Morphological and Mechanical Characterization of DNA SAMs Combining Nanolithography with AFM and Optical Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Samples Preparation
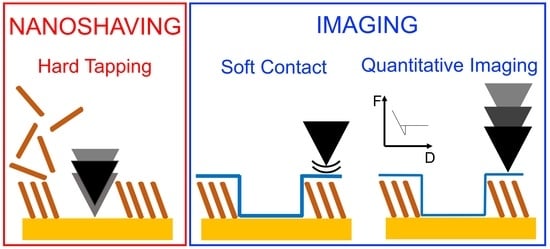
2.3. Atomic Force Microscopy (AFM)
2.4. Spectroscopic Ellipsometry (SE)
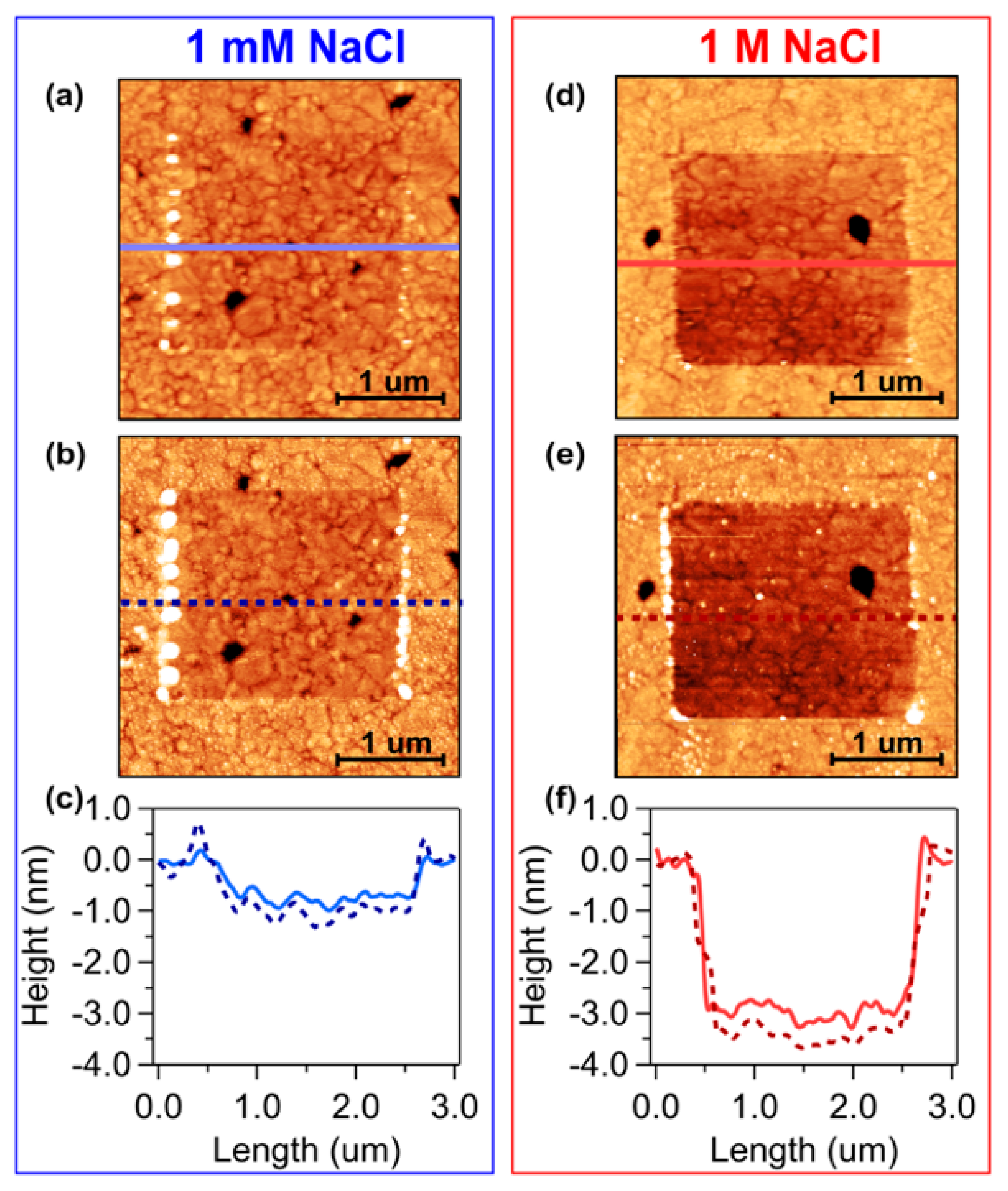
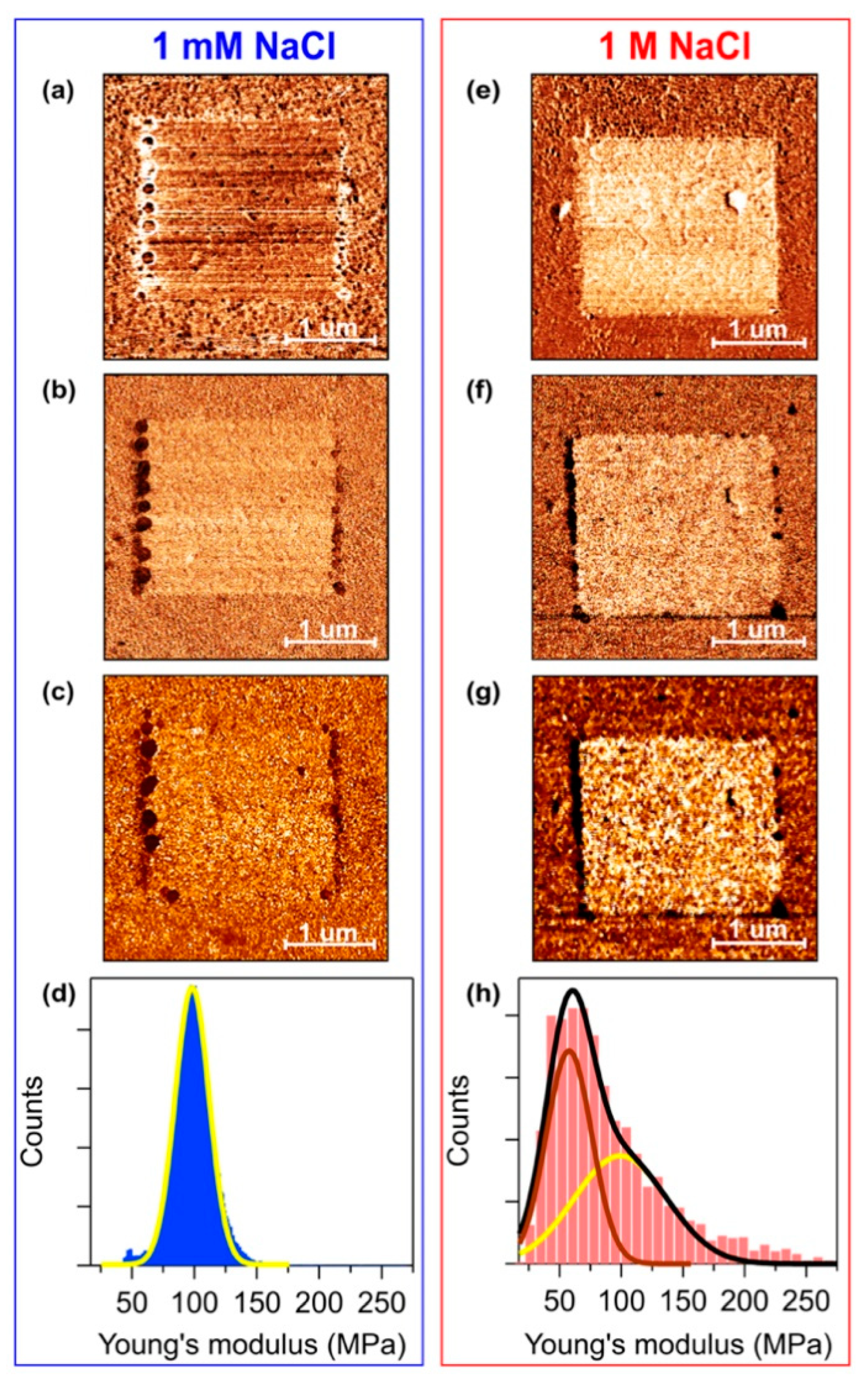
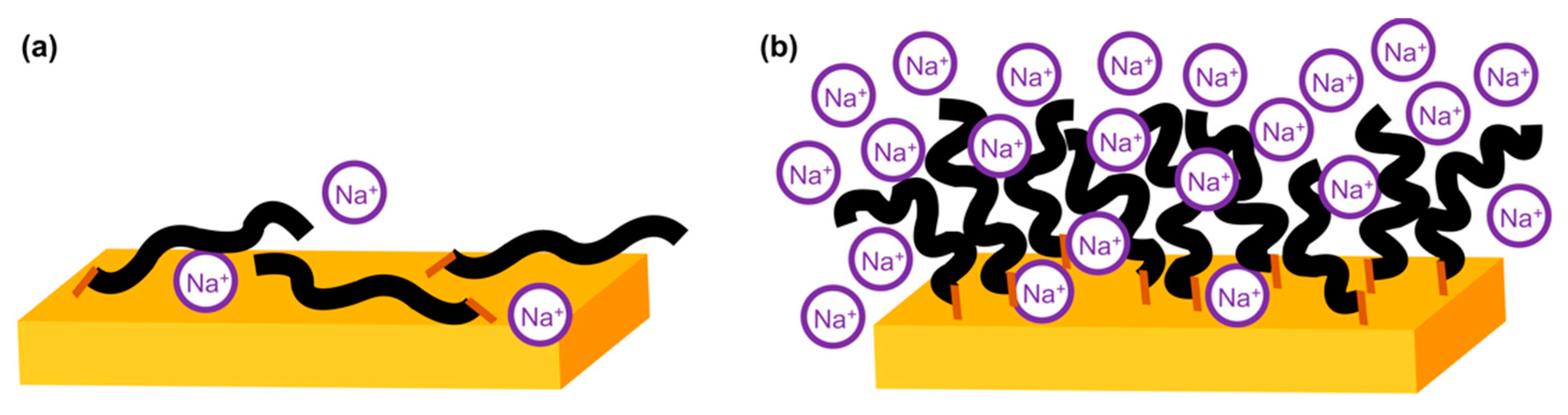
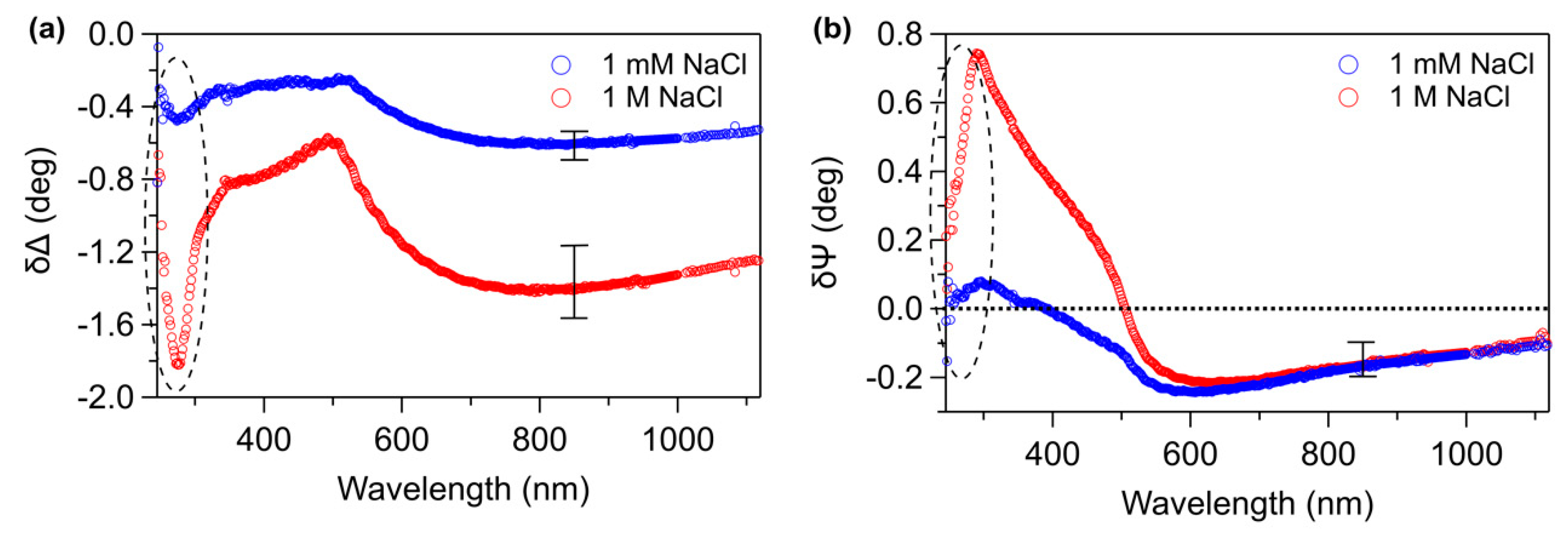
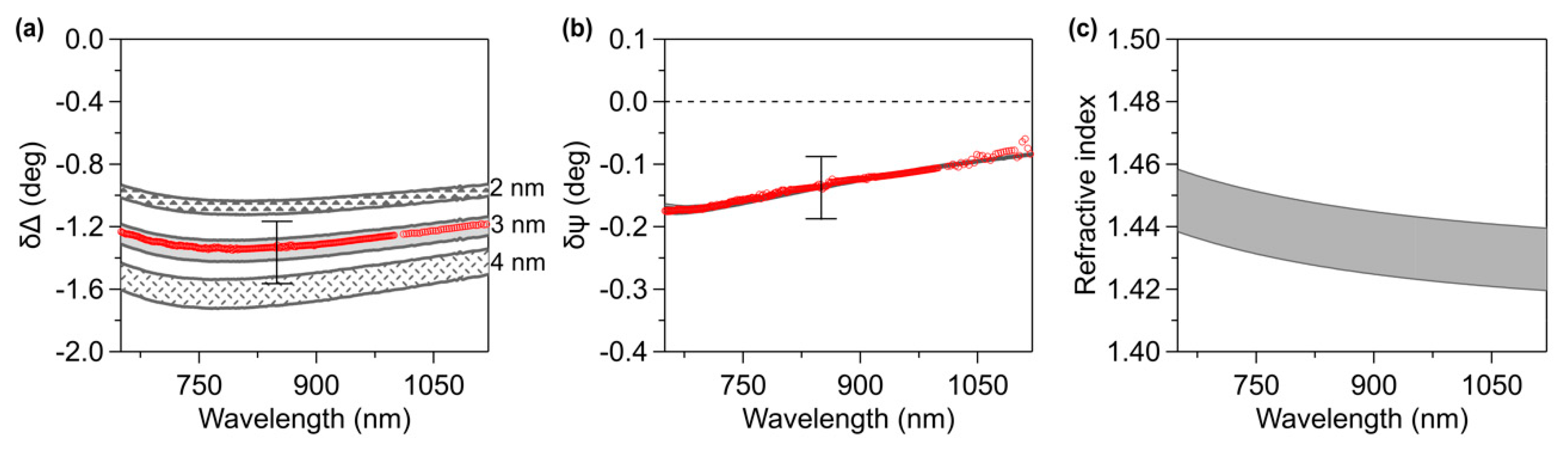
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brittain, W.J.; Brandsetter, T.; Prucker, O.; Rühe, J. The Surface Science of Microarray Generation—A Critical Inventory. ACS Appl. Mater. Interfaces 2019, 11, 39397–39409. [Google Scholar] [CrossRef]
- Xiao, Q.; Wu, J.; Dang, P.; Ju, H. Multiplexed chemiluminescence imaging assay of protein biomarkers using DNA microarray with proximity binding-induced hybridization chain reaction amplification. Anal. Chim. Acta 2018, 1032, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Chen, T.; Zhang, Q.; Zhou, M.; Zhu, X. Application of DNA nanodevices for biosensing. Analyst 2020, 145, 3481–3489. [Google Scholar] [CrossRef] [PubMed]
- MacAskill, A.; Crawford, D.; Graham, D.; Faulds, K. DNA Sequence Detection Using Surface-Enhanced Resonance Raman Spectroscopy in a Homogeneous Multiplexed Assay. Anal. Chem. 2009, 81, 8134–8140. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Lin, M.; Ji, W.; Yuan, H.; Zhang, Y.; Jing, Z.; Zhao, J. Boronic Acid Functionalized Au Nanoparticles for Selective MicroRNA Signal Amplification in Fiber-Optic Surface Plasmon Resonance Sensing System. ACS Sens. 2018, 7, 929–935. [Google Scholar] [CrossRef]
- Caneira, C.R.F.; Soares, R.R.G.; Pinto, I.F.; Mueller-Landau, H.S.; Azevedo, A.M.; Chu, V.; Conde, J.P. Development of a rapid bead-based microfluidic platform for DNA hybridization using single- and multi-mode interactions for probe immobilization. Sens. Actuators B Chem. 2019, 286, 328–336. [Google Scholar] [CrossRef]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Park, M.; Hwang, J.; Kim, J.H.; Chung, D.-R.; Lee, K.; Kang, M. Development of Label-Free Colorimetric Assay for MERS-CoV Using Gold Nanoparticles. ACS Sens. 2019, 4, 1306–1312. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [Green Version]
