Atmospheric Biodetection Part I: Study of Airborne Bacterial Concentrations from January 2018 to May 2020 at Saclay, France
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of the Observation Site
2.2. Collection and Sample Analysis
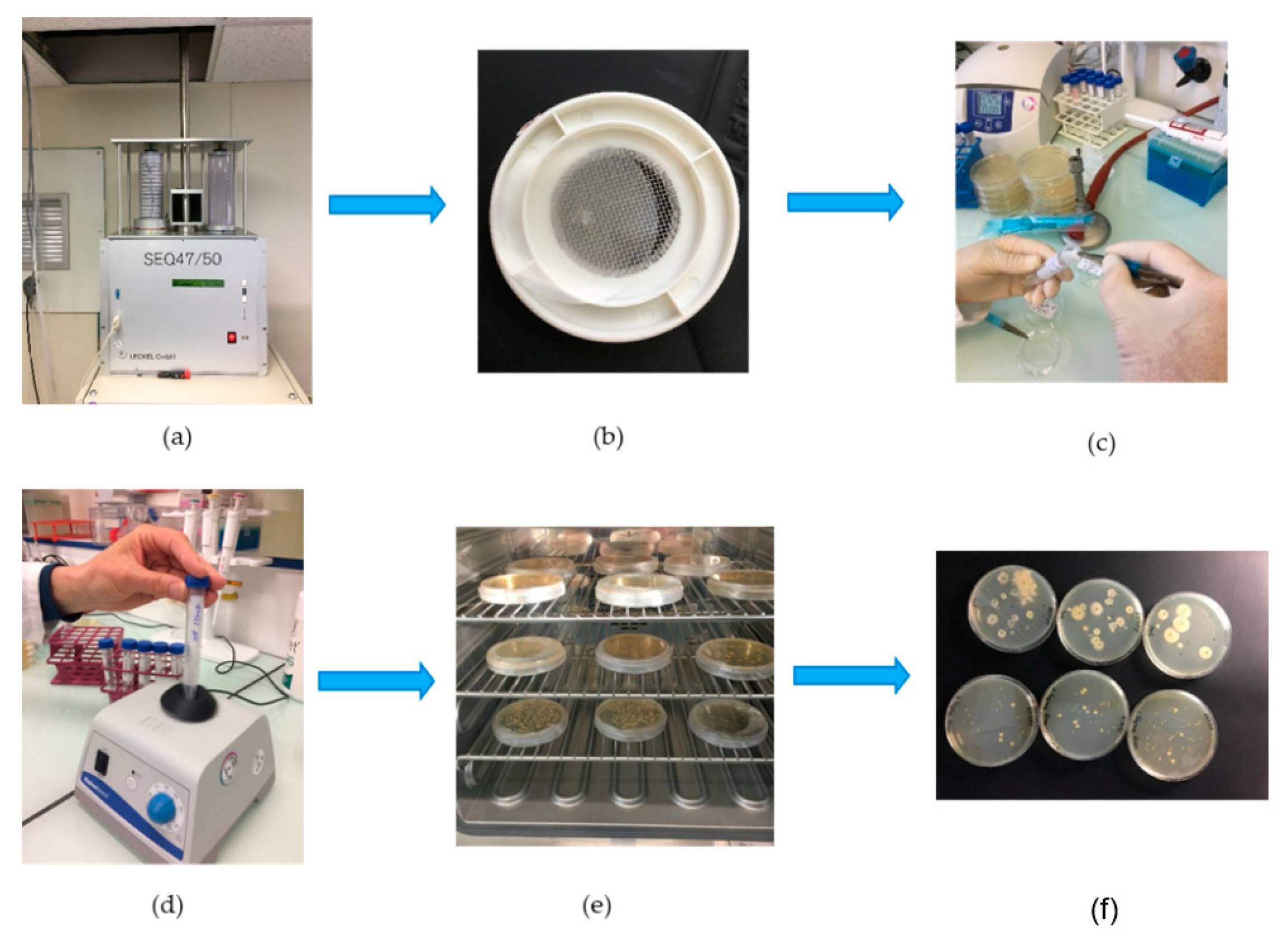
2.2.1. Collection, Extraction and Culture Procedure
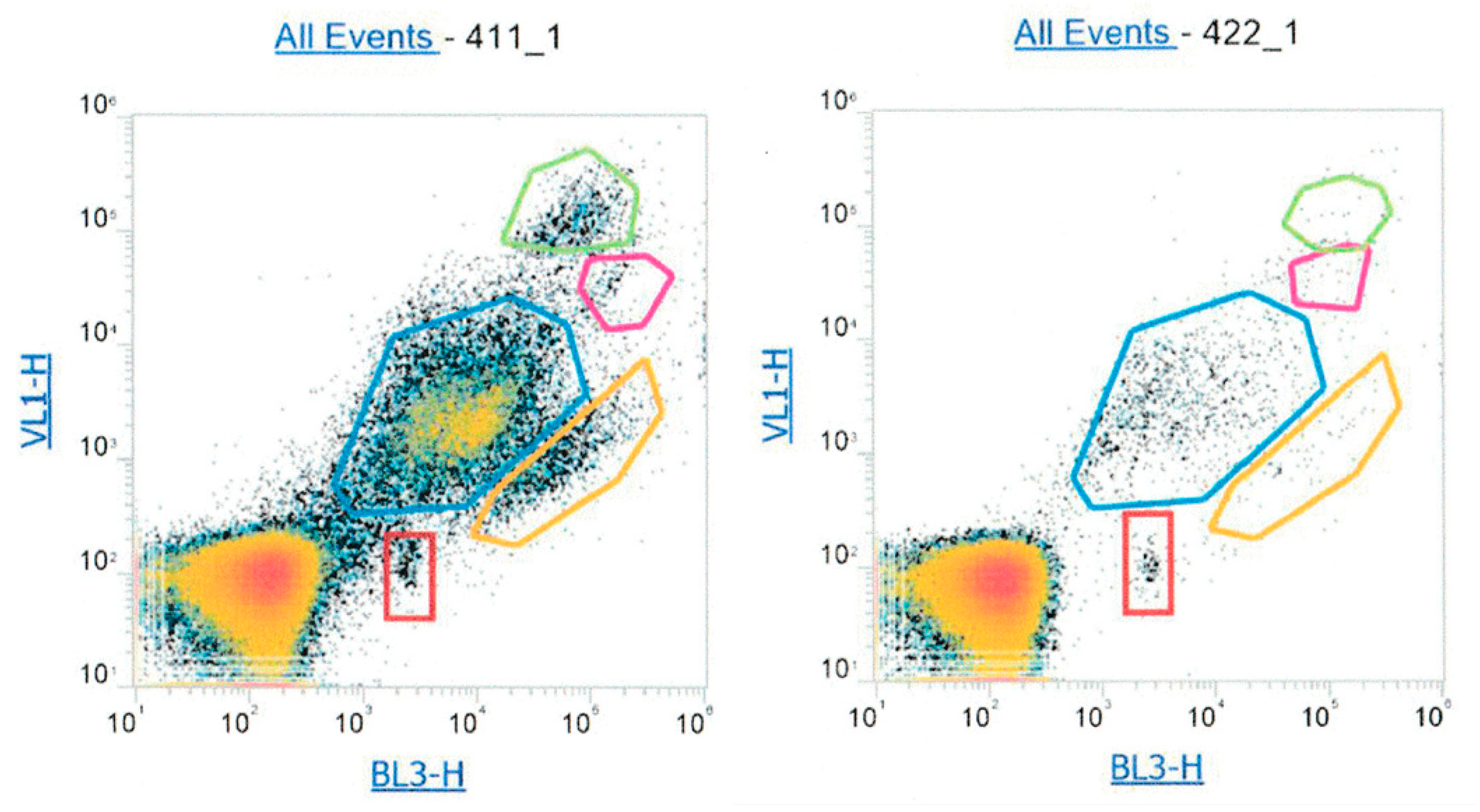
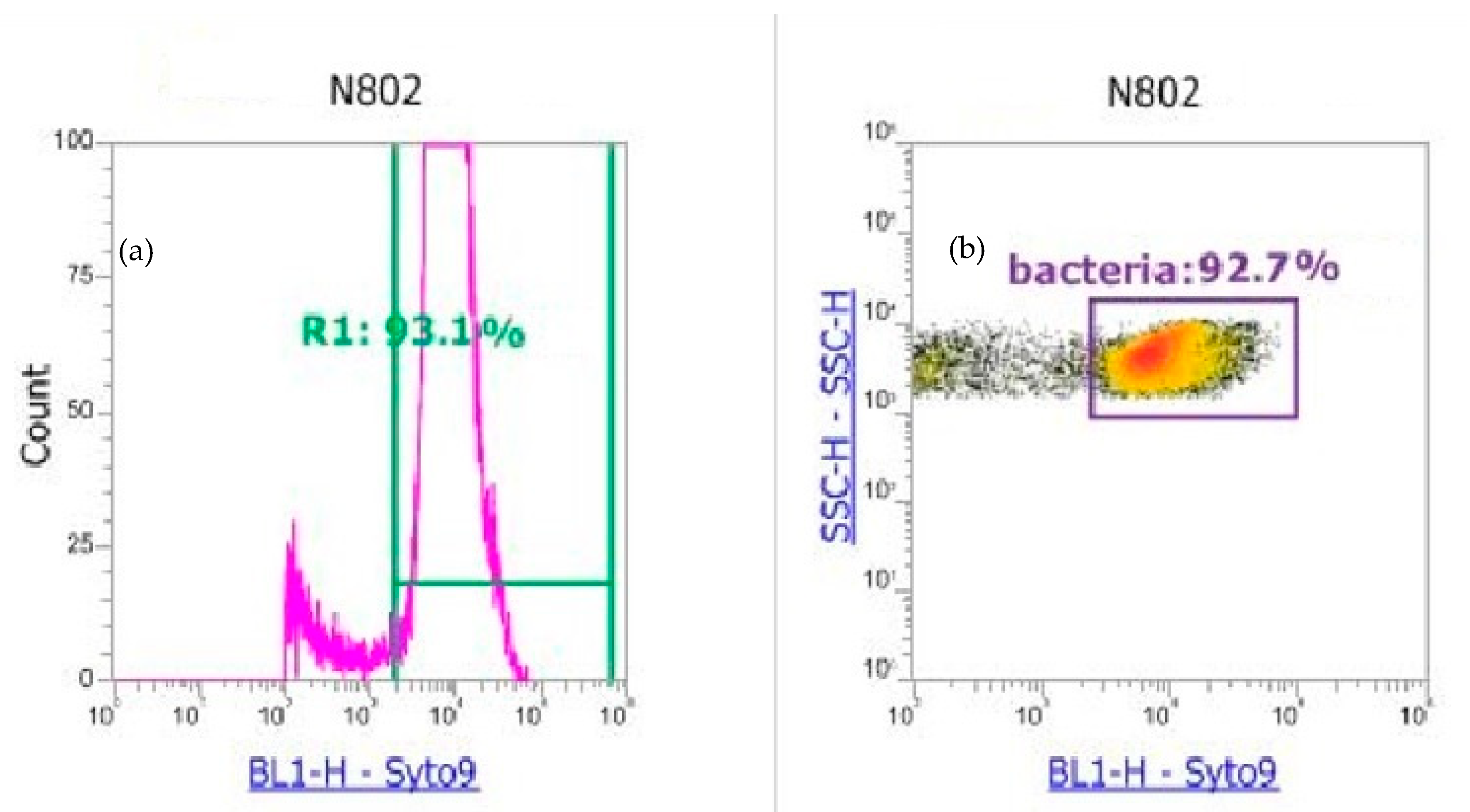
2.2.2. Flow Cytometry Analysis of Atmospheric Samples
2.2.3. Geographical Origins of Total Airborne Bacteria Affecting Saclay
3. Results
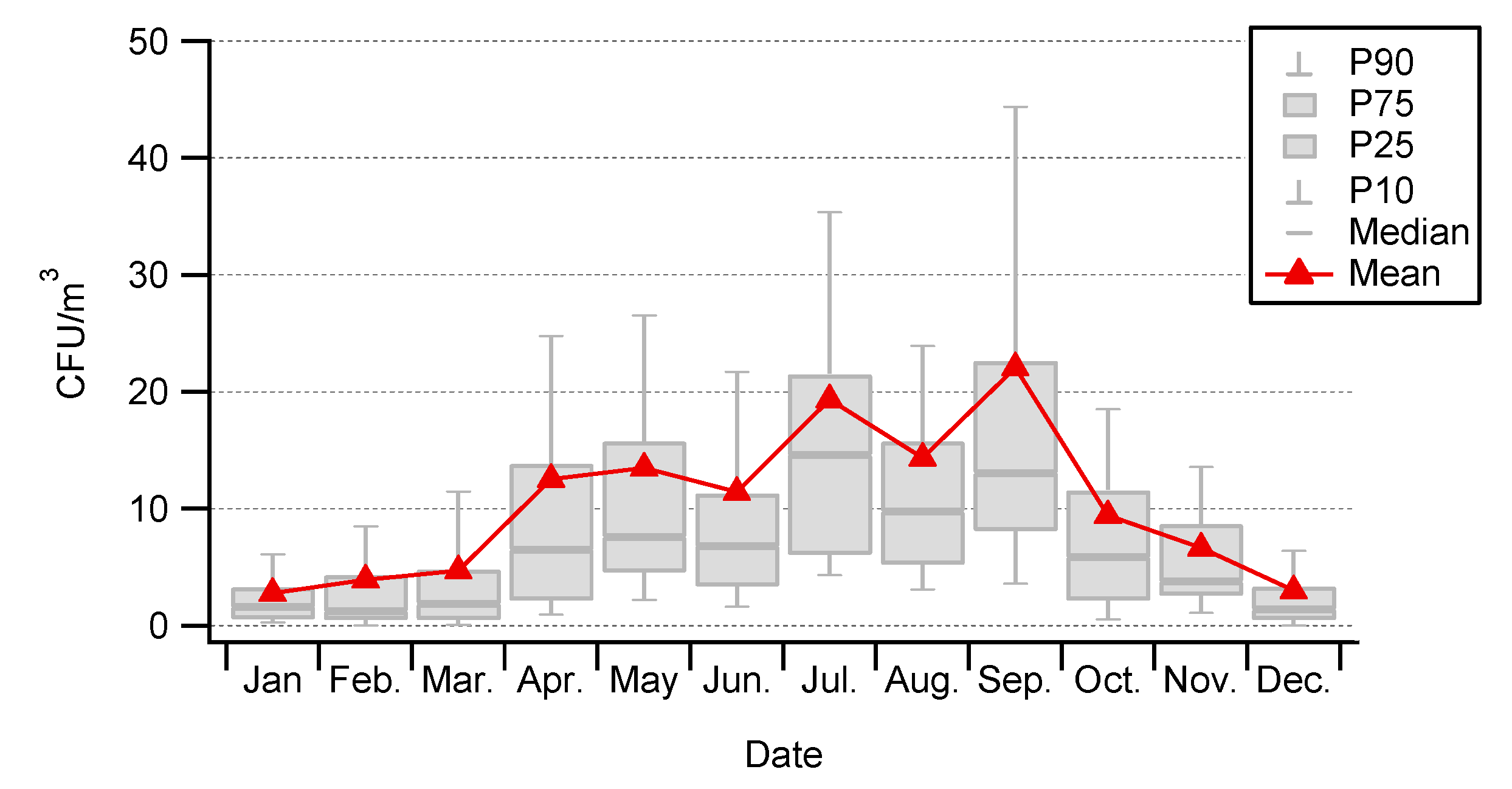
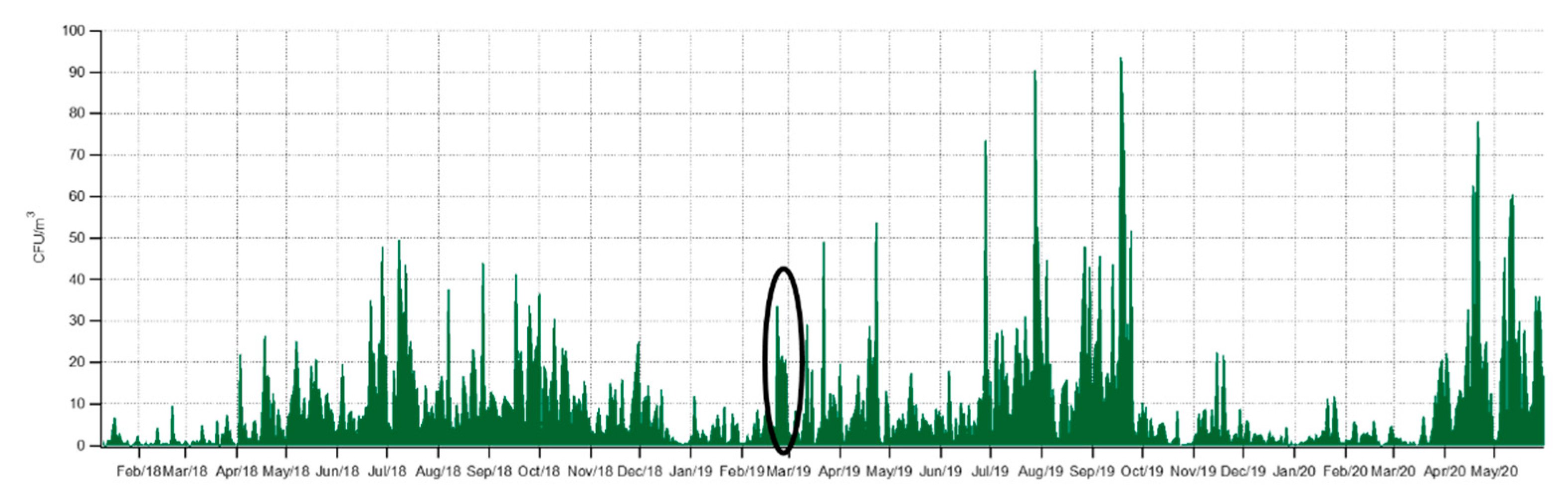
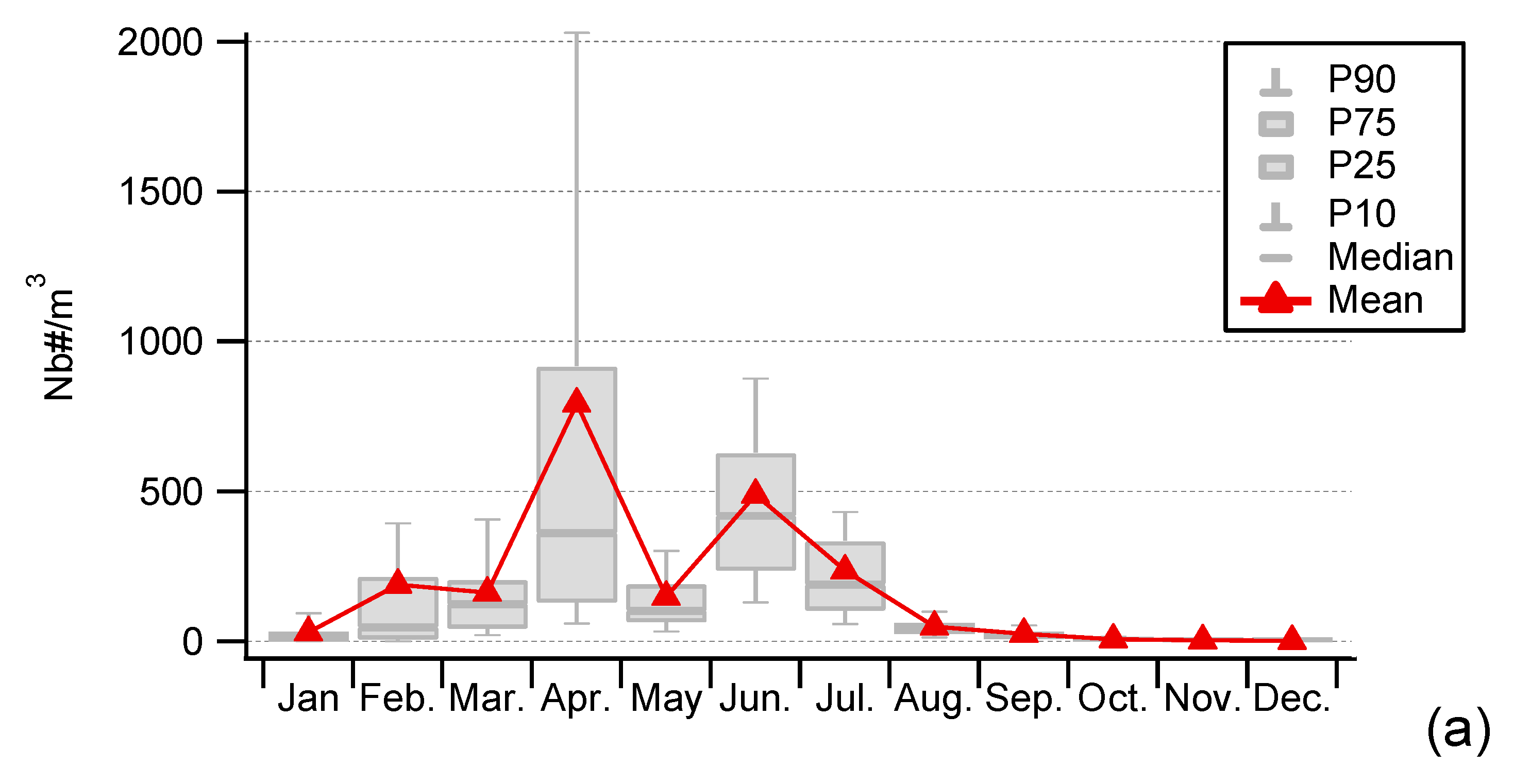
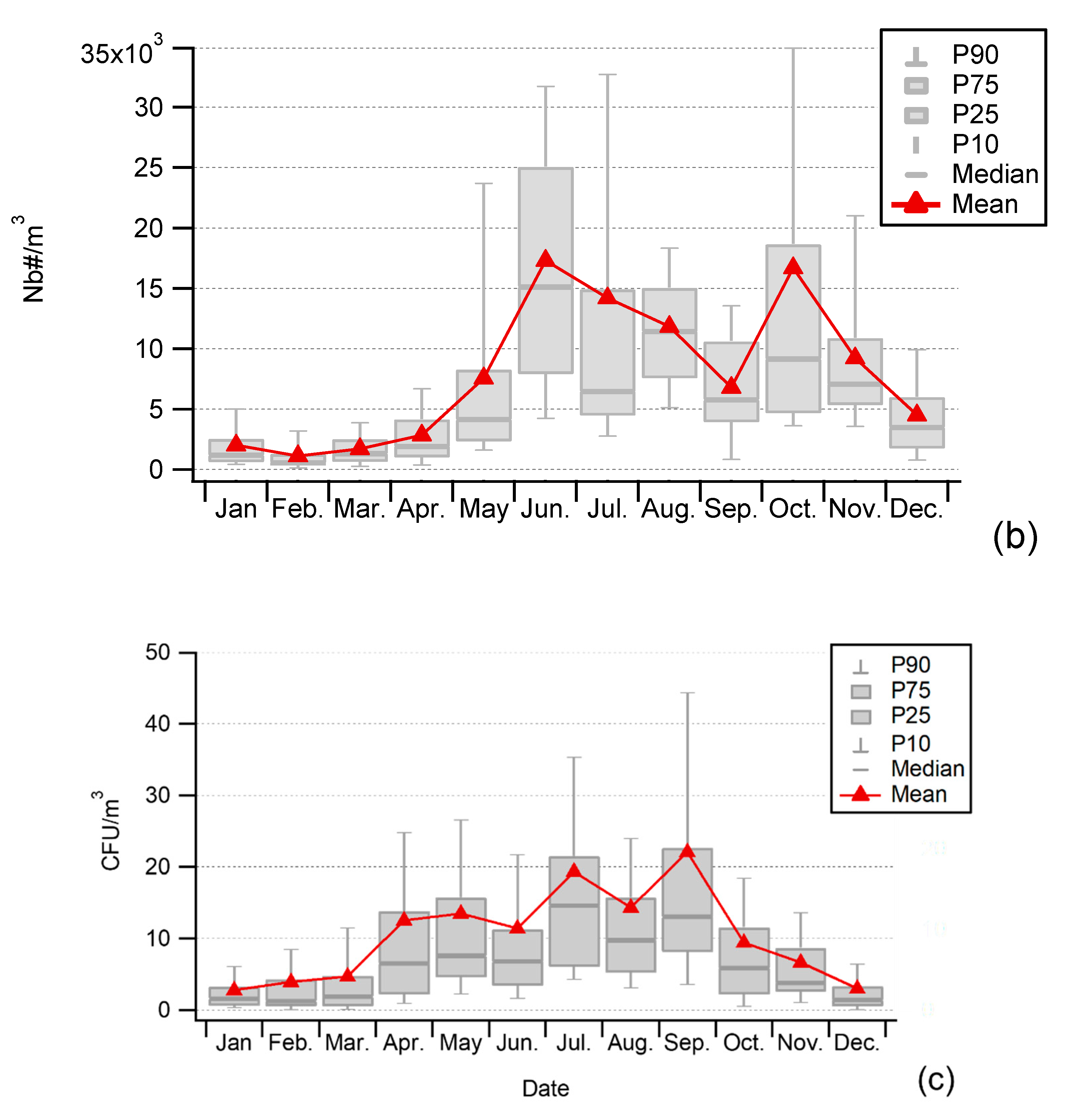
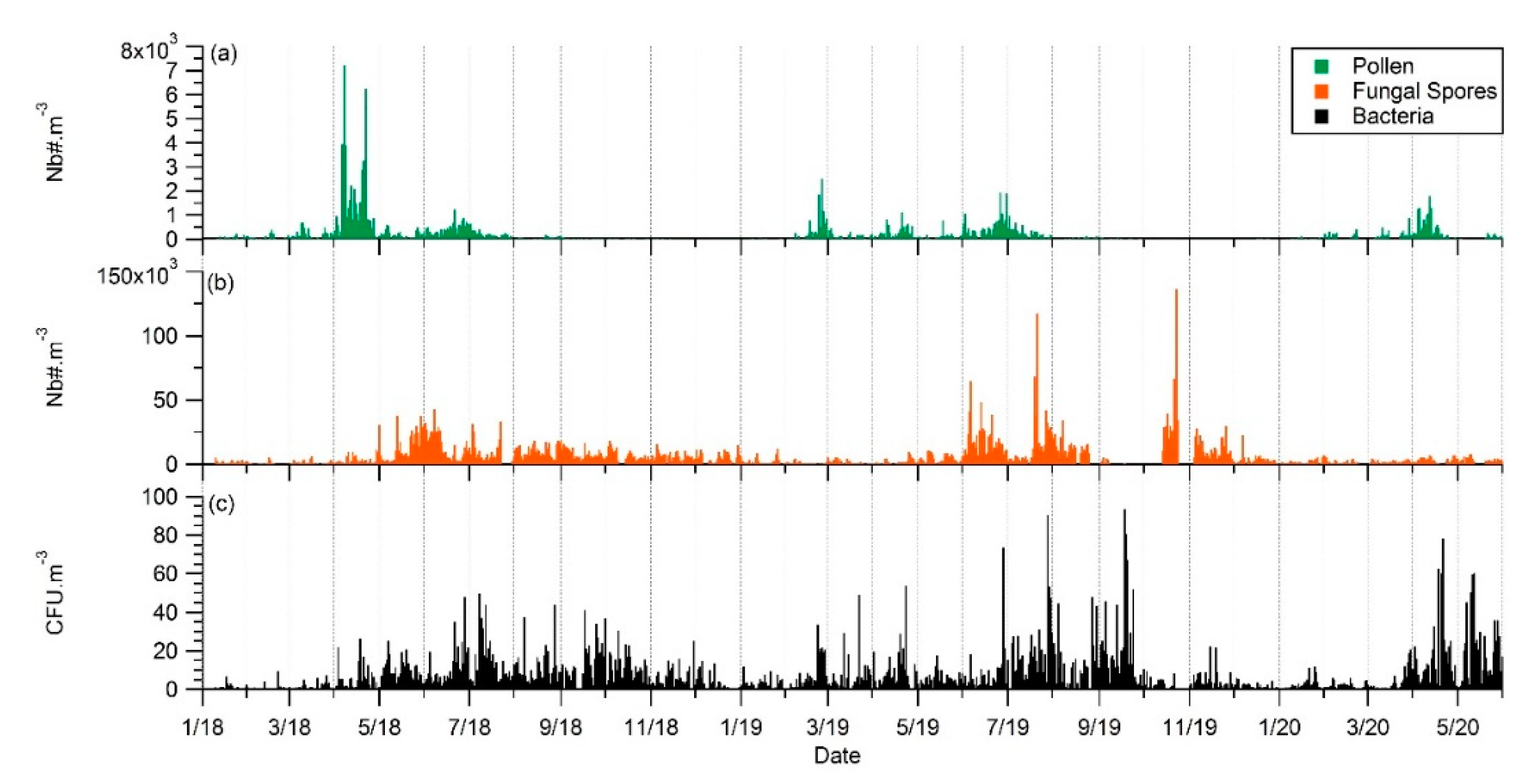
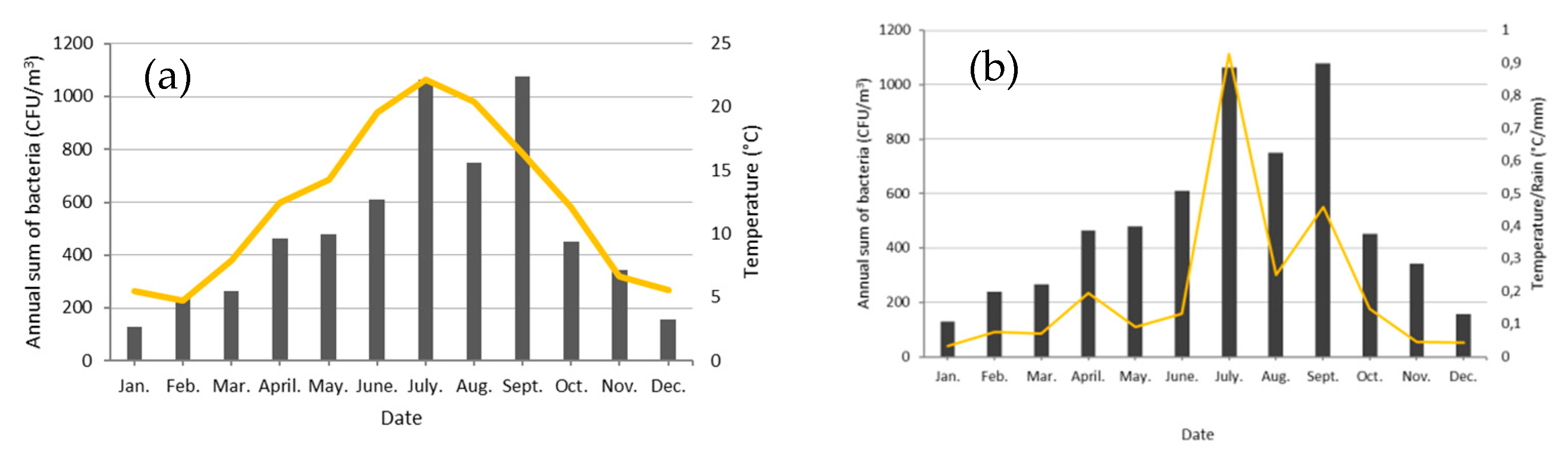
3.1. Seasonality of Cultivable Bacterial Concentrations at Saclay
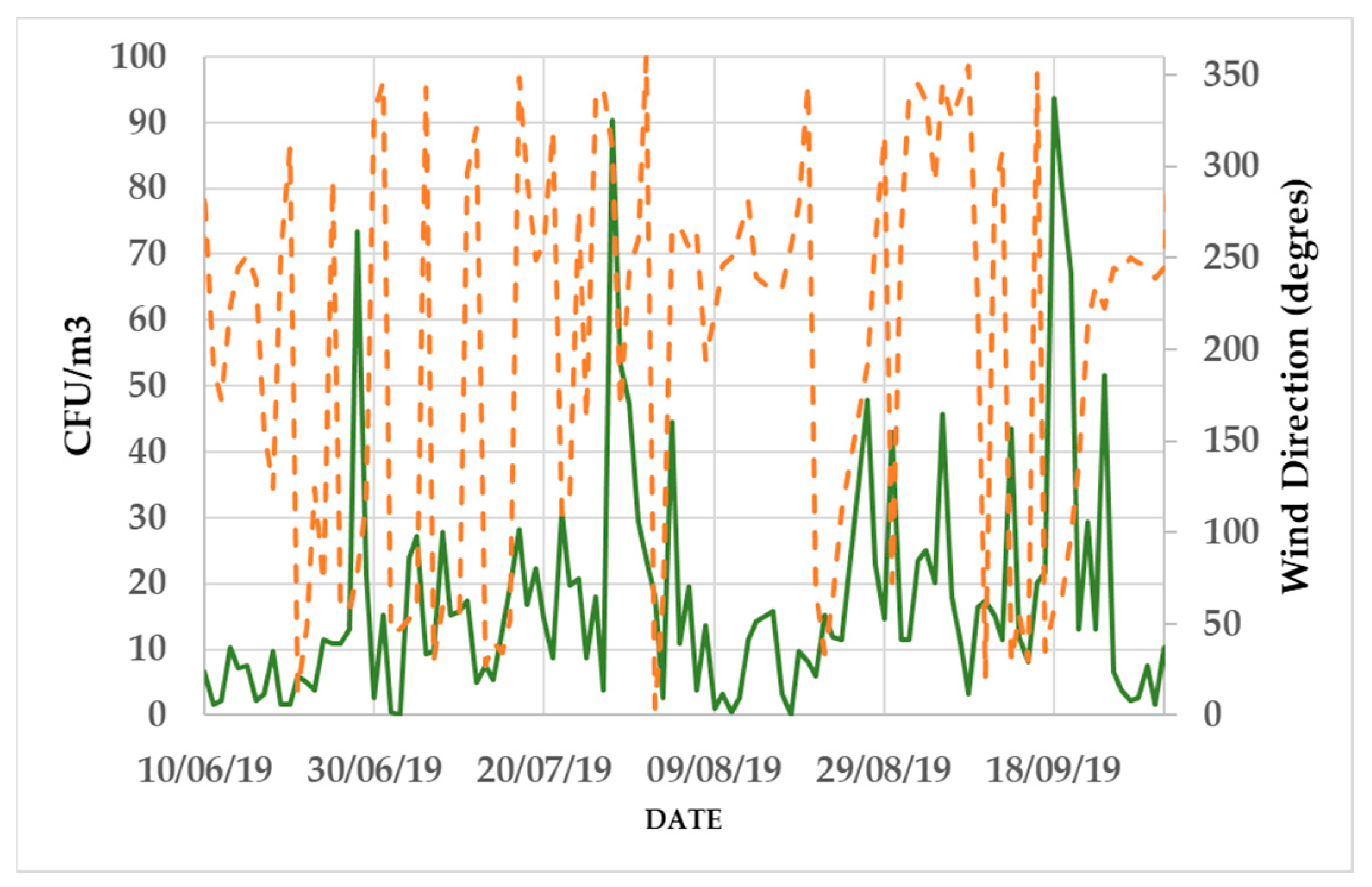
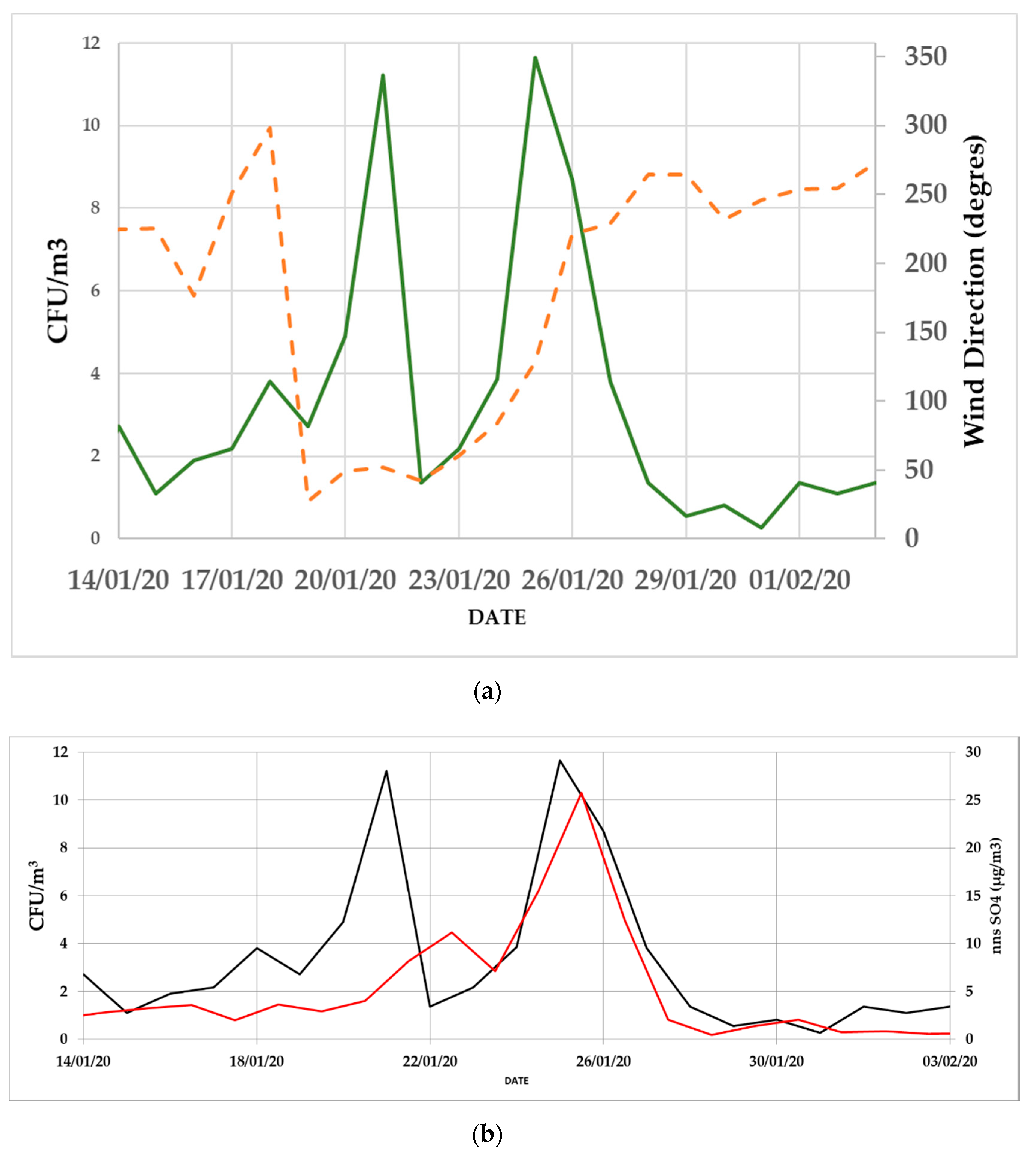
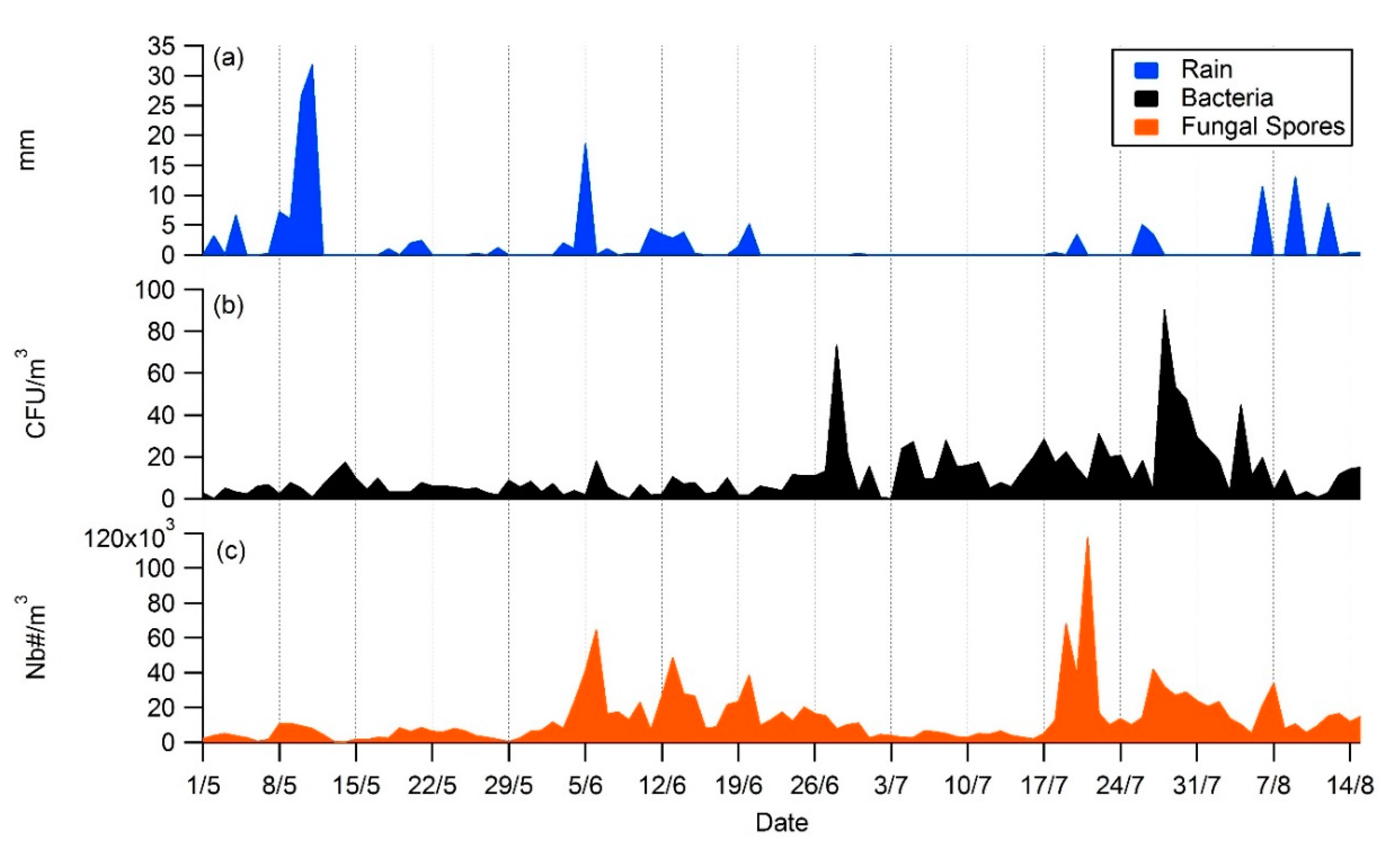
3.2. Daily Variability of Bacterial Concentrations at Saclay
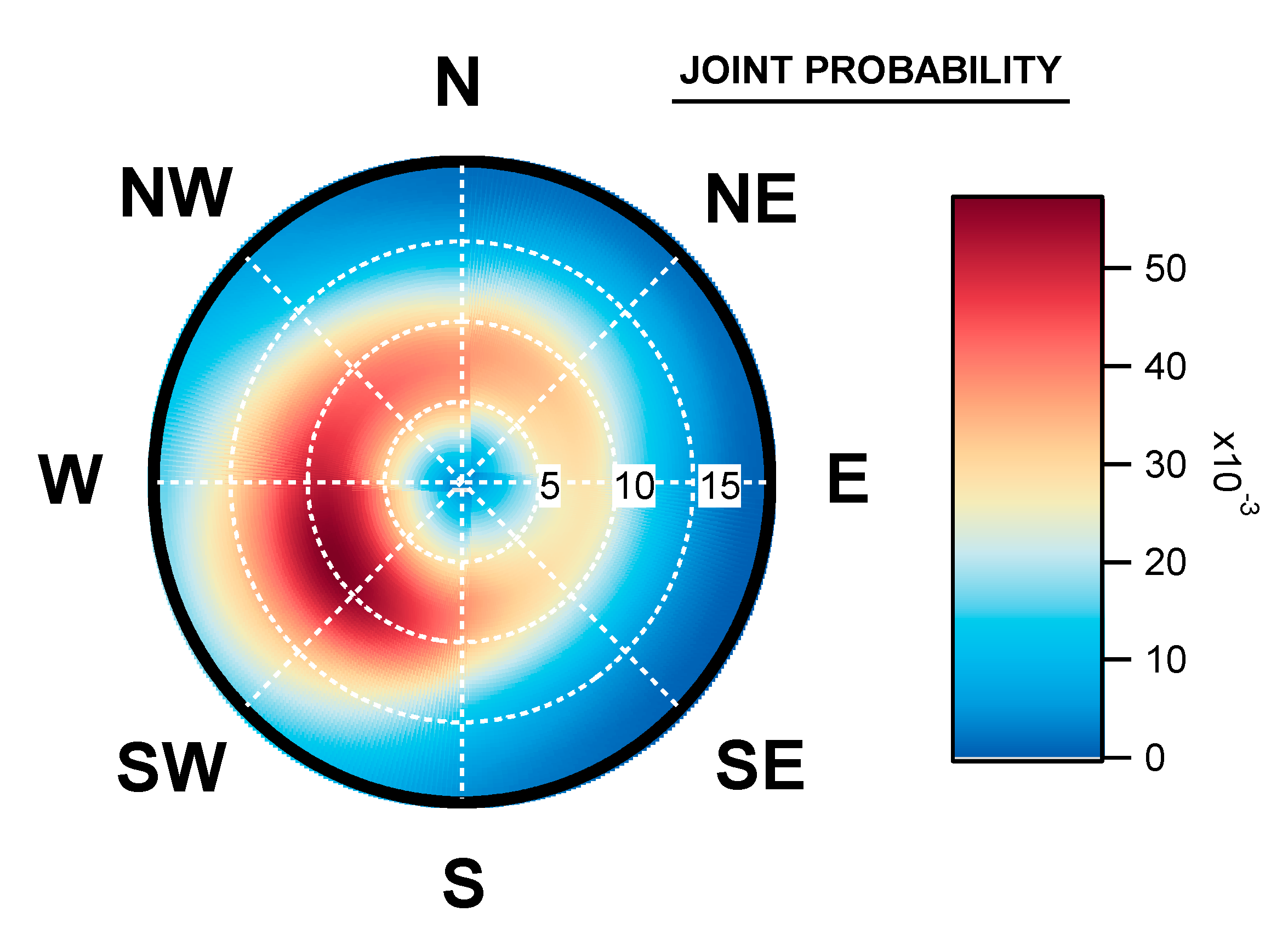
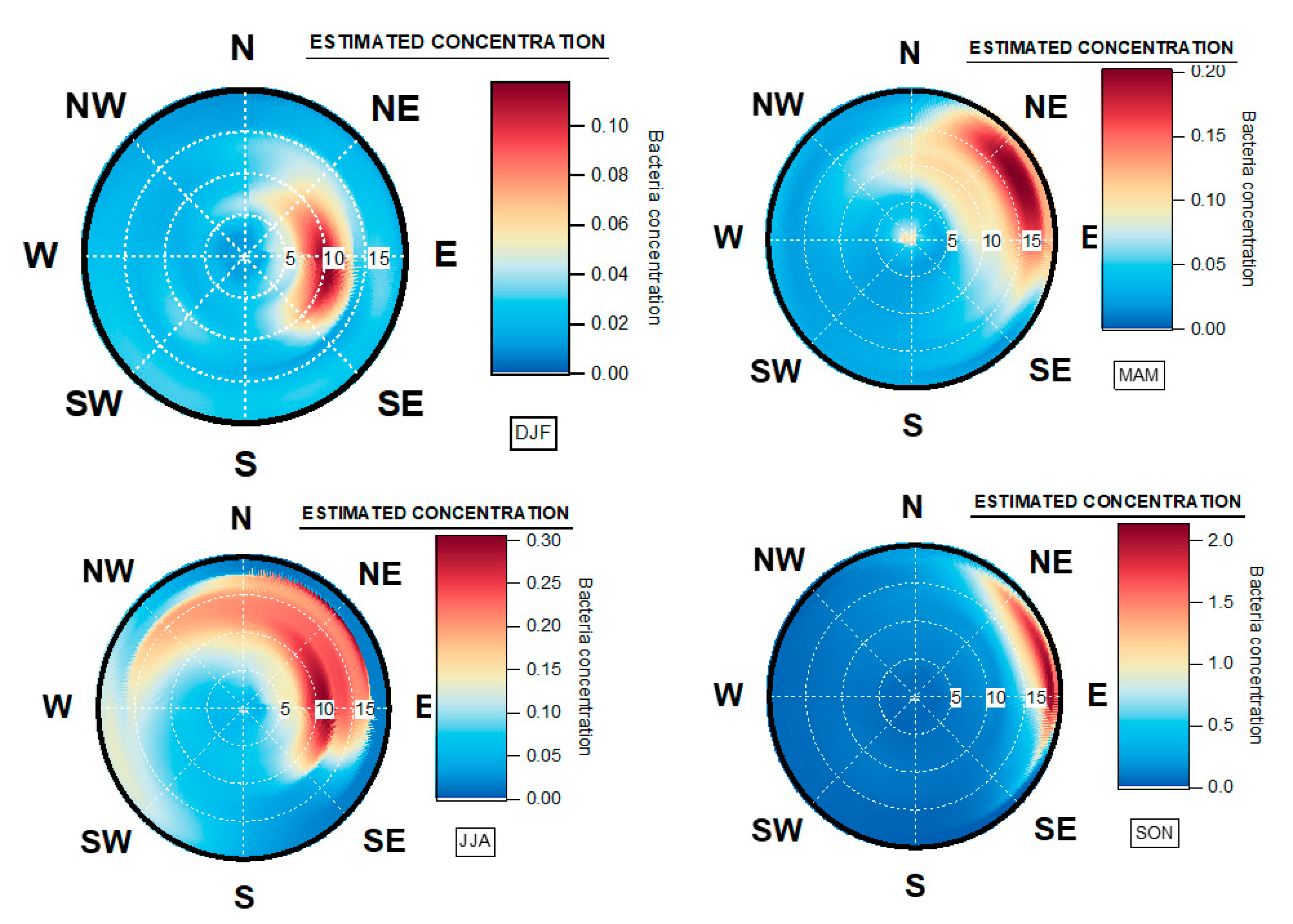
3.3. Geographical Origin of Bacterial Concentrations
4. Discussion
4.1. Seasonality and Comparisons of the Major Primary Bioaerosols Present at Saclay
4.2. Long-Term Observations and Identification of Potential Sources
4.3. Daily Variability, Meteorological Parameters, and Transport
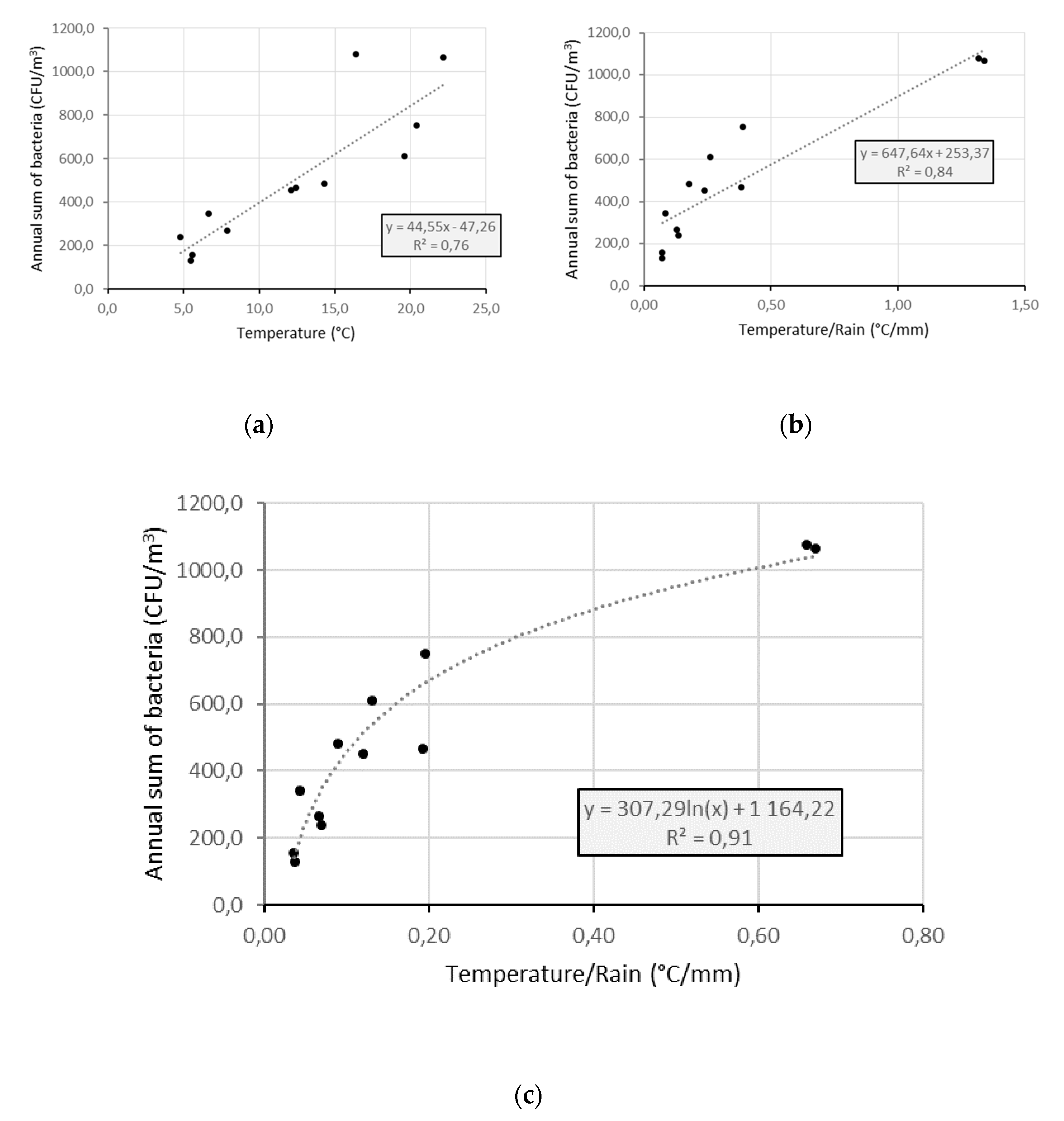
4.4. Factors Controlling the Seasonal Cycle of Airborne Bacterial Concentrations at Saclay
4.5. Seasonal Geographical Origins of Bacteria Concentrations Measured at Saclay
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Sample | Date | TB | R1 | R2 | R3 | R4 | R5 | TC CFU/m3 | R3 CFU/m3 | LOSSES% |
|---|---|---|---|---|---|---|---|---|---|---|
| N408 | 21 February 19 | 240 | 12,000 | 381,300 | 31,800 | 2400 | 1200 | 4 | 578 | 99.2 |
| N409 | 22 February 19 | 1850 | 34,500 | 553.8 | 46,800 | 450 | 450 | 34 | 851 | 96.0 |
| N410 | 23 February 19 | 580 | 7200 | 219,600 | 21,000 | 3900 | 5280 | 11 | 382 | 97.2 |
| N411 | 24 February 19 | 1150 | 7200 | 276,600 | 30,300 | 3000 | 13,500 | 21 | 551 | 96.2 |
| N412 | 25 February 19 | 1190 | 33,240 | 701,259 | 69,141 | 6321 | 20,760 | 22 | 1257 | 98.3 |
| N413 | 26 February 19 | 760 | 41,940 | 563,820 | 55,839 | 5241 | 3921 | 14 | 1015 | 98.6 |
| N414 | 27 February 19 | 540 | 36,699 | 486,519 | 33,399 | 4761 | 9159 | 10 | 607 | 98.4 |
