, Beijing, China, 2010. (In Chinese). [Google Scholar]
- Li, Z.F. Biomass and Carbon Storage of the Larix gmelinii Forest’s Research. Master of Thesis, Inner Mongolia Agricultural University, Huhehaote, China, 2013. (In Chinese). [Google Scholar]
- Sturtz, S.; Ligges, U.; Gelman, A. R2WinBUGS: A package for running WinBUGS from R.J. Stat. Softw. 2005, 12, 1–16. [Google Scholar]
- Ketterings, Q.A.; Coe, R.; Van Noordwijk, M.; Ambagau, Y.; Palm, C.A. Reducing uncertainty in the use of allometric biomass equations for predicting above-ground tree biomass in mixed secondary forests. For. Ecol. Manag. 2001, 146, 199–209. [Google Scholar] [CrossRef]
- West, G.B.; Brown, J.H.; Enquist, B.J. A general model for the structure and allometry of plant vascular systems. Nature 1999, 400, 664–667. [Google Scholar]
- Huang, X.Z.; Chen, D.S.; Sun, X.M.; Zhang, S.G. Estimation of above-ground tree biomass based on probability distribution of allometric parameters. Sci. Silvae Sin. 2014, 50, 34–41. (In Chinese) [Google Scholar]
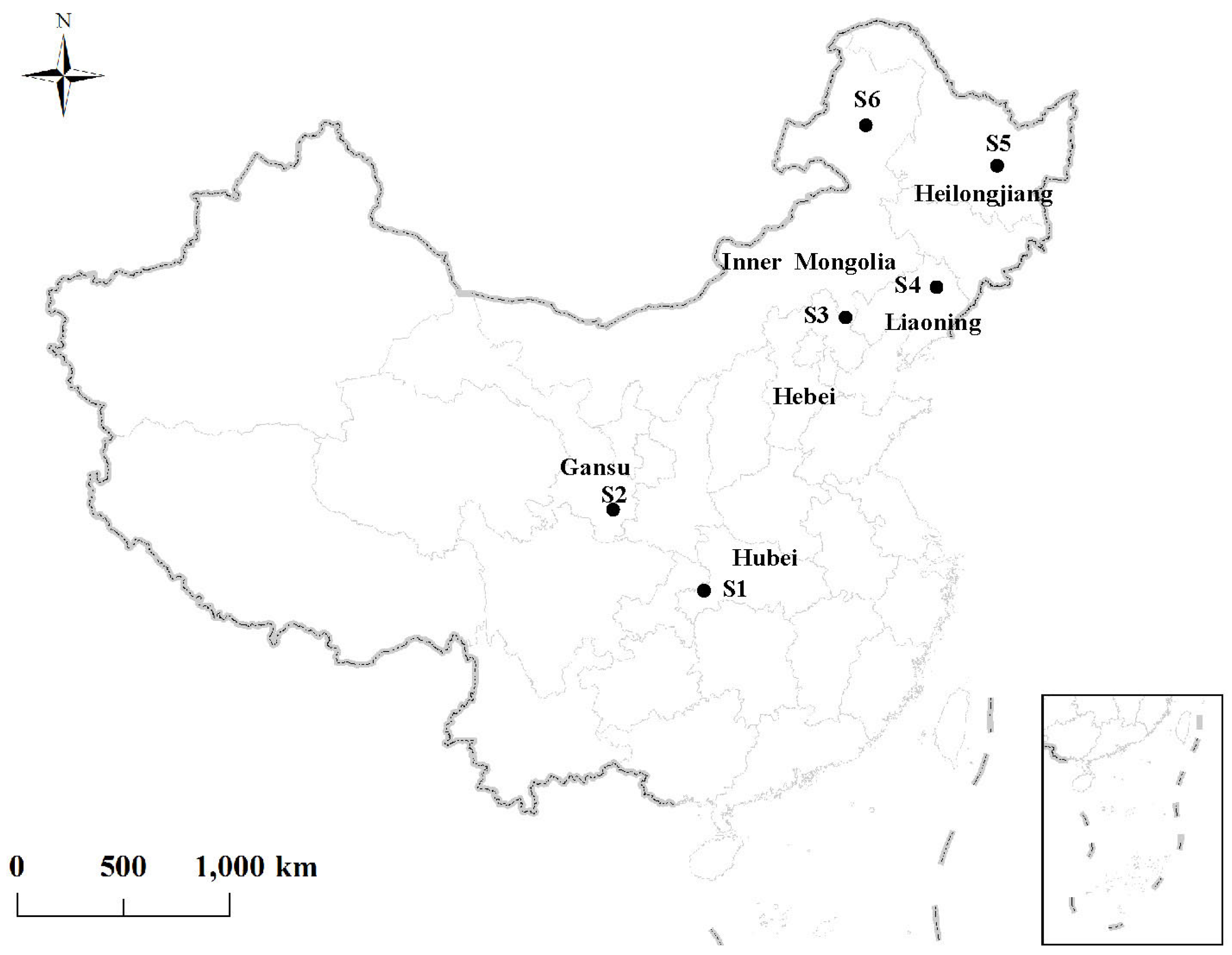
| Code | Site | Province | Latitude and Longitude | Climate Zone | Annual Average Temperature (°C) | Annual Precipitation (mm) | Larch Species |
|---|---|---|---|---|---|---|---|
| S1 | Changlinggang farm | Hubei province | 30.48° N, 110.02° E | Subtropics | 11.7 | 1884 | Larix kaempferi |
| S2 | Tianshui city | Gansu province | 34.09° N, 105.52° E | Warm temperate | 11 | 800 | Larix kaempferi |
| S3 | Weichang county | Hebei province | 41.43° N, 118.7° E | Temperate | 3.3 | 445 | Larix principis-rupprechtii |
| S4 | Dagujia farm | Liaoning province | 42.21° N, 124.52° E | Temperate | 6 | 806 | Larix kaempferi |
| S5 | Mengjiagang farm | Heilongjiang province | 46.32° N, 129.10° E | Cold temperate | 2.7 | 550 | Larix olgensis |
| S6 | Wuerqihan Forestry bureau | Inner Mongolia province | 49.34° N, 121.25° E | Cold temperate | 2.6 | 560 | Larix gmelinii |
| Region | Sample Trees | Biomass Component | Min–Max | Mean(S.D.) | Region | Sample Trees | Biomass Component | Min–Max | Mean(S.D.) |
|---|---|---|---|---|---|---|---|---|---|
| S1 | 60 | branch | 0.81–16.97 | 10.66(0.84) | S4 | 60 | branch | 1.88–25.99 | 10.93(1.09) |
| needle | 0.46–5.7 | 3.31(0.20) | needle | 0.77–11.12 | 3.78(0.53) | ||||
| stem | 1.34–158.47 | 87.11(9.23) | stem | 0.82–181.25 | 68.96(9.78) | ||||
| bark | 0.28–16.97 | 9.49(0.84) | bark | 0.26–31.90 | 9.92(1.41) | ||||