- Wold, E.D.; McBride, R.; Axup, J.Y.; Kazane, S.A.; Smider, V.V. Antibody Microarrays Utilizing Site-Specific Antibody–Oligonucleotide Conjugates. Bioconjug. Chem. 2015, 26, 807–811. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, S.; Zhou, Z.; Zhang, R.; Shen, H.; Song, J.; Su, P.; Yang, Y. Enhanced reusability and activity: DNA directed immobilization of enzyme on polydopamine modified magnetic nanoparticles. Biochem. Eng. J. 2018, 137, 108–115. [Google Scholar] [CrossRef]
- Wang, H.; Liu, M.; Bai, W.; Sun, H.; Li, Y.; Deng, H. A convenient electrogenerated chemiluminescence biosensing method for selective detection of 5-hydroxymethylcytosine in genomic DNA. Sens. Actuators B Chem. 2019, 284, 236–242. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, J. Improving the Signal Sensitivity and Photostability of DNA Hybridizations on Microarrays by Using Dye-Doped Core−Shell Silica Nanoparticles. Anal. Chem. 2004, 76, 5302–5312. [Google Scholar] [CrossRef] [PubMed]
- Seymour, E.; Daaboul, G.G.; Zhang, X.; Scherr, S.M.; Ünlü, N.L.; Connor, J.H.; Ünlü, M.S. DNA-Directed Antibody Immobilization for Enhanced Detection of Single Viral Pathogens. Anal. Chem. 2015, 87, 10505–10512. [Google Scholar] [CrossRef]
- Chen, H.; Meisburger, S.P.; Pabit, S.A.; Sutton, J.L.; Webb, W.W.; Pollack, L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. USA 2012, 109, 799–804. [Google Scholar] [CrossRef] [Green Version]
- Guilbaud, S.; Salomé, L.; Destainville, N.; Manghi, M.; Tardin, C. Dependence of DNA Persistence Length on Ionic Strength and Ion Type. Phys. Rev. Lett. 2019, 122, 028102. [Google Scholar] [CrossRef] [Green Version]
- Herne, T.M.; Tarlov, M.J. Characterization of DNA Probes Immobilized on Gold Surfaces. J. Am. Chem. Soc. 1997, 119, 8916–8920. [Google Scholar] [CrossRef]
- Li, Z.; Niu, T.; Zhang, Z.; Feng, G.; Bi, S. Effect of monovalent cations (Li+, Na+, K+, Cs+) on self-assembly of thiol-modified double-stranded and single-stranded DNA on gold electrode. Analyst 2012, 137, 1680–1691. [Google Scholar] [CrossRef]
- Castelino, K.; Kannan, B.; Majumdar, A. Characterization of Grafting Density and Binding Efficiency of DNA and Proteins on Gold Surfaces. Langmuir 2005, 21, 1956–1961. [Google Scholar] [CrossRef]
