SAXS Studies of the Endoglucanase Cel12A from Gloeophyllum trabeum Show Its Monomeric Structure and Reveal the Influence of Temperature on the Structural Stability of the Enzyme
Abstract
:1. Introduction
2. Results and Discussion
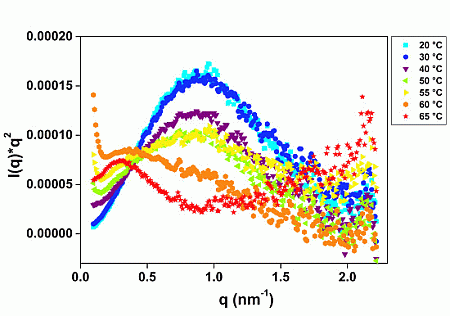
2.1. GtCel12A Is One of the Most Thermal Stable among Other Characterized Cel12A

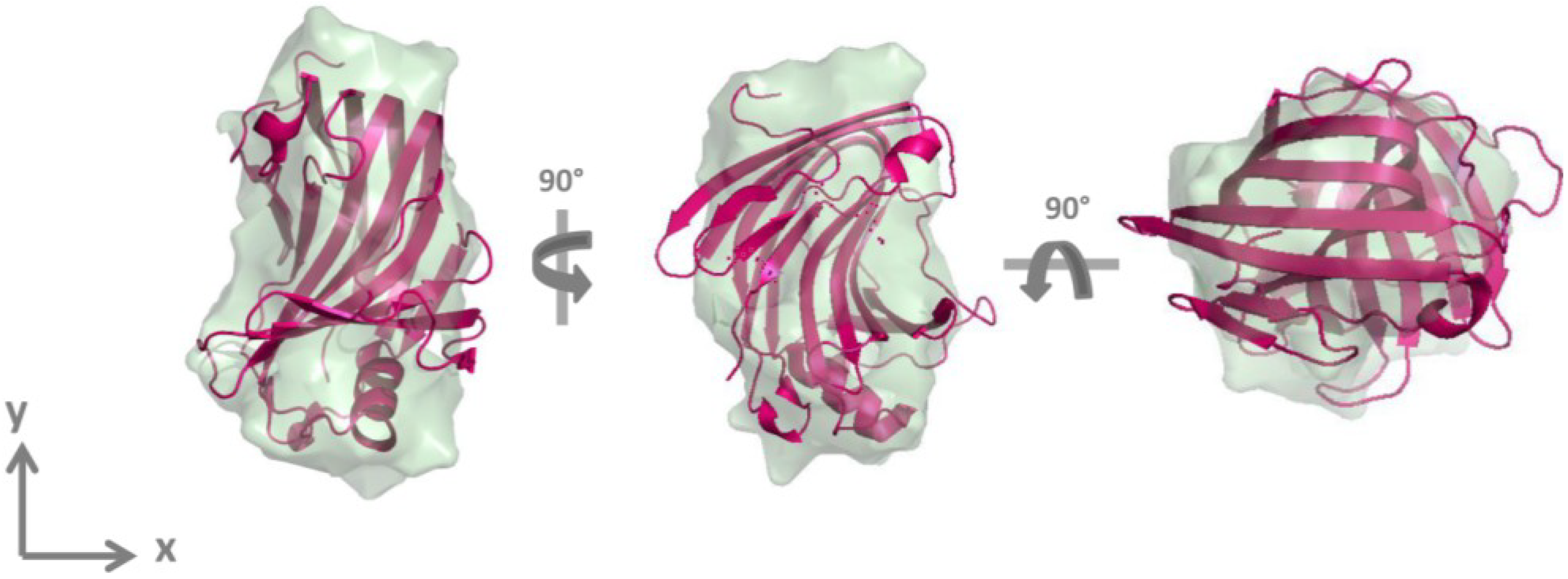
2.2. GtCel12A Forms Monomers in Solution
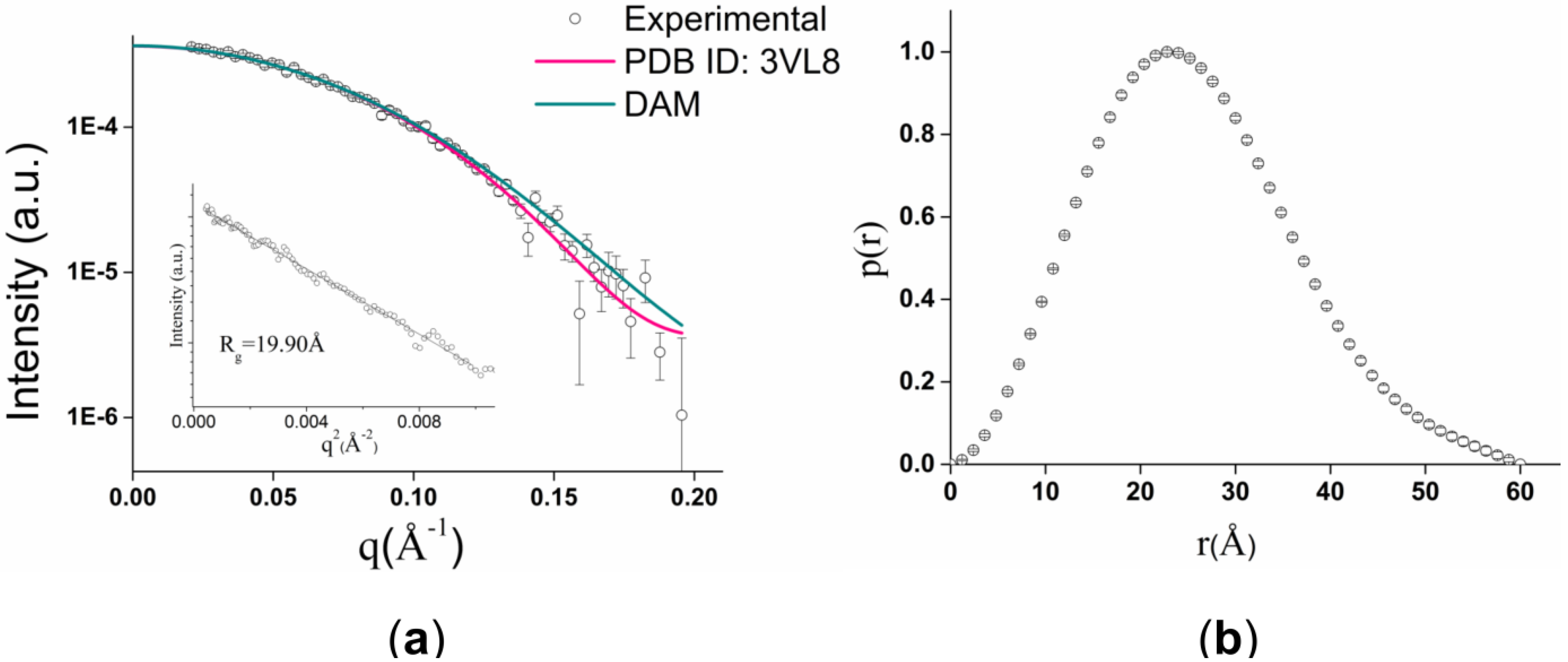
| Parameters | Experimental | PDB ID: 3VL8 (AaXEG) | DAM |
|---|---|---|---|
| Rg (Å) | 19.4 ± 0.1 | 18.25 | 18.57 |
| Dmax (Å) | 60.00 | 53.09 | 56.78 |
| Resolution (Å) | 32.14 | – | 32.14 |
| SAXS MW (kDa) (theoretical MW = 26.17) | 28.30 | – | – |
| χ | – | 1.40 | 1.47 |

3. Experimental Section
3.1. Gene Cloning, Recombinant Expression, Protein Production, Purification and Identity Confirmation
3.2. CD
3.3. Native PAGE
3.4. SAXS Studies
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goodell, B. Brown-rot fungal degradation of wood: Our evolving view. In Wood Deterioration and Preservation: Advances in Our Changing World; Goodell, B., Nicholas, D.D., Schultz, T.P., Eds.; American Chemical Society Publication: Washington, DC, USA, 2003; pp. 97–118. [Google Scholar]
- Eriksson, K.E.L.; Blanchette, R.A. Microbial and Enzymatic Degradation of Wood and Wood Components; Springer-Verlag: Berlin, Germany, 1990. [Google Scholar]
- Kerem, Z.; Jensen, K.A.; Hammel, K.E. Biodegradative mechanism of the brown rot basidiomycete Gloeophyllum trabeum: Evidence for an extracellular hydroquinone-driven fenton reaction. FEBS Lett. 1999, 446, 49–54. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Gilkes, N.R.; Henrissat, B.; Kilburn, D.G.; Miller, R.C.; Warren, R.A. Domains in microbial beta-1,4-glycanases: Sequence conservation, function, and enzyme families. Microbiol. Rev. 1991, 55, 303–315. [Google Scholar]