| root | 0.63–42.57 | 20.46(2.35) | root | 2.32–50.28 | 23.59(2.25) | ||||
| total | 3.52–236.53 | 131.23(13.27) | total | 7.00–270.73 | 117.17(14.54) | ||||
| S2 | 60 | branch | 0.92–25.94 | 8.44(1.03) | S5 | 60 | branch | 0.4–26.09 | 6.28(1.13) |
| needle | 0.47–13.58 | 3.54(0.4) | needle | 0.06–7.07 | 1.64(0.26) | ||||
| stem | 2.42–306.17 | 78.41(12.24) | stem | 0.7–236.42 | 55.96(10.30) | ||||
| bark | 0.66–29.20 | 8.52(1.09) | bark | 0.35–22.96 | 6.47(1.06) | ||||
| root | 0.95–82.85 | 18.35(2.96) | root | 0.33–76.23 | 20.32(3.56) | ||||
| total | 5.44–457.74 | 117.25(17.25) | total | 2.2–362.41 | 90.68(16.07) | ||||
| S3 | 60 | branch | 0.48–27.39 | 7.01(0.96) | S6 | 60 | branch | 0.58–58.13 | 13.23(2.01) |
| needle | 0.2–8.7 | 2.28(0.31) | needle | 0.13–14.65 | 3.37(0.51) | ||||
| stem | 0.56–204.79 | 38.75(6.56) | stem | 1.53–144.4 | 35.14(5.00) | ||||
| bark | 0.22–20.19 | 5.65(0.83) | bark | 0.47–20.68 | 5.28(0.73) | ||||
| root | 0.23–76.79 | 13.20(2.33) | root | 0.41–59.34 | 11.23(1.81) | ||||
| total | 1.80–337.45 | 66.89(10.65) | total | 4.07–276.64 | 68.25(9.84) |
| Biomass Component | Parameter | Estimate | S.D. | p-Value |
|---|---|---|---|---|
| Branch | a | −2.6813 | 0.1489 | <0.01 |
| b | 1.7831 | 0.0583 | <0.01 | |
| Needle | a | −3.2790 | 0.1748 | <0.01 |
| b | 1.5779 | 0.0685 | <0.01 | |
| Stemwood | a | −3.5205 | 0.0811 | <0.01 |
| b | 2.7278 | 0.0318 | <0.01 | |
| Stembark | a | −3.9267 | 0.0924 | <0.01 |
| b | 2.1516 | 0.0362 | <0.01 | |
| Root | a | −3.9618 | 0.0912 | <0.01 |
| b | 2.4586 | 0.0357 | <0.01 | |
| Total | a | −2.1166 | 0.0537 | <0.01 |
| b | 2.4199 | 0.0210 | <0.01 |
| Region | Biomass Component | Parameter | Estimate | S.D. | Region | Biomass Component | Parameter | Estimate | S.D. |
|---|---|---|---|---|---|---|---|---|---|
| S1 | Stem wood | a | −2.903 | 0.427 | S4 | Stem wood | a | −2.489 | 0.411 |
| b | 2.473 | 0.326 | b | 2.473 | 0.326 | ||||
| Stem bark | a | −3.541 | 0.449 | Stem bark | a | −3.499 | 0.405 | ||
| b | 2.049 | 0.167 | b | 2.049 | 0.167 | ||||
| Branch | a | −4.026 | 0.507 | Branch | a | −4.027 | 0.582 | ||
| b | 2.195 | 0.198 | b | 2.195 | 0.198 | ||||
| Needle | a | −4.288 | 0.656 | Needle | a | −4.214 | 0.633 | ||
| b | 1.954 | 0.207 | b | 1.954 | 0.207 | ||||
| Root | a | −3.793 | 0.482 | Root | a | −3.621 | 0.407 | ||
| b | 2.396 | 0.150 | b | 2.396 | 0.150 | ||||
| Total biomass | a | −2.084 | 0.221 | Total biomass | a | −1.780 | 0.207 | ||