- Peterson, A.W.; Heaton, R.J.; Georgiadis, R.M. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001, 29, 5163–5168. [Google Scholar] [CrossRef] [Green Version]
- Steel, A.B.; Levicky, R.L.; Herne, T.M.; Tarlov, M.J. Immobilization of Nucleic Acids at Solid Surfaces: Effect of Oligonucleotide Length on Layer Assembly. Biophys. J. 2000, 79, 975–981. [Google Scholar] [CrossRef] [Green Version]
- Moses, S.; Brewer, S.H.; Lowe, L.B.; Lappi, S.E.; Gilvey, L.B.G.; Sauthier, M.; Tenent, R.C.; Feldheim, D.L.; Franzen, S. Characterization of Single- and Double-Stranded DNA on Gold Surfaces. Langmuir 2004, 20, 11134–11140. [Google Scholar] [CrossRef] [PubMed]
- Steel, A.B.; Herne, T.M.; Tarlov, M.J. Electrochemical Quantitation of DNA Immobilized on Gold. Anal. Chem. 1998, 70, 4670–4677. [Google Scholar] [CrossRef] [PubMed]
- Keighley, S.D.; Li, P.; Estrela, P.; Migliorato, P. Optimization of DNA immobilization on gold electrodes for label-free detection by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2008, 23, 1291–1297. [Google Scholar] [CrossRef]
- Georgiadis, R.; Peterlinz, K.P.; Peterson, A.W. Quantitative Measurements and Modeling of Kinetics in Nucleic Acid Monolayer Films Using SPR Spectroscopy. J. Am. Chem. Soc. 2000, 122, 3166–3173. [Google Scholar] [CrossRef]
- Wolf, L.K.; Gao, Y.; Georgiadis, R.M. Sequence-Dependent DNA Immobilization: Specific versus Nonspecific Contributions. Langmuir 2004, 20, 3357–3361. [Google Scholar] [CrossRef]
- Kumar, K.S.; Naaman, R. Quantitative Analysis and Characterization of Self-Assembled DNA on a Silver Surface. Langmuir 2012, 28, 14514–14517. [Google Scholar] [CrossRef]
- Petrovykh, D.Y.; Kimura-Suda, H.; Whitman, L.J.; Tarlov, M.J. Quantitative Analysis and Characterization of DNA Immobilized on Gold. J. Am. Chem. Soc. 2003, 125, 5219–5226. [Google Scholar] [CrossRef]
- Howell, C.; Hamoudi, H.; Zharnikov, M. Thymine/adenine diblock-oligonucleotide monolayers and hybrid brushes on gold: A spectroscopic study. Biointerphases 2013, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Charrier, E.E.; Pogoda, K.; Wells, R.G.; Janmey, P.A. Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation. Nat. Commun. 2018, 9, 449. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Maoz, R.; Schmid, G.; Sagiv, J. Template Guided Self-Assembly of [Au55] Clusters on Nanolithographically Defined Monolayer Patterns. Nano Lett. 2002, 2, 1055–1060. [Google Scholar] [CrossRef] [Green Version]