| N415 | 28 February 19 | 300 | 9339 | 73,860 | 5241 | 1419 | 2520 | 5 | 95 | 94.3 |
| N416 | 1 March 19 | 130 | 2460 | 94,101 | 4059 | 960 | 381 | 2 | 74 | 96.8 |
| N417 | 2 March 19 | 150 | 1461 | 50,061 | 1959 | 1599 | 1599 | 3 | 36 | 92.3 |
| N418 | 3 March 19 | 100 | 8799 | 54,639 | 1821 | 1920 | 2541 | 2 | 33 | 94.5 |
| N419 | 4 March 19 | 200 | 4641 | 30,561 | 1320 | 741 | 921 | 4 | 24 | 84.8 |
| N420 | 5 March 19 | 460 | 4140 | 43,401 | 1980 | 180 | 180 | 8 | 36 | 76.8 |
| N423 | 8 March 19 | 20 | 2001 | 20,739 | 1161 | 381 | 420 | 3 | 21 | 86.2 |
| N424 | 9 March 19 | 110 | 1680 | 31,581 | 2601 | 201 | 159 | 0 | 47 | 99.2 |
| N425 | 10 March 19 | 420 | 6501 | 30,120 | 1311 | 459 | 141 | 2 | 24 | 91.6 |
| Month | Rain (mm) | T (°C) | CFU/m3 | T/Rain (°C/mm) |
|---|---|---|---|---|
| January | 167.1 | 5.5 | 128.8 | 0.07 |
| February | 63.7 | 4.8 | 238.2 | 0.14 |
| March | 113.3 | 7.9 | 265.6 | 0.13 |
| April | 63.8 | 12.5 | 465.2 | 0.38 |
| May | 157.5 | 14.3 | 480.4 | 0.18 |
| June | 148 | 19.6 | 608.9 | 0.26 |
| July | 23.9 | 22.2 | 1064.0 | 1.34 |
| August | 81.7 | 20.4 | 750.8 | 0.39 |
| September | 35.8 | 16.4 | 1077.3 | 1.32 |
| October | 82.6 | 12.1 | 452.1 | 0.24 |
| November | 142.9 | 6.7 | 342.7 | 0.09 |
| December | 133.9 | 5.6 | 155.3 | 0.07 |


References
- Després, V.R.; Huffman, J.A.; Burrows, S.M.; Hoose, C.; Safatov, A.S.; Buryak, G.; Fröhlich-Nowoisky, J.; Elbert, W.; Andreae, M.O.; Pöschl, U.; et al. Primary biological aerosol particles in the atmosphere: A review. Tellus B Chem. Phys. Meteorol. 2012, 64, 15598. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich-Nowoisky, J.; Kampf, C.J.; Weber, B.; Huffman, J.A.; Pöhlker, C.; Andreae, M.O.; Lang-Yona, N.; Burrows, S.M.; Gunthe, S.S.; Elbert, W.; et al. Bioaerosols in the earth system: Climate, health, and ecosystem interactions. Atmos. Res. 2016, 182, 346–376. [Google Scholar] [CrossRef] [Green Version]
- Núñez, A.; Amo de Paz, G.; Rastrojo, A.; García, A.M.; Alcamí, A.; Gutiérrez-Bustillo, A.M.; Moreno, D.A. Monitoring of the airborne biological particles in outdoor atmosphere. Part 1: Importance, variability and ratios. Int. Microbiol. 2016, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Núñez, A.; Amo de Paz, G.; Rastrojo, A.; García, A.M.; Alcamí, A.; Gutiérrez-Bustillo, A.M.; Moreno, D.A. Monitoring of airborne biological particles in outdoor atmosphere. Part 2: Metagenomics applied to urban environments. Int. Microbiol. 2016, 69–80. [Google Scholar] [CrossRef]