| b | 2.419 | 0.105 | b | 2.419 | 0.105 | ||||
| S2 | Stem wood | a | −2.924 | 0.455 | S5 | Stem wood | a | −2.667 | 0.458 |
| b | 2.473 | 0.326 | b | 2.473 | 0.326 | ||||
| Stem bark | a | −3.637 | 0.602 | Stem bark | a | −3.651 | 0.423 | ||
| b | 2.049 | 0.167 | b | 2.049 | 0.167 | ||||
| Branch | a | −3.905 | 0.519 | Branch | a | −3.997 | 0.566 | ||
| b | 2.195 | 0.196 | b | 2.195 | 0.206 | ||||
| Needle | a | −4.332 | 0.639 | Needle | a | −4.510 | 0.701 | ||
| b | 1.954 | 0.207 | b | 1.954 | 0.207 | ||||
| Root | a | −3.698 | 0.356 | Root | a | −3.783 | 0.408 | ||
| b | 2.396 | 0.150 | b | 2.396 | 0.150 | ||||
| Total biomass | a | −2.078 | 0.233 | Total biomass | a | −1.943 | 0.208 | ||
| b | 2.419 | 0.105 | b | 2.419 | 0.105 | ||||
| S3 | Stem wood | a | −2.924 | 0.398 | S6 | Stem wood | a | −2.728 | 0.410 |
| b | 2.473 | 0.326 | b | 2.473 | 0.326 | ||||
| Stem bark | a | −3.571 | 0.308 | Stem bark | a | −3.643 | 0.305 | ||
| b | 2.049 | 0.167 | b | 2.049 | 0.167 | ||||
| Branch | a | −2.983 | 0.592 | Branch | a | −4.097 | 0.444 | ||
| b | 2.195 | 0.198 | b | 2.195 | 0.198 | ||||
| Needle | a | −3.683 | 0.621 | Needle | a | −4.793 | 0.655 | ||
| b | 1.954 | 0.207 | b | 1.954 | 0.207 | ||||
| Root | a | −3.641 | 0.396 | Root | a | −3.451 | 0.428 | ||
| b | 2.396 | 0.150 | b | 2.396 | 0.150 | ||||
| Total biomass | a | −1.864 | 0.207 | Total biomass | a | −1.931 | 0.197 | ||
| b | 2.419 | 0.105 | b | 2.419 | 0.105 |
| Biomass Component | Parameter | Estimate Values | S.D. | T-Value | p-Value |
|---|---|---|---|---|---|
| Stem wood | a | −3.684 | 0.284 | 12.983 | <0.01 |
| b | 2.783 | 0.102 | 27.233 | <0.01 | |
| σa | 0.068 | ||||
| σb | 0.008 | ||||
| σab | −0.024 | ||||
| ε | 0.223 | ||||
| Stem bark | a | −3.971 | 0.145 | 27.256 | <0.01 |
| b | 2.166 | 0.053 | 40.534 | <0.01 | |
| σa | 0.068 | ||||
| σb | 0.008 | ||||
| σab | −0.024 | ||||
| ε | 0.304 | ||||
| Branch | a | −2.668 | 0.314 | 8.485 | <0.01 |
| b | 1.783 | 0.141 | 12.662 | <0.01 | |
| σa | 0.491 | ||||
| σb | 0.104 | ||||
| σab | −0.207 | ||||
| ε | 0.384 | ||||
| Needle | a | −3.091 | 0.553 | 5.592 | <0.01 |
| b | 1.515 | 0.207 | 7.331 | <0.01 | |
| σa | 1.673 | ||||
| σb | 0.233 | ||||
| σab | −0.604 | ||||
| ε | 0.475 | ||||
| Root | a | −3.484 | 0.433 | 8.509 | <0.01 |
| b | 2.370 | 0.150 | 15.820 | <0.01 | |
| σa | 1.079 | ||||
| σb | 0.128 | ||||
| σab | −0.370 | ||||
| ε | 0.247 | ||||
| Total | a | −2.110 | 0.206 | 10.225 | <0.01 |