- Ryu, Y.K.; Garcia, R. Advanced oxidation scanning probe lithography. Nanotechnology 2017, 28, 142003–142020. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Liu, G.-Y. Nanometer-Scale Fabrication by Simultaneous Nanoshaving and Molecular Self-Assembly. Langmuir 1997, 13, 127–129. [Google Scholar] [CrossRef]
- Liu, G.-Y.; Xu, S.; Qian, Y. Nanofabrication of Self-Assembled Monolayers Using Scanning Probe Lithography. Acc. Chem. Res. 2000, 33, 457–466. [Google Scholar] [CrossRef]
- Unruh, D.A.; Malduin, C.; Pastine, S.J.; Rolandi, M.; Fréchet, J.M.J. Bifunctional Patterning of Mixed Monolayer Surfaces Using Scanning Probe Lithography for Multiplexed Directed Assembly. J. Am. Chem. Soc. 2010, 132, 6890–6891. [Google Scholar] [CrossRef]
- Ambrosetti, E.; Paoletti, P.; Bosco, A.; Parisse, P.; Scaini, D.; Tagliabue, E.; de Marco, A.; Casalis, L. Quantification of Circulating Cancer Biomarkers via Sensitive Topographic Measurements on Single Binder Nanoarrays. ACS Omega 2017, 2, 2618–2629. [Google Scholar] [CrossRef] [Green Version]
- Bano, F.; Fruk, L.; Sanavio, B.; Glettenberg, M.; Casalis, L.; Niemeyer, C.M.; Scoles, G. Toward Multiprotein Nanoarrays Using Nanografting and DNA Directed Immobilization of Proteins. Nano Lett. 2009, 9, 2614–2618. [Google Scholar] [CrossRef]
- Solano, I.; Parisse, P.; Gramazio, F.; Ianeselli, L.; Medagli, B.; Cavalleri, O.; Casalis, L.; Canepa, M. Atomic Force Microscopy and Spectroscopic Ellipsometry combined analysis of Small Ubiquitin-like Modifier adsorption on functional monolayers. Appl. Surf. Sci. 2017, 421, 722–727. [Google Scholar] [CrossRef]
- Zhou, D.; Sinniah, K.; Abell, C.; Rayment, T. Use of Atomic Force Microscopy for Making Addresses in DNA Coatings. Langmuir 2002, 18, 8278–8281. [Google Scholar] [CrossRef]
- Solano, I.; Parisse, P.; Gramazio, F.; Cavalleri, O.; Bracco, G.; Castronovo, M.; Casalis, L.; Canepa, M. Spectroscopic ellipsometry meets AFM nanolithography: About hydration of bio-inert oligo(ethylene glycol)-terminated self assembled monolayers on gold. Phys. Chem. Chem. Phys. 2015, 17, 28774–28781. [Google Scholar] [CrossRef] [Green Version]