- Górny, R.L. Microbial aerosols: Sources, properties, health effects, exposure assessment—A review. Kona Powder Part. J. 2020, 37, 64–84. [Google Scholar] [CrossRef] [Green Version]
- DeMott, P.J.; Prenni, A.J.; Liu, X.; Kreidenweis, S.M.; Petters, M.D.; Twohy, C.H.; Richardson, M.S.; Eidhammer, T.; Rogers, D.C. Predicting global atmospheric ice nuclei distributions and their impacts on climate. Proc. Natl. Acad. Sci. USA 2010, 107, 11217–11222. [Google Scholar] [CrossRef] [Green Version]
- Delort, A.-M.; Vaïtilingom, M.; Amato, P.; Sancelme, M.; Parazols, M.; Mailhot, G.; Laj, P.; Deguillaume, L. A short overview of the microbial population in clouds: Potential roles in atmospheric chemistry and nucleation processes. Atmos. Res. 2010, 98, 249–260. [Google Scholar] [CrossRef]
- Morris, C.E.; Conen, F.; Alex Huffman, J.; Phillips, V.; Pöschl, U.; Sands, D.C. Bioprecipitation: A feedback cycle linking Earth history, ecosystem dynamics and land use through biological ice nucleators in the atmosphere. Glob. Chang. Biol. 2014, 20, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Delort, A.-M.; Amato, P. Microbiology of Aerosols; Delort, A.-M., Amato, P., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2018; ISBN 978-1-119-13229-5. [Google Scholar]
- Douwes, J.; Thorne, P.; Pearce, N.; Heederik, D. Bioaerosol Health Effects and Exposure Assessment: Progress and Prospects. Ann. Occup. Hyg. 2003, 47, 187–200. [Google Scholar] [CrossRef] [Green Version]
- Gao, M.; Jia, R.; Qiu, T.; Han, M.; Song, Y.; Wang, X. Seasonal size distribution of airborne culturable bacteria and fungi and preliminary estimation of their deposition in human lungs during non-haze and haze days. Atmos. Environ. 2015, 118, 203–210. [Google Scholar] [CrossRef]
- Andualem, Z.; Gizaw, Z.; Bogale, L.; Dagne, H. Indoor bacterial load and its correlation to physical indoor air quality parameters in public primary schools. Multidiscip. Respir. Med. 2019, 14, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holty, J.-E.C.; Bravata, D.M.; Liu, H.; Olshen, R.A.; McDonald, K.M.; Owens, D.K. Systematic Review: A century of inhalational anthrax cases from 1900 to 2005. Ann. Intern. Med. 2006, 144, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.U.; Hong, I.G.; Lee, D.H.; Chong, E.-S.; Jung, J.H.; Lee, J.H.; Kim, H.J.; Lee, I.-S. Bacterial bioaerosol concentrations in public restroom environments. Aerosol Air Qual. Res. 2012, 12, 251–255. [Google Scholar] [CrossRef] [Green Version]
