Populus Callus Cell Lines: A Novel Source of Extracellular Vesicles with Nanocarrier Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Callus Induction and Maintenance
2.2. Extracellular Vesicles Extraction
2.3. Transmission Electron Microscopy
2.4. Nanoparticle Track Analysis
2.5. In Vitro Transcription of Fluorescent RNA
2.6. Loading Assays
2.7. Uptake Assays
3. Results
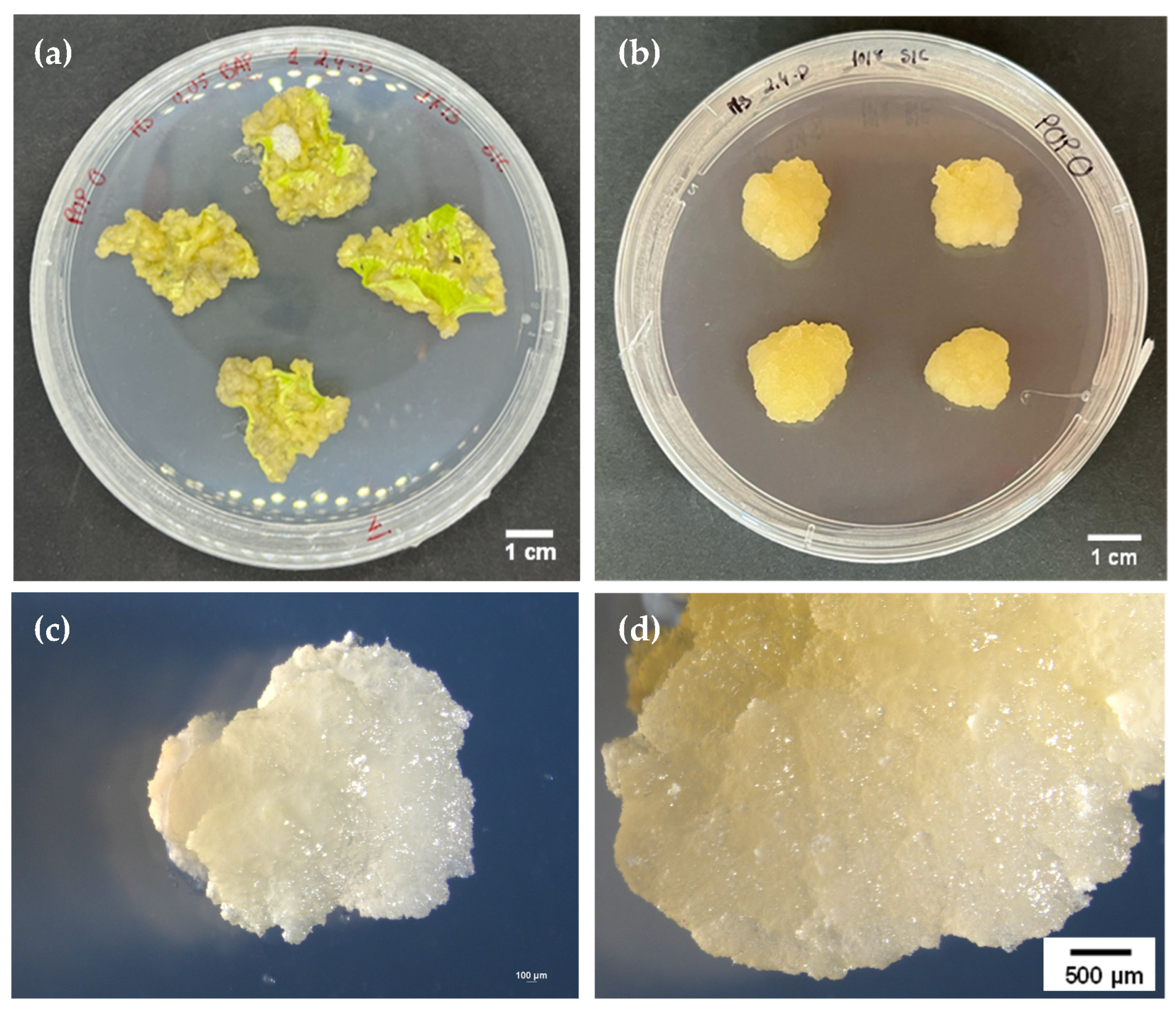
3.1. Callus Induction and Mass Propagation
3.2. EV Isolation and Characterization
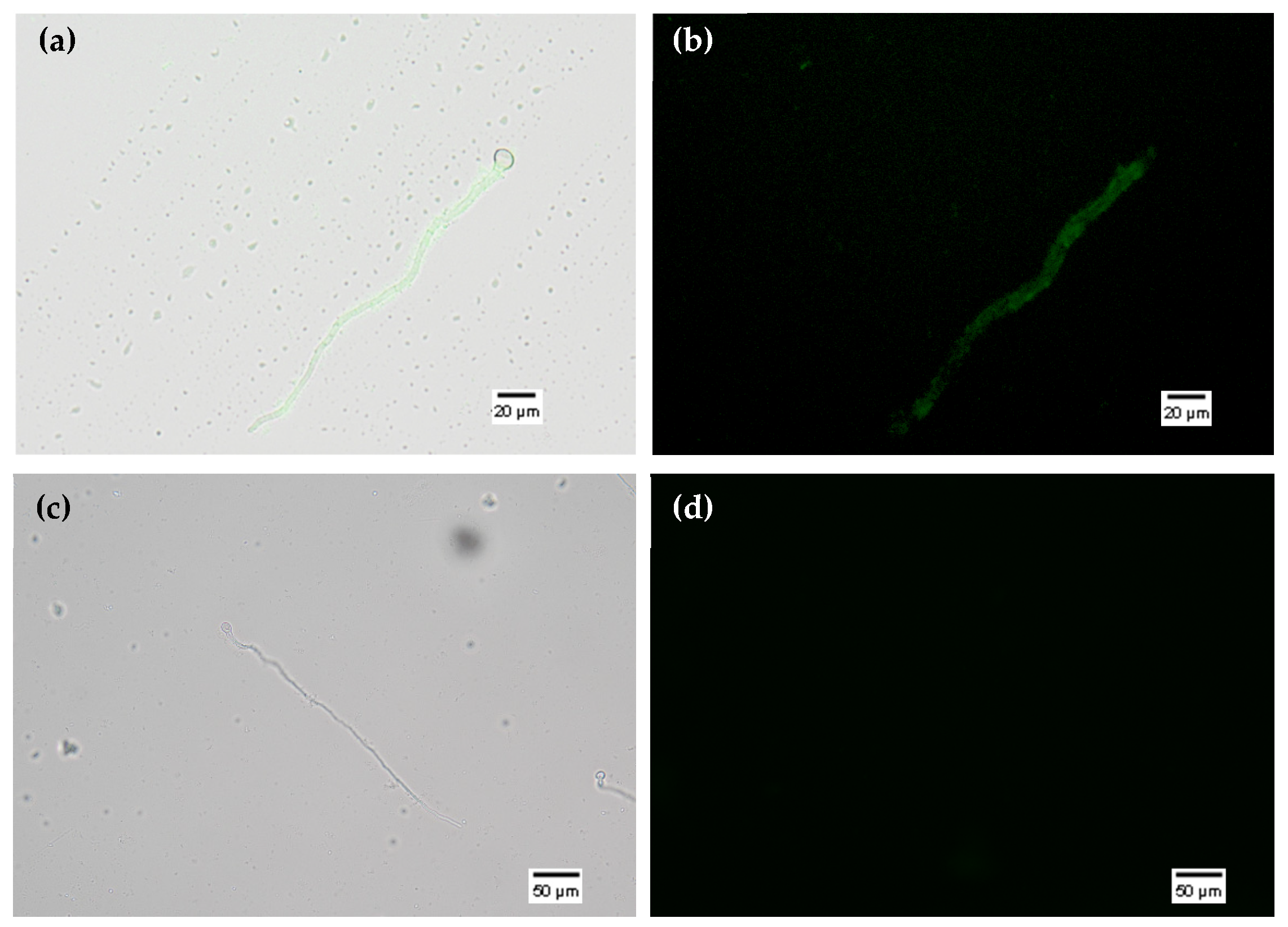
3.3. Loading and Uptake Assays
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2,4-D | 2,4-dichlorophenoxyacetic acid |
| AWF | Apoplastic Washing Fluid |
| BAP | 6-Benzylaminopurine |
| miRNA | micro RNAs |
| MNase | Micrococcal Nuclease |
| NTA | Nanoparticle Track analysis |
| RNAi | RNA interference |
| siRNA | small interfering RNAs |
| TEM | Transmission Electron Microscopy |
| VIB | Vesicle Isolation Buffer |
References
- Efferth, T. Biotechnology applications of plant callus cultures. Engineering 2019, 5, 50–59. [Google Scholar] [CrossRef]
- Long, Y.; Yang, Y.; Pan, G.; Shen, Y. New insights into tissue culture plant-regeneration mechanisms. Front. Plant Sci. 2022, 13, 926752. [Google Scholar] [CrossRef]
- Nic-Can, G.I.; Avilez-Montalvo, J.R.; Aviles-Montalvo, R.N.; Márquez-López, R.E.; Mellado-Mojica, E.; Galaz-Ávalos, R.M.; Loyola-Vargas, V.M. The relationship between stress and somatic embryogenesis. In Somatic Embryogenesis: Fundamental Aspects and Applications, 1st ed.; Loyola-Vargas, V., Ochoa-Alejo, N., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 151–170. [Google Scholar] [CrossRef]
- Wawrosch, C.; Zotchev, S.B. Production of bioactive plant secondary metabolites through in vitro technologies—Status and outlook. Appl. Microbiol. Biotechnol. 2021, 105, 6649–6668. [Google Scholar] [CrossRef]
- Kankaanpää, S.; Väisänen, E.; Goeminne, G.; Soliymani, R.; Desmet, S.; Samoylenko, A.; Vainio, S.; Wingsle, G.; Boerjan, W.; Vanholme, R.; et al. Extracellular vesicles of Norway spruce contain precursors and enzymes for lignin formation and salicylic acid. Plant Physiol. 2024, 196, 788–809. [Google Scholar] [CrossRef] [PubMed]
- Kırbaş, O.K.; Sağraç, D.; Çiftçi, Ö.C.; Özdemir, G.; Öztürkoğlu, D.; Bozkurt, B.T.; Derman, Ü.C.; Taşkan, E.; Taşlı, P.N.; Özdemir, B.S.; et al. Unveiling the potential: Extracellular vesicles from plant cell suspension cultures as a promising source. BioFactors 2025, 51, e2090. [Google Scholar] [CrossRef] [PubMed]