- Canepa, P.; Gonella, G.; Pinto, G.; Grachev, V.; Canepa, M.; Cavalleri, O. Anchoring of Aminophosphonates on Titanium Oxide for Biomolecular Coupling. J. Phys. Chem. C 2019, 123, 16843–16850. [Google Scholar] [CrossRef] [Green Version]
- Patron, A.M.; Bramer, A.M.; Santavicca, D.F.; Mullen, T.J. Nanopatterning of Cu-Ligated Mercaptoalkanoic Acid Multilayers on Si Substrates via Atomic Force Lithography. J. Phys. Chem. C 2020, 124, 1214–1219. [Google Scholar] [CrossRef]
- Liang, J.; Scoles, G. Nanografting of Alkanethiols by Tapping Mode Atomic Force Microscopy. Langmuir 2007, 23, 6142–6147. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F.; Martínez-Martín, D.; Medalsy, I.; Alsteens, D.; Müller, D.J. Multiparametric imaging of biological systems by force-distance curve–based AFM. Nat. Methods 2013, 10, 847–854. [Google Scholar] [CrossRef]
- Pinto, G.; Parisse, P.; Solano, I.; Canepa, P.; Canepa, M.; Casalis, L.; Cavalleri, O. Functionalizing gold with single strand DNA: Novel insight into optical properties via combined spectroscopic ellipsometry and nanolithography measurements. Soft Matter 2019, 15, 2463–2468. [Google Scholar] [CrossRef]
- Gupta, P.; Loos, K.; Korniakov, A.; Spagnoli, C.; Cowman, M.; Ulman, A. Facile Route to Ultraflat SAM-Protected Gold Surfaces by “Amphiphile Splitting”. Angew. Chem. Int. Ed. 2004, 43, 520–523. [Google Scholar] [CrossRef]
- Toccafondi, C.; Cavalleri, O.; Bisio, F.; Canepa, M. Yeast Cytochrome c adsorption on SiO2/Si substrates studied by in situ spectroscopic ellipsometry. Thin Solid Film. 2013, 543, 78–82. [Google Scholar] [CrossRef]
- Solano, I.; Parisse, P.; Cavalleri, O.; Gramazio, F.; Casalis, L.; Canepa, M. Investigating organic multilayers by spectroscopic ellipsometry: Specific and non-specific interactions of polyhistidine with NTA self-assembled monolayers. Beilstein J. Nanotechnol. 2016, 7, 544–553. [Google Scholar] [CrossRef] [Green Version]
- Toccafondi, C.; Prato, M.; Maidecchi, G.; Penco, A.; Bisio, F.; Cavalleri, O.; Canepa, M. Optical properties of Yeast Cytochrome c monolayer on gold: An in situ spectroscopic ellipsometry investigation. J. Colloid Interface Sci. 2011, 364, 125–132. [Google Scholar] [CrossRef]
- Kaiser, W.; Rant, U. Conformations of End-Tethered DNA Molecules on Gold Surfaces: Influences of Applied Electric Potential, Electrolyte Screening, and Temperature. J. Am. Chem. Soc. 2010, 132, 7935–7945. [Google Scholar] [CrossRef]