- Norbäck, D.; Cai, G.-H. Microbial Agents in the Indoor Environment: Associations with Health. In Indoor Environmental Quality and Health Risk toward Healthier Environment for All; Springer: Berlin, Germany, 2020; pp. 179–198. [Google Scholar]
- Mirhoseini, S.H.; Didehdar, M.; Akbari, M.; Moradzadeh, R.; Jamshidi, R.; Torabi, S. Indoor exposure to airborne bacteria and fungi in sensitive wards of an academic pediatric hospital. Aerobiologia 2020, 36, 225–232. [Google Scholar] [CrossRef]
- Blachowicz, A.; Venkateswaran, K.; Wang, C.C.C. Persistence of Fungi in Atypical, Closed Environments: Cultivation to Omics. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 45, pp. 67–86. [Google Scholar]
- Schnell, R.; Vali, G. Biogenic ice nuclei: Part I. Terrestrial and marine sources. J. Atmos. Sci. 1976, 33, 1554–1564. [Google Scholar] [CrossRef] [Green Version]
- Vali, G.; Christensen, M.; Fresh, R.; Galyan, E.; Maki, L.; Schnell, R. Biogenic ice nuclei. Part II: Bacterial sources. J. Atmos. Sci. 1976, 33, 1565–1570. [Google Scholar] [CrossRef] [Green Version]
- Jones, B.L.; Cookson, J.T. Natural atmospheric microbial conditions in a typical suburban area. Appl. Environ. Microbiol. 1983, 45, 919–934. [Google Scholar] [CrossRef] [Green Version]
- Crane, S.; Moore, J.; Grismer, M.; Miner, J. Bacterial pollution from agricultural sources: A review. Trans. ASAE 1983, 26, 858–866. [Google Scholar] [CrossRef]
- Shaffer, B.T.; Lighthart, B. Survey of culturable airborne bacteria at four diverse locations in Oregon: Urban, rural, forest, and coastal. Microb. Ecol. 1997, 34, 167–177. [Google Scholar] [CrossRef]
- Burrows, S.M.; Elbert, W.; Lawrence, M.G.; Pöschl, U. Bacteria in the global atmosphere—Part 1: Review and synthesis of literature data for different ecosystems. Atmos. Chem. Phys. 2009, 9, 9263–9280. [Google Scholar] [CrossRef] [Green Version]
- Burrows, S.M.; Butler, T.; Jöckel, P.; Tost, H.; Kerkweg, A.; Pöschl, U.; Lawrence, M.G. Bacteria in the global atmosphere—Part 2: Modeling of emissions and transport between different ecosystems. Atmos. Chem. Phys. 2009, 9, 9281–9297. [Google Scholar] [CrossRef] [Green Version]
- Bowers, R.M.; McLetchie, S.; Knight, R.; Fierer, N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 2011, 5, 601–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallawicha, K.; Lung, S.-C.C.; Chuang, Y.-C.; Wu, C.-D.; Chen, T.-H.; Tsai, Y.-J.; Chao, H.J. Spatiotemporal distributions and land-use regression models of ambient bacteria and endotoxins in the greater Taipei area. Aerosol Air Qual. Res. 2015, 15, 1448–1459. [Google Scholar] [CrossRef] [Green Version]