| b | 2.419 | 0.079 | 30.455 | <0.01 | |
| σa | 0.241 | ||||
| σb | 0.189 | ||||
| σab | −0.091 | ||||
| ε | 0.142 |
| Region | Biomass Component | Parameter | Estimate Value | S.D. | Region | Biomass Component | Parameter | Estimate Value | S.D. |
|---|---|---|---|---|---|---|---|---|---|
| S1 | Stem wood | a | −4.284 | 0.210 | S4 | Stem wood | a | −3.843 | 0.153 |
| b | 3.061 | 0.080 | b | 2.836 | 0.061 | ||||
| Stem bark | a | −3.916 | 0.118 | Stem bark | a | −3.965 | 0.111 | ||
| b | 2.166 | 0.046 | b | 2.142 | 0.043 | ||||
| Branch | a | −2.354 | 0.322 | Branch | a | −3.277 | 0.255 | ||
| b | 1.605 | 0.123 | b | 1.835 | 0.101 | ||||
| Needle | a | −2.274 | 0.421 | Needle | a | −4.225 | 0.339 | ||
| b | 1.263 | 0.161 | b | 1.717 | 0.135 | ||||
| Root | a | −4.412 | 0.222 | Root | a | −4.030 | 0.186 | ||
| b | 2.600 | 0.084 | b | 2.551 | 0.074 | ||||
| Total biomass | a | −2.650 | 0.137 | Total biomass | a | −2.540 | 0.104 | ||
| b | 2.649 | 0.052 | b | 2.554 | 0.041 | ||||
| S2 | Stem wood | a | −3.356 | 0.152 | S5 | Stem wood | a | −3.007 | 0.195 |
| b | 2.671 | 0.063 | b | 2.597 | 0.071 | ||||
| Stem bark | a | −3.904 | 0.106 | Stem bark | a | −3.897 | 0.115 | ||
| b | 2.150 | 0.043 | b | 2.153 | 0.042 | ||||
| Branch | a | −3.387 | 0.282 | Branch | a | −1.989 | 0.343 | ||
| b | 2.287 | 0.118 | b | 1.543 | 0.125 | ||||
| Needle | a | −4.702 | 0.337 | Needle | a | −1.659 | 0.422 | ||
| b | 2.268 | 0.141 | b | 1.020 | 0.154 | ||||
| Root | a | −4.653 | 0.170 | Root | a | −3.135 | 0.219 | ||
| b | 2.705 | 0.071 | b | 2.138 | 0.080 | ||||
| Total biomass | a | −2.298 | 0.095 | Total biomass | a | −1.451 | 0.134 | ||
| b | 2.519 | 0.040 | b | 2.208 | 0.049 | ||||
| S3 | Stem wood | a | −3.114 | 0.122 | S6 | Stem wood | a | −4.394 | 0.25 |
| b | 2.525 | 0.051 | b | 2.971 | 0.091 | ||||
| Stem bark | a | −3.927 | 0.102 | Stem bark | a | −3.971 | 0.134 | ||
| b | 2.146 | 0.042 | b | 2.151 | 0.048 | ||||
| Branch | a | −2.757 | 0.192 | Branch | a | −2.374 | 0.381 | ||
| b | 1.794 | 0.081 | b | 1.673 | 0.138 | ||||
| Needle | a | −2.958 | 0.258 | Needle | a | −2.867 | 0.477 | ||
| b | 1.429 | 0.108 | b | 1.434 | 0.173 | ||||
| Root | a | −3.929 | 0.133 | Root | a | −2.041 | 0.314 | ||
| b | 2.434 | 0.056 | b | 1.829 | 0.114 | ||||
| Total biomass | a | −1.849 | 0.076 | Total biomass | a | −1.878 | 0.159 | ||
| b | 2.277 | 0.032 | b | 2.307 | 0.058 |
| Model | Biomass Component | R2 | MAB | RMSE | F-Value | p-Value |