- Gil, P.S.; Lacks, D.J.; Parisse, P.; Casalis, L.; Nkoua Ngavouka, M.D. Single-stranded DNA oligomer brush structure is dominated by intramolecular interactions mediated by the ion environment. Soft Matter 2018, 14, 9675–9680. [Google Scholar] [CrossRef] [PubMed]
- Hill, H.D.; Millstone, J.E.; Banholzer, M.J.; Mirkin, C.A. The Role Radius of Curvature Plays in Thiolated Oligonucleotide Loading on Gold Nanoparticles. ACS Nano 2009, 3, 418–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prato, M.; Moroni, R.; Bisio, F.; Rolandi, R.; Mattera, L.; Cavalleri, O.; Canepa, M. Optical Characterization of Thiolate Self-Assembled Monolayers on Au(111). J. Phys. Chem. C 2008, 112, 3899–3906. [Google Scholar] [CrossRef]
- Prato, M.; Alloisio, M.; Jadhav, S.A.; Chincarini, A.; Svaldo-Lanero, T.; Bisio, F.; Cavalleri, O.; Canepa, M. Optical Properties of Disulfide-Functionalized Diacetylene Self-Assembled Monolayers on Gold: A Spectroscopic Ellipsometry Study. J. Phys. Chem. C 2009, 113, 20683–20688. [Google Scholar] [CrossRef]
- Zahn, D.R.T.; Silaghi, S.D. Biomolecular Layers on Silicon Studied by Optical Spectroscopy. In Advances in Solid State Physics; Kramer, B., Ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2005; Volume 45, pp. 45–57. ISBN 978-3-540-26041-7. [Google Scholar]
- Nizioł, J.; Makyła-Juzak, K.; Marzec, M.M.; Ekiert, R.; Marzec, M.; Gondek, E. Thermal stability of the solid DNA as a novel optical material. Opt. Mater. 2017, 66, 344–350. [Google Scholar] [CrossRef]
- Paulson, B.; Shin, I.; Jeong, H.; Kong, B.; Khazaeinezhad, R.; Dugasani, S.R.; Jung, W.; Joo, B.; Lee, H.-Y.; Park, S.; et al. Optical dispersion control in surfactant-free DNA thin films by vitamin B2 doping. Sci. Rep. 2018, 8, 9358. [Google Scholar] [CrossRef] [Green Version]
- Parisse, P.; Solano, I.; Magnozzi, M.; Bisio, F.; Casalis, L.; Cavalleri, O.; Canepa, M. Thickness and Beyond. Exploiting Spectroscopic Ellipsometry and Atomic Force Nanolithography for the Investigation of Ultrathin Interfaces of Biologic Interest. In Ellipsometry of Functional Organic Surfaces and Films; Hinrichs, K., Eichhorn, K.-J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 52, pp. 63–93. ISBN 978-3-319-75894-7. [Google Scholar]