- Tignat-Perrier, R.; Dommergue, A.; Thollot, A.; Magand, O.; Amato, P.; Joly, M.; Sellegri, K.; Vogel, T.M.; Larose, C. Seasonal shift in airborne microbial communities. Sci. Total Environ. 2020, 716, 137129. [Google Scholar] [CrossRef]
- Reche, I.; D’Orta, G.; Mladenov, N.; Winget, D.M.; Suttle, C.A. Deposition rates of viruses and bacteria above the atmospheric boundary layer. ISME J. 2018, 12, 1154–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyler, M.; Shipe, E. Bacterial aerosol samplers: I. Development and evaluation of the all-glass impinger. Appl. Microbiol. 1959, 7, 337–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.S.; Heinsohn, P.A. Sampling and Analysis of Indoor Microorganisms; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Mainelis, G. Collection of airborne mcroorganisms by electrostatic precipitation. Aerosol Sci. Technol. 1999, 30, 127–144. [Google Scholar] [CrossRef]
- Roux, J.M.; Sarda-Estève, R.; Delapierre, G.; Nadal, M.H.; Bossuet, C.; Olmedo, L. Development of a new portable air sampler based on electrostatic precipitation. Environ. Sci. Pollut. Res. 2016, 23, 8175–8183. [Google Scholar] [CrossRef]
- Haddrell, A.E.; Thomas, R.J. Aerobiology: Experimental considerations, observations, and future tools. Appl. Environ. Microbiol. 2017, 83, e00809-17. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, R.M.W.; Garcia-Alcega, S.; Coulon, F.; Dumbrell, A.J.; Whitby, C.; Colbeck, I. Bioaerosol biomonitoring: Sampling optimization for molecular microbial ecology. Mol. Ecol. Resour. 2019, 19, 672–690. [Google Scholar] [CrossRef]
- Fennelly, M.; Sewell, G.; Prentice, M.; O’Connor, D.; Sodeau, J. Review: The use of real-time fluorescence instrumentation to monitor ambient Primary Biological Aerosol Particles (PBAP). Atmosphere 2018, 9, 1. [Google Scholar] [CrossRef] [Green Version]
- Huffman, J.A.; Perring, A.E.; Savage, N.J.; Clot, B.; Crouzy, B.; Tummon, F.; Shoshanim, O.; Damit, B.; Schneider, J.; Sivaprakasam, V. Real-time sensing of bioaerosols: Review and current perspectives. Aerosol Sci. Technol. 2020, 54, 465–495. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.S.; Kim, H.R.; Ko, H.S.; Jeong, S.B.; Kim, B.C.; Jung, J. Continuous surveillance of bioaerosols on-site by an automated bioaerosol monitoring system. ACS Sens. 2020, 5, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cheng, X.; Wu, W.; Wang, Q.; Tong, Z.; Zhang, X.; Deng, D.; Li, Y. Forecasting of bioaerosol concentration by a back propagation neural network model. Sci. Total Environ. 2020, 698, 134315. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Qi, J.; Beibei, E.; Yin, Y.; Gao, D. Concentration, viability and size distribution of bacteria in atmospheric bioaerosols under different types of pollution. Environ. Pollut. 2020, 257, 113485. [Google Scholar] [CrossRef] [PubMed]
- Petit, J.-E.; Favez, O.; Albinet, A.; Canonaco, F. A user-friendly tool for comprehensive evaluation of the geographical origins of atmospheric pollution: Wind and trajectory analyses. Environ. Model. Softw. 2017, 88, 183–187. [Google Scholar] [CrossRef] [Green Version]
- Sarda-Estève, R.; Baisnée, D.; Guinot, B.; Petit, J.-E.; Sodeau, J.; O’Connor, D.; Besancenot, J.-P.; Thibaudon, M.; Gros, V. Temporal variability and geographical origins of airborne pollen grains concentrations from 2015 to 2018 at Saclay, France. Remote Sens. 2018, 10, 1932. [Google Scholar] [CrossRef] [Green Version]
- Sarda-Estève, R.; Baisnée, D.; Guinot, B.; Sodeau, J.; O’Connor, D.; Belmonte, J.; Besancenot, J.-P.; Petit, J.-E.; Thibaudon, M.; Oliver, G.; et al. Variability and geographical origin of five years airborne fungal spore concentrations measured at Saclay, France from 2014 to 2018. Remote Sens. 2019, 11, 1671. [Google Scholar] [CrossRef] [Green Version]
- Freney, E.; Zhang, Y.; Croteau, P.; Amodeo, T.; Williams, L.; Truong, F.; Petit, J.-E.; Sciare, J.; Sarda-Esteve, R.; Bonnaire, N. The second ACTRIS inter-comparison (2016) for Aerosol Chemical Speciation Monitors (ACSM): Calibration protocols and instrument performance evaluations. Aerosol Sci. Technol. 2019, 53, 830–842. [Google Scholar] [CrossRef]
- Roszak, D.B.; Colwell, R.R. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 1987, 51, 365–379. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoefel, D.; Grooby, W.L.; Monis, P.T.; Andrews, S.; Saint, C.P. Enumeration of water-borne bacteria using viability assays and flow cytometry: A comparison to culture-based techniques. J. Microbiol. Methods 2003, 55, 585–597. [Google Scholar] [CrossRef]