- Park, H.Y.; Kang, M.H.; Lee, G.; Kim, J.W. Enhancement of skin regeneration through activation of TGF-β/SMAD signaling pathway by Panax ginseng meyer non-edible callus-derived extracellular vesicles. J. Ginseng Res. 2024, 49, 34–41. [Google Scholar] [CrossRef]
- Yugay, Y.; Tsydeneshieva, Z.; Rusapetova, T.; Grischenko, O.; Mironova, A.; Bulgakov, D.; Silant’ev, V.; Tchernoded, G.; Bulgakov, V.; Shkryl, Y. Isolation and Characterization of Extracellular Vesicles from Arabidopsis thaliana Cell Culture and Investigation of the Specificities of Their Biogenesis. Plants 2023, 12, 3604. [Google Scholar] [CrossRef]
- Kocholatá, M.; Prusova, M.; Malinska, H.A.; Maly, J.; Janouskova, O. Comparison of two isolation methods of tobacco-derived extracellular vesicles, their characterization and uptake by plant and rat cells. Sci. Rep. 2022, 12, 19896. [Google Scholar] [CrossRef]
- Vader, P.; Mol, E.A.; Pasterkamp, G.; Schiffelers, R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016, 106, 148–156. [Google Scholar] [CrossRef]
- Paganini, C.; Capasso Palmiero, U.; Pocsfalvi, G.; Touzet, N.; Bongiovanni, A.; Arosio, P. Scalable production and isolation of extracellular vesicles: Available sources and lessons from current industrial bioprocesses. Biotechnol. J. 2019, 14, 1800528. [Google Scholar] [CrossRef]
- Giancaterino, S.; Boi, C. Alternative biological sources for extracellular vesicles production and purification strategies for process scale-up. Biotechnol. Adv. 2023, 63, 108092. [Google Scholar] [CrossRef]
- Rome, S. Biological properties of plant-derived extracellular vesicles. Food Funct. 2019, 10, 529–538. [Google Scholar] [CrossRef]
- van Niel, G.; d’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Gao, J.; He, Y.; Jiang, L. Plant extracellular vesicles. Protoplasma 2020, 257, 3–12. [Google Scholar] [CrossRef]
- Lian, M.Q.; Chng, W.H.; Liang, J.; Yeo, H.Q.; Lee, C.K.; Belaid, M.; Tollemeto, M.; Wacker, M.G.; Czarny, B.; Pastorin, G. Plant-derived extracellular vesicles: Recent advancements and current challenges on their use for biomedical applications. J. Extracell. Vesicles 2022, 11, 12283. [Google Scholar] [CrossRef]
- Nemati, M.; Singh, B.; Mir, R.A.; Nemati, M.; Babaei, A.; Ahmadi, M.; Rasmi, Y.; Golezani, A.G.; Rezaie, J. Plant-derived extracellular vesicles: A novel nanomedicine approach with advantages and challenges. Cell Commun. Signal 2022, 20, 69. [Google Scholar] [CrossRef]