- Canepa, M.; Maidecchi, G.; Toccafondi, C.; Cavalleri, O.; Prato, M.; Chaudhari, V.; Esaulov, V.A. Spectroscopic ellipsometry of self assembled monolayers: Interface effects. The case of phenyl selenide SAMs on gold. Phys. Chem. Chem. Phys. 2013, 15, 11559–11565. [Google Scholar] [CrossRef]
- Hamoudi, H.; Prato, M.; Dablemont, C.; Cavalleri, O.; Canepa, M.; Esaulov, V.A. Self-Assembly of 1,4-Benzenedimethanethiol Self-Assembled Monolayers on Gold. Langmuir 2010, 26, 7242–7247. [Google Scholar] [CrossRef]
- Bordi, F.; Prato, M.; Cavalleri, O.; Cametti, C.; Canepa, M.; Gliozzi, A. Azurin Self-Assembled Monolayers Characterized by Coupling Electrical Impedance Spectroscopy and Spectroscopic Ellipsometry. J. Phys. Chem. B 2004, 108, 20263–20272. [Google Scholar] [CrossRef]
- Krivacic, J.R.; Urry, D.W. Ultraviolet and Visible Refractive Indices of Spectro-Quality Solvents. Anal. Chem. 1970, 42, 596–599. [Google Scholar] [CrossRef]
- Berlind, T.; Pribil, G.K.; Thompson, D.; Woollam, J.A.; Arwin, H. Effects of ion concentration on refractive indices of fluids measured by the minimum deviation technique. Phys. Stat. Sol. C 2008, 5, 1249–1252. [Google Scholar] [CrossRef]
- Maartensson, J.; Arwin, H. Interpretation of Spectroscopic Ellipsometry Data on Protein Layers on Gold Including Substrate-Layer Interactions. Langmuir 1995, 11, 963–968. [Google Scholar] [CrossRef]
- Fujiwara, H. Spectroscopic Ellipsometry: Principles and Applications; John Wiley & Sons: New York, NY, USA, 2007; ISBN 978-0-470-06019-3. [Google Scholar]
- Gray, D.E.; Case-Green, S.C.; Fell, T.S.; Dobson, P.J.; Southern, E.M. Ellipsometric and Interferometric Characterization of DNA Probes Immobilized on a Combinatorial Array. Langmuir 1997, 13, 2833–2842. [Google Scholar] [CrossRef]
- Elhadj, S.; Singh, G.; Saraf, R.F. Optical Properties of an Immobilized DNA Monolayer from 255 to 700 nm. Langmuir 2004, 20, 5539–5543. [Google Scholar] [CrossRef]
- Legay, G.; Markey, L.; Meunier-Prest, R.; Finot, E. Measurements of thickness dispersion in biolayers by scanning force microscopy and comparison with spectroscopic ellipsometry analysis. Ultramicroscopy 2007, 107, 1111–1117. [Google Scholar] [CrossRef]
- Inagaki, T.; Hamm, R.N.; Arakawa, E.T.; Painter, L.R. Optical and dielectric properties of DNA in the extreme ultraviolet. J. Chem. Phys. 1974, 61, 4246–4250. [Google Scholar] [CrossRef]