- Peccia, J.; Hernandez, M. Incorporating polymerase chain reaction-based identification, population characterization, and quantification of microorganisms into aerosol science: A review. Atmos. Environ. 2006, 40, 3941–3961. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y. Diurnal distribution of total and culturable atmospheric bacteria at a rural site. Aerosol Sci. Technol. 1999, 30, 246–254. [Google Scholar] [CrossRef]
- Tong, Y.; Lighthart, B. The annual bacterial particle concentration and size distribution in the ambient atmosphere in a rural area of the Willamette Valley, Oregon. Aerosol Sci. Technol. 2000, 32, 393–403. [Google Scholar] [CrossRef]
- Gandolfi, I.; Bertolini, V.; Bestetti, G.; Ambrosini, R.; Innocente, E.; Rampazzo, G.; Papacchini, M.; Franzetti, A. Spatio-temporal variability of airborne bacterial communities and their correlation with particulate matter chemical composition across two urban areas. Appl. Microbiol. Biotechnol. 2015, 99, 4867–4877. [Google Scholar] [CrossRef] [PubMed]
- Cahill, T.A.; Ashbaugh, L.L.; Barone, J.B.; Eldred, R.A.; Feeney, P.J.; Flocchini, R.G.; Goodart, C.; Shadoan, D.J.; Wolfe, G.W. Analysis of respirable fractions in atmospheric particulates via sequential filtration. J. Air Pollut. Control Assoc. 1977, 27, 675–678. [Google Scholar] [CrossRef] [Green Version]
- Zhen, H.; Han, T.; Fennell, D.E.; Mainelis, G. Release of Free DNA by Membrane-Impaired Bacterial Aerosols Due to Aerosolization and Air Sampling. Appl. Environ. Microbiol. 2013, 79, 7780–7789. [Google Scholar] [CrossRef] [Green Version]
- John, W.; Hering, S.; Reischl, G.; Sasaki, G.; Goren, S. Characteristics of nuclepore filters with large pore size—II. Filtration properties. Atmos. Environ. 1983, 17, 373–382. [Google Scholar] [CrossRef]
- Sciare, J.; Oikonomou, K.; Cachier, H.; Mihalopoulos, N.; Andreae, M.O.; Maenhaut, W.; Sarda-Estève, R. Aerosol mass closure and reconstruction of the light scattering coefficient over the Eastern Mediterranean Sea during the MINOS campaign. Atmos. Chem. Phys. 2005, 5, 2253–2265. [Google Scholar] [CrossRef] [Green Version]
- Stetzenbach, L.D.; Buttner, M.P.; Cruz, P. Detection and enumeration of airborne biocontaminants. Curr. Opin. Biotechnol. 2004, 15, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Marchand, G.; Barette, M.-C.; Lesage, J. Dénombrement des Bactéries et Moisissures Viables de l’air Prélevées sur Filtre de Polycarbonate. Available online: https://www.irsst.qc.ca/publications-et-outils/publication/i/100372/n/denombrement-des-bacteries-et-moisissures-viables-de-l-air-prelevees-sur-cassette-de-polycarbonate-methode-ma-368 (accessed on 30 January 2020).