- Woith, E.; Guerriero, G.; Hausman, J.F.; Renaut, J.; Leclercq, C.C.; Weise, C.; Legay, S.; Weng, A.; Melzig, M.F. Plant extracellular vesicles and nanovesicles: Focus on secondary metabolites, proteins and lipids with perspectives on their potential and sources. Int. J. Mol. Sci. 2021, 22, 3719. [Google Scholar] [CrossRef]
- Qiang, W.; Li, J.; Ruan, R.; Li, Q.; Zhang, X.; Yan, A.; Zhu, H. Plant-derived extracellular vesicles as a promising anti-tumor approach: A comprehensive assessment of effectiveness, safety, and mechanisms. Phytomedicine 2024, 130, 155750. [Google Scholar] [CrossRef]
- Lykogianni, M.; Bempelou, E.; Karamaouna, F.; Aliferis, K.A. Do pesticides promote or hinder sustainability in agriculture? The challenge of sustainable use of pesticides in modern agriculture. Sci. Total Environ. 2021, 795, 148625. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Hamby, R.; Sanchez, J.N.; Cai, Q.; Yan, Q.; Jin, H. RNAs—A new frontier in crop protection. Curr. Opin. Biotechnol. 2021, 70, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Yang, M.; Le, B.H.; He, W.; Hou, Y. The master role of siRNAs in plant immunity. Mol. Plant Pathol. 2022, 23, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Mishra, S.K.; Sridharan, K.; Barnes, E.R.; Alyokhin, A.; Tuttle, R.; Kokulapalan, W.; Garby, D.; Skizim, N.J.; Tang, Y.W.; et al. First sprayable double-stranded RNA-based biopesticide product targets proteasome subunit beta type-5 in Colorado potato beetle (Leptinotarsa decemlineata). Front. Plant Sci. 2021, 12, 728652. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.H.; Liu, Q.Y.; Xie, Z.M.; Guo, H.S. Exploring the challenges of RNAi-based strategies for crop protection. Adv. Biotechnol. 2024, 2, 23. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, Y.; Xu, J.; Fan, S.; Zhu, N.; Meng, Q.; Dai, S.; Yuan, X. Cross-Kingdom RNA Transport Based on Extracellular Vesicles Provides Innovative Tools for Plant Protection. Plants 2024, 13, 2712. [Google Scholar] [CrossRef] [PubMed]
- Niño-Sánchez, J.; Sambasivam, P.T.; Sawyer, A.; Hamby, R.; Chen, A.; Czislowski, E.; Li, P.; Manzie, N.; Gardiner, D.M.; Ford, R.; et al. BioClay™ prolongs RNA interference-mediated crop protection against Botrytis cinerea. J. Integr. Plant Biol. 2022, 64, 2187–2198. [Google Scholar] [CrossRef]