- Palonen, H. Role of Lignin in the Enzymatic Hydrolysis of Lignocellulose; VTT Biotechnology: Espoo, Finland, 2004. [Google Scholar]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Bauer, M.W.; Driskill, L.E.; Callen, W.; Snead, M.A.; Mathur, E.J.; Kelly, R.M. An endoglucanase, EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes beta-1,4 bonds in mixed-linkage (1-->3),(1-->4)-beta-d-glucans and cellulos. J. Bacteriol. 1999, 181, 284–290. [Google Scholar]
- Bok, J.D.; Yernool, D.A.; Eveleigh, D.E. Purification, characterization, and molecular analysis of thermostable cellulases CelA and CelB from Thermotoga neapolitan. Appl. Environ. Microbiol. 1998, 64, 4774–4781. [Google Scholar]
- Wicher, K.B.; Abou-Hachem, M.; Halldórsdóttir, S.; Thorbjarnadóttir, S.H.; Eggertsson, G.; Hreggvidsson, G.O.; Nordberg Karlsson, E.; Holst, O. Deletion of a cytotoxic, N-terminal putatitive signal peptide results in a significant increase in production yields in Escherichia coli and improved specific activity of Cel12A from Rhodothermus marinus. Appl. Microbiol. Biotechnol. 2001, 55, 578–584. [Google Scholar] [CrossRef]
- Ding, S.; Adney, W.S.; Vinzant, T.B.; Decker, S.R.; Baker, J.O.; Thomas, S.R.; Himmel, M.E. Glycoside hydrolase gene cluster of Acidothermus cellulolyticus. In Applications of enzymes to lignocellulosics; Mansfield, S.D., Saddler, J.N., Eds.; American Chemical Society: Washington, DC, USA, 2003; pp. 332–360. [Google Scholar]
- Sandgren, M.; Gualfetti, P.J.; Shaw, A.; Gross, L.S.; Saldajeno, M.; Day, A.G.; Jones, T.A.; Mitchinson, C. Comparison of family 12 glycoside hydrolases and recruited substitutions important for thermal stability. Protein Sci. 2003, 12, 848–860. [Google Scholar] [CrossRef]
- Fischer, H.; de Oliveira Neto, M.; Napolitano, H.B.; Polikarpov, I.; Craievich, A.F. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 2010, 43, 101–109. [Google Scholar] [CrossRef]
- ExPASy-ProtParam tool. Available online: http://web.expasy.org/protparam/ (accessed on 2 April 2013).