- Baisnee, D.; Sarda-Esteve, R.; Fenaille, F.; Le, M.T.; Sciare, J.; Stavroulas, I.; Thibaudon, M.; Peyraut, M.; Bossuet, C. Fine Variability of Pollens, Fungal Spores and Bacteria during Spring 2016 in Cyprus Island. In Proceedings of the 35th Annual Conference of American Association for Aerosol Research(AAAR), Oregon Convention Center, OR, USA, 16–21 October 2016. [Google Scholar]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Etcheverry, S.; Etcheverry, S.; Faridi, A.; Ramachandraiah, H.; Kumar, T.; Margulis, W.; Laurell, F.; Russom, A. High performance micro-flow cytometer based on optical fibres. Sci. Rep. 2017, 7, 5628. [Google Scholar] [CrossRef] [Green Version]
- Clarke, R.G.; Pinder, A.C. Improved detection of bacteria by flow cytometry using a combination of antibody and viability markers. J. Appl. Microbiol. 1998, 84, 577–584. [Google Scholar] [CrossRef]
- Berney, M.; Hammes, F.; Bosshard, F.; Weilenmann, H.-U.; Egli, T. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 2007, 73, 3283–3290. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.M.T.; Pyle, B.H.; Camper, A.K. Specific and rapid enumeration of viable but nonculturable and viable-culturable gram-negative bacteria by using flow cytometry. Appl. Environ. Microbiol. 2010, 76, 5088–5096. [Google Scholar] [CrossRef] [Green Version]
- Lighthart, B. The ecology of bacteria in the alfresco atmosphere. Fems Microbiol. Ecol. 1997, 23, 263–274. [Google Scholar] [CrossRef]
- Negron, A.; DeLeon-Rodriguez, N.; Waters, S.M.; Ziemba, L.D.; Anderson, B.; Bergin, M.; Konstantinidis, K.T.; Nenes, A. Using flow cytometry and light-induced fluorescence to characterize the variability and characteristics of bioaerosols in springtime in Metro Atlanta, Georgia. Atmos. Chem. Phys. 2020, 20, 1817–1838. [Google Scholar] [CrossRef] [Green Version]
- Lighthart, B.; Shaffer, B.T. Viable bacterial aerosol particle size distributions in the midsummer atmosphere at an isolated location in the high desert chaparral. Aerobiologia 1995, 11, 19–25. [Google Scholar] [CrossRef]
- King, M.D.; McFarland, A.R. Use of an Andersen bioaerosol sampler to simultaneously provide culturable particle and culturable organism size distributions. Aerosol Sci. Technol. 2012, 46, 852–861. [Google Scholar] [CrossRef]
- Henry, R.; Norris, G.A.; Vedantham, R.; Turner, J.R. Source region identification using kernel smoothing. Environ. Sci. Technol. 2009, 43, 4090–4097. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.A.; Vedantham, R.; Norris, G.A.; Brown, S.G.; Roberts, P. Determining source impacts near roadways using wind regression and organic source markers. Atmos. Environ. 2012, 47, 261–268. [Google Scholar] [CrossRef]
- Yamartino, R.J. A comparison of several “single-pass” estimators of the standard deviation of wind direction. J. Clim. Appl. Meteorol. 1984, 23, 1362–1366. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.U.; Lee, G.; Heo, K.J. Concentration of culturable bioaerosols during winter. J. Aerosol Sci. 2016, 94, 1–8. [Google Scholar] [CrossRef]
- Galán, C.; Ariatti, A.; Bonini, M.; Clot, B.; Crouzy, B.; Dahl, A.; Fernandez-González, D.; Frenguelli, G.; Gehrig, R.; Isard, S.; et al. Recommended terminology for aerobiological studies. Aerobiologia 2017, 33, 293–295. [Google Scholar] [CrossRef]
- Elbert, W.; Taylor, P.E.; Andreae, M.O.; Pöschl, U. Contribution of fungi to primary biogenic aerosols in the atmosphere: Wet and dry discharged spores, carbohydrates, and inorganic ions. Atmos. Chem. Phys. 2007, 7, 4569–4588. [Google Scholar] [CrossRef] [Green Version]
- Spracklen, D.V.; Heald, C.L. The contribution of fungal spores and bacteria to regional and global aerosol number and ice nucleation immersion freezing rates. Atmos. Chem. Phys. 2014, 14, 9051–9059. [Google Scholar] [CrossRef] [Green Version]
- Lindemann, J.; Upper, C.D. Aerial dispersal of epiphytic bacteria over bean plants. Appl. Environ. Microbiol. 1985, 50, 1229–1232. [Google Scholar] [CrossRef] [Green Version]
- Lindow, S.E.; Brandl, M.T. Microbiology of the Phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef] [Green Version]
- Kellogg, C.A.; Griffin, D.W. Aerobiology and the global transport of desert dust. Trends Ecol. Evol. 2006, 21, 638–644. [Google Scholar] [CrossRef]
- Núñez, A.; Moreno, D.A. The differential vertical distribution of the airborne biological particles reveals an atmospheric reservoir of microbial pathogens and aeroallergens. Microb. Ecol. 2020, 80, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, V.; Gandolfi, I.; Ambrosini, R.; Bestetti, G.; Innocente, E.; Rampazzo, G.; Franzetti, A. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl. Microbiol. Biotechnol. 2013, 97, 6561–6570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Ahn, J.; Liu, D.; Chen, S.; Ye, X.; Ding, T. Evaluation of ultrasound-induced dmage to Escherichia coli and Staphylococcus aureus by flow cytometry and transmission electron microscopy. Appl. Environ. Microbiol. 2016, 82, 1828–1837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cáliz, J.; Triadó-Margarit, X.; Camarero, L.; Casamayor, E.O. A long-term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc. Natl. Acad. Sci. USA 2018, 115, 12229–12234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horner-Devine, M.C.; Carney, K.M.; Bohannan, B.J.M. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. B Biol. Sci. 2004, 271, 113–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhamme, D.T.; Prosser, J.I.; Nicol, G.W. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. Isme J. 2011, 5, 1067–1071. [Google Scholar] [CrossRef] [Green Version]
- Smets, W.; Moretti, S.; Denys, S.; Lebeer, S. Airborne bacteria in the atmosphere: Presence, purpose, and potential. Atmos. Environ. 2016, 139, 214–221. [Google Scholar] [CrossRef]
- Jones, A.M.; Harrison, R.M. The effects of meteorological factors on atmospheric bioaerosol concentrations—A review. Sci. Total Environ. 2004, 326, 151–180. [Google Scholar] [CrossRef]
- Huffman, J.A.; Prenni, A.J.; DeMott, P.J.; Pöhlker, C.; Mason, R.H.; Robinson, N.H.; Fröhlich-Nowoisky, J.; Tobo, Y.; Després, V.R.; Garcia, E. High concentrations of biological aerosol particles and ice nuclei during and after rain. Atmos. Chem. Phys. 2013, 13, 6151–6164. [Google Scholar] [CrossRef] [Green Version]
- Uetake, J.; Tobo, Y.; Uji, Y.; Hill, T.C.J.; DeMott, P.J.; Kreidenweis, S.M.; Misumi, R. Seasonal changes of airborne bacterial communities over Tokyo and influence of local meteorology. Front. Microbiol. 2019, 10, 1572. [Google Scholar] [CrossRef] [Green Version]
- Butterworth, J.; McCartney, H.A. The dispersal of bacteria from leaf surfaces by water splash. J. Appl. Bacteriol. 1991, 71, 484–496. [Google Scholar] [CrossRef]
- Gao, M.; Yan, X.; Qiu, T.; Han, M.; Wang, X. Variation of correlations between factors and culturable airborne bacteria and fungi. Atmos. Environ. 2016, 128, 10–19. [Google Scholar] [CrossRef]
- Li, Y.; Lu, R.; Li, W.; Xie, Z.; Song, Y. Concentrations and size distributions of viable bioaerosols under various weather conditions in a typical semi-arid city of Northwest China. J. Aerosol Sci. 2017, 106, 83–92. [Google Scholar] [CrossRef]
- Walklate, P.; McCartney, H.; Fitt, B.D. Vertical dispersal of plant pathogens by splashing. Part II: Experimental study of the relationship between raindrop size and the maximum splash height. Plant Pathol. 1989, 38, 64–70. [Google Scholar] [CrossRef]
- Lovell, D.J.; Parker, S.R.; Van Peteghem, P.; Webb, D.A.; Welham, S.J. Quantification of Raindrop Kinetic Energy for Improved Prediction of Splash-Dispersed Pathogens. Phytopathology 2002, 92, 497–503. [Google Scholar] [CrossRef]
- Pruvost, O.; Boher, B.; Brocherieux, C.; Nicole, M.; Chiroleu, F. Survival of Xanthomonas axonopodis pv. citri in leaf lesions under tropical environmental conditions and simulated splash dispersal of inoculum. Phytopathology 2002, 92, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Innocente, E.; Squizzato, S.; Visin, F.; Facca, C.; Rampazzo, G.; Bertolini, V.; Gandolfi, I.; Franzetti, A.; Ambrosini, R.; Bestetti, G. Influence of seasonality, air mass origin and particulate matter chemical composition on airborne bacterial community structure in the Po Valley, Italy. Sci. Total Environ. 2017, 593–594, 677–687. [Google Scholar] [CrossRef]
- Wei, M.; Xu, C.; Xu, X.; Zhu, C.; Li, J.; Lv, G. Characteristics of atmospheric bacterial and fungal communities in PM2.5 following biomass burning disturbance in a rural area of North China Plain. Sci. Total Environ. 2019, 651, 2727–2739. [Google Scholar] [CrossRef]
- Bowers, R.M.; Clements, N.; Emerson, J.B.; Wiedinmyer, C.; Hannigan, M.P.; Fierer, N. Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ. Sci. Technol. 2013, 47, 12097–12106. [Google Scholar] [CrossRef]
- Tignat-Perrier, R.; Dommergue, A.; Thollot, A.; Keuschnig, C.; Magand, O.; Vogel, T.M.; Larose, C. Global airborne microbial communities controlled by surrounding landscapes and wind conditions. Sci. Rep. 2019, 9, 14441. [Google Scholar] [CrossRef]
- De Leon-Rodriguez, N. Microbes in the Atmosphere: Prevalence, Species Composition, and Relevance to Cloud Formation—Georgia Institute of Technology August 2015. Available online: https://smartech.gatech.edu/handle/ (accessed on 20 April 2020).
- Zheng, X.-Y.; Tong, L.; Shen, D.; Yu, J.-E.; Hu, Z.-Q.; Li, Y.-J.; Zhang, L.-J.; Xue, E.-F.; Tang, H.-F. Airborne bacteria enriched PM 2.5 enhances the inflammation in an allergic adolescent mouse model induced by Ovalbumin. Inflammation 2020, 43, 32–43. [Google Scholar] [CrossRef]


| Actions | Steps | Function |
|---|---|---|
| Performance Test | 1 | Internal Instrument Verification |
| Sanitize | 2 | Microfluidic System Cleaning |
| 5 µm Filtered Bacteria Sample with SY9 | 3 | Verifications or Adjustment of the PM |
| Sanitize | 4 | Microfluidic System Cleaning |
| Ultra-Pure Water with SY9 | 5 | Identification of the Background |
| Sanitize | 6 | Microfluidic System Cleaning |
| 5 µm Filtered Atmospheric sample with SY9 | 7 | Identification and counting of total bacteria |
| Sanitize | 8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarda-Estève, R.; Baisnée, D.; Guinot, B.; Mainelis, G.; Sodeau, J.; O’Connor, D.; Besancenot, J.P.; Thibaudon, M.; Monteiro, S.; Petit, J.-E.; et al. Atmospheric Biodetection Part I: Study of Airborne Bacterial Concentrations from January 2018 to May 2020 at Saclay, France. Int. J. Environ. Res. Public Health 2020, 17, 6292. https://doi.org/10.3390/ijerph17176292
Sarda-Estève R, Baisnée D, Guinot B, Mainelis G, Sodeau J, O’Connor D, Besancenot JP, Thibaudon M, Monteiro S, Petit J-E, et al. Atmospheric Biodetection Part I: Study of Airborne Bacterial Concentrations from January 2018 to May 2020 at Saclay, France. International Journal of Environmental Research and Public Health. 2020; 17(17):6292. https://doi.org/10.3390/ijerph17176292
Chicago/Turabian StyleSarda-Estève, Roland, Dominique Baisnée, Benjamin Guinot, Gediminas Mainelis, John Sodeau, David O’Connor, Jean Pierre Besancenot, Michel Thibaudon, Sara Monteiro, Jean-Eudes Petit, and et al. 2020. "Atmospheric Biodetection Part I: Study of Airborne Bacterial Concentrations from January 2018 to May 2020 at Saclay, France" International Journal of Environmental Research and Public Health 17, no. 17: 6292. https://doi.org/10.3390/ijerph17176292