- Bi, K.; Liang, Y.; Mengiste, T.; Sharon, A. Killing softly: A roadmap of Botrytis cinerea pathogenicity. Trends Plant Sci. 2023, 28, 211–222. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.P.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2017, 173, 728–741. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Hamby, R.; Wang, M.; Qiao, L.; Jin, H. Synthesizing Fluorescently Labeled dsRNAs and sRNAs to Visualize Fungal RNA Uptake. In RNA Tagging: Methods and Protocols, 1st ed.; Heinlein, M., Ed.; Springer: New York, NY, USA, 2020; Volume 2166, pp. 215–225. [Google Scholar] [CrossRef]
- Aranda, P.S.; LaJoie, D.M.; Jorcyk, C.L. Bleach Gel: A Simple Agarose Gel for Analyzing RNA Quality. Electrophoresis 2012, 33, 366–369. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Yang, P.; Zhao, Z.; Virag, A.; Becker, T.; Zhao, L.; Liu, W.; Xia, Y. Botrytis cinerea in vivo Inoculation Assays for Early-, Middle-and Late-stage Strawberries. Bio-protocol 2023, 13, e4859. [Google Scholar] [CrossRef] [PubMed]
- Michler, C.H.; Bauer, E.O. High frequency somatic embryogenesis from leaf tissue of Populus spp. Plant Sci. 1991, 77, 111–118. [Google Scholar] [CrossRef]
- Lelu-Walter, M.A.; Thompson, D.; Harvengt, L.; Sanchez, L.; Toribio, M.; Pâques, L.E. Somatic embryogenesis in forestry with a focus on Europe: State-of-the-art, benefits, challenges and future direction. Tree Genet. Genomes 2013, 9, 883–899. [Google Scholar] [CrossRef]
- Arencibia, A.D.; Gómez, A.; Poblete, M.; Vergara, C. High-performance micropropagation of dendroenergetic poplar hybrids in photomixotrophic Temporary Immersion Bioreactors (TIBs). Ind. Crops Prod. 2017, 96, 102–109. [Google Scholar] [CrossRef]
- Kim, S.; Kim, Y.O.; Lee, Y.; Choi, I.; Joshi, C.P.; Lee, K.; Bae, H.J. The transgenic poplar as an efficient bioreactor system for the production of xylanase. Biosci. Biotechnol. Biochem. 2012, 76, 1140–1145. [Google Scholar] [CrossRef] [PubMed]
- Verdú-Navarro, F.; Moreno-Cid, J.A.; Weiss, J.; Egea-Cortines, M. The advent of plant cells in bioreactors. Front. Plant Sci. 2023, 14, 1310405. [Google Scholar] [CrossRef]
- Martínez, M.; Corredoira, E. Recent Advances in Plant Somatic Embryogenesis: Where We Stand and Where to Go? Int. J. Mol. Sci. 2024, 25, 8912. [Google Scholar] [CrossRef] [PubMed]
- Kocholatá, M.; Malý, J.; Kříženecká, S.; Janoušková, O. Diversity of extracellular vesicles derived from calli, cell culture and apoplastic fluid of tobacco. Sci. Rep. 2024, 14, 30111. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Growing pains: Addressing the pitfalls of plant extracellular vesicle research. New Phytol. 2020, 228, 1505–1510. [Google Scholar] [CrossRef]