- Svergun, D.I.; Petoukhov, M.V.; Koch, M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001, 80, 2946–2953. [Google Scholar] [CrossRef]
- Kozin, M.B.; Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001, 34, 33–41. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Shimizu, T.; Hirano, H.; Sato, M.; Hashimoto, H. Structural basis for inhibition of xyloglucan-specific endo-β-1,4-glucanase (XEG) by XEG-protein inhibitor. J. Biol. Chem. 2012, 287, 18710–18716. [Google Scholar]
- Glatter, O.; Kratky, O. Small Angle X-ray Scattering; Academic Press: London, UK, 1982. [Google Scholar]
- Doniach, S.; Bascle, J.; Garel, T.; Orland, H. Partially folded states of proteins: Characterization by X-ray scattering. J. Mol. Biol. 1995, 254, 960–967. [Google Scholar] [CrossRef]
- Rambo, R.P.; Tainer, J.A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 2011, 95, 559–571. [Google Scholar] [CrossRef]
- Storms, R.; Zheng, Y.; Li, H.; Sillaots, S.; Martinez-Perez, A.; Tsang, A. Plasmid vectors for protein production, gene expression and molecular manipulations in Aspergillus niger. Plasmid 2005, 53, 191–204. [Google Scholar] [CrossRef]
- Aslanidis, C.; de Jong, P. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 1990, 18, 6069–6074. [Google Scholar] [CrossRef]
- Debets, A.J.M.; Bos, C.J. Isolation of small protoplasts from Aspergillus niger. Fungal Genet. Newsl. 1986, 33, 24. [Google Scholar]
- Käfer, E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977, 19, 33–131. [Google Scholar]
- Chen, Y.H.; Yang, J.T. A new approach to the calculation of secondary structures of globular proteins by optical rotatory dispersion and circular dichroism. Biochem. Biophys. Res. Commun. 1971, 44, 1285–1291. [Google Scholar]
- Hammersley, A.P. FIT2D: An Introduction and Overview; ESRF Internal Report, ESRF97HA02T; European Synchrotron Radiation Facility: Grenoble, France, 1997. [Google Scholar]
- Guinier, A.; Fournet, G. Small-Angle Scattering of X-rays; John Wiley and Sons: New York, NY, USA, 1955; pp. 1–268. [Google Scholar]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using percentual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Svergun, D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999, 76, 2879–2886. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Miotto, L.S.; Dos Reis, C.V.; De Oliveira Neto, M.; Polikarpov, I. SAXS Studies of the Endoglucanase Cel12A from Gloeophyllum trabeum Show Its Monomeric Structure and Reveal the Influence of Temperature on the Structural Stability of the Enzyme. Materials 2014, 7, 5202-5211. https://doi.org/10.3390/ma7075202
Miotto LS, Dos Reis CV, De Oliveira Neto M, Polikarpov I. SAXS Studies of the Endoglucanase Cel12A from Gloeophyllum trabeum Show Its Monomeric Structure and Reveal the Influence of Temperature on the Structural Stability of the Enzyme. Materials. 2014; 7(7):5202-5211. https://doi.org/10.3390/ma7075202
Chicago/Turabian StyleMiotto, Lis S., Caio V. Dos Reis, Mario De Oliveira Neto, and Igor Polikarpov. 2014. "SAXS Studies of the Endoglucanase Cel12A from Gloeophyllum trabeum Show Its Monomeric Structure and Reveal the Influence of Temperature on the Structural Stability of the Enzyme" Materials 7, no. 7: 5202-5211. https://doi.org/10.3390/ma7075202