- Samoc, A.; Miniewicz, A.; Samoc, M.; Grote, J.G. Refractive-index anisotropy and optical dispersion in films of deoxyribonucleic acid. J. Appl. Polym. Sci. 2007, 105, 236–245. [Google Scholar] [CrossRef]
- Peterson, A.W.; Wolf, L.K.; Georgiadis, R.M. Hybridization of Mismatched or Partially Matched DNA at Surfaces. J. Am. Chem. Soc. 2002, 124, 14601–14607. [Google Scholar] [CrossRef]
- Rodenhausen, K.B.; Kasputis, T.; Pannier, A.K.; Gerasimov, J.Y.; Lai, R.Y.; Solinsky, M.; Tiwald, T.E.; Wang, H.; Sarkar, A.; Hofmann, T.; et al. Combined optical and acoustical method for determination of thickness and porosity of transparent organic layers below the ultra-thin film limit. Rev. Sci. Instrum. 2011, 82, 103111. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.W.; Byun, J.S.; Kim, Y.D.; Hemzal, D.; Humliček, J. Study of the interaction between HSA and oligo-DNA using total internal reflection ellipsometry. J. Korean Phys. Soc. 2012, 60, 1288–1291. [Google Scholar] [CrossRef]
- Shi, J.; Hong, B.; Parikh, A.N.; Collins, R.W.; Allara, D.L. Optical characterization of electronic transitions arising from the Au/S interface of self-assembled n-alkanethiolate monolayers. Chem. Phys. Lett. 1995, 246, 90–94. [Google Scholar] [CrossRef]
- Peterlinz, K.A.; Georgiadis, R. Two-color approach for determination of thickness and dielectric constant of thin films using surface plasmon resonance spectroscopy. Opt. Commun. 1996, 130, 260–266. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, G.; Canepa, P.; Canale, C.; Canepa, M.; Cavalleri, O. Morphological and Mechanical Characterization of DNA SAMs Combining Nanolithography with AFM and Optical Methods. Materials 2020, 13, 2888. https://doi.org/10.3390/ma13132888
Pinto G, Canepa P, Canale C, Canepa M, Cavalleri O. Morphological and Mechanical Characterization of DNA SAMs Combining Nanolithography with AFM and Optical Methods. Materials. 2020; 13(13):2888. https://doi.org/10.3390/ma13132888
Chicago/Turabian StylePinto, Giulia, Paolo Canepa, Claudio Canale, Maurizio Canepa, and Ornella Cavalleri. 2020. "Morphological and Mechanical Characterization of DNA SAMs Combining Nanolithography with AFM and Optical Methods" Materials 13, no. 13: 2888. https://doi.org/10.3390/ma13132888