|---|---|---|---|---|---|---|
| General model | Stem wood | 0.967 | 0.213 | 0.275 | ||
| Dummy variable model | 0.977 | 0.169 | 0.230 | 18.610 | <0.01 | |
| Mixed effect model | 0.979 | 0.165 | 0.222 | 12.733 | <0.01 | |
| Bayesian hierarchical model | 0.987 | 0.166 | 0.219 | 11.532 | <0.01 | |
| General model | Stem bark | 0.934 | 0.224 | 0.313 | ||
| Dummy variable model | 0.935 | 0.217 | 0.310 | 2.955 | <0.01 | |
| Mixed effect model | 0.938 | 0.213 | 0.305 | 1.339 | <0.01 | |
| Bayesian hierarchical model | 0.937 | 0.217 | 0.303 | 1.256 | <0.01 | |
| General model | Branch | 0.790 | 0.361 | 0.505 | ||
| Dummy variable model | 0.857 | 0.279 | 0.392 | 25.911 | <0.01 | |
| Mixed effect model | 0.880 | 0.269 | 0.382 | 17.713 | <0.01 | |
| Bayesian hierarchical model | 0.879 | 0.272 | 0.378 | 16.321 | <0.01 | |
| General model | Needle | 0.682 | 0.456 | 0.592 | ||
| Dummy variable model | 0.776 | 0.372 | 0.487 | 20.019 | <0.01 | |
| Mixed effect model | 0.799 | 0.355 | 0.471 | 13.713 | <0.01 | |
| Bayesian hierarchical model | 0.798 | 0.356 | 0.465 | 12.722 | <0.01 | |
| General model | Root | 0.950 | 0.236 | 0.309 | ||
| Dummy variable model | 0.967 | 0.186 | 0.247 | 20.237 | <0.01 | |
| Mixed effect model | 0.968 | 0.184 | 0.246 | 13.785 | <0.01 | |
| Bayesian hierarchical model | 0.968 | 0.185 | 0.242 | 12.695 | <0.01 | |
| General model | Total biomass | 0.982 | 0.140 | 0.182 | ||
| Dummy variable model | 0.988 | 0.106 | 0.141 | 23.388 | <0.01 | |
| Mixed effect model | 0.989 | 0.105 | 0.141 | 16.056 | <0.01 | |
| Bayesian hierarchical model | 0.989 | 0.105 | 0.139 | 15.472 | <0.01 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Huang, X.; Zhang, S.; Sun, X. Biomass Modeling of Larch (Larix spp.) Plantations in China Based on the Mixed Model, Dummy Variable Model, and Bayesian Hierarchical Model. Forests 2017, 8, 268. https://doi.org/10.3390/f8080268
Chen D, Huang X, Zhang S, Sun X. Biomass Modeling of Larch (Larix spp.) Plantations in China Based on the Mixed Model, Dummy Variable Model, and Bayesian Hierarchical Model. Forests. 2017; 8(8):268. https://doi.org/10.3390/f8080268
Chicago/Turabian StyleChen, Dongsheng, Xingzhao Huang, Shougong Zhang, and Xiaomei Sun. 2017. "Biomass Modeling of Larch (Larix spp.) Plantations in China Based on the Mixed Model, Dummy Variable Model, and Bayesian Hierarchical Model" Forests 8, no. 8: 268. https://doi.org/10.3390/f8080268