- Rikkert, L.G.; Nieuwland, R.; Terstappen, L.W.M.M.; Coumans, F.A.W. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles 2019, 8, 1555419. [Google Scholar] [CrossRef]
- Duan, X.; Chen, L.; Liu, Y.; Chen, H.; Wang, F.; Hu, Y. Integrated physicochemical, hormonal, and transcriptomic analysis reveals the underlying mechanism of callus formation in Pinellia ternata hydroponic cuttings. Front. Plant Sci. 2023, 14, 1189499. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.S.; Ge, X.L.; Wang, R.; Yang, H.F.; Bai, Y.E.; Guo, Y.H.; Zhang, J.; Lu, M.Z.; Zhao, S.T.; Wang, L.Q. An efficient Agrobacterium-mediated transformation method for hybrid poplar 84K (Populus alba × P. glandulosa) using calli as explants. Int. J. Mol. Sci. 2022, 23, 2216. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Hu, H.; Huang, X.; Zhang, Y.; Wang, Y.; Li, Z.; Zou, C.; Peng, H.; Li, L.; Gao, S.; et al. Metabolomic and proteomic analysis of maize embryonic callus induced from immature embryo. Sci. Rep. 2017, 7, 1004. [Google Scholar] [CrossRef]
- Kumari, A.; Ray, K.; Sadhna, S.; Pandey, A.K.; Sreelakshmi, Y.; Sharma, R. Metabolomic homeostasis shifts after callus formation and shoot regeneration in tomato. PLoS ONE 2017, 12, e0176978. [Google Scholar] [CrossRef]
- Amiri, A.; Bagherifar, R.; Ansari Dezfouli, E.; Kiaie, S.H.; Jafari, R.; Ramezani, R. Exosomes as bio-inspired nanocarriers for RNA delivery: Preparation and applications. J. Transl. Med. 2022, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. State-of-the-art exosome loading and functionalization techniques for enhanced therapeutics: A review. Crit. Rev. Biotechnol. 2020, 40, 804–820. [Google Scholar] [CrossRef]
- Nordmeier, S.; Ke, W.; Afonin, K.A.; Portnoy, V. Exosome mediated delivery of functional nucleic acid nanoparticles (NANPs). Nanomedicine 2020, 30, 102285. [Google Scholar] [CrossRef]
- Iqbal, Z.; Rehman, K.; Mahmood, A.; Shabbir, M.; Liang, Y.; Duan, L.; Zeng, H. Exosome for mRNA delivery: Strategies and therapeutic applications. J. Nanobiotechnology 2024, 22, 395. [Google Scholar] [CrossRef] [PubMed]
- de Voogt, W.S.; Tanenbaum, M.E.; Vader, P. Illuminating RNA trafficking and functional delivery by extracellular vesicles. Adv. Drug Deliv. Rev. 2021, 174, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Chen, J.; Zhao, M.; Shen, H.; Jin, Q.; Xiao, D.; Peng, Z.; Chen, T.; Zhang, Y.; Rao, D.; et al. Plant-derived extracellular vesicles: Composition, function and clinical potential. J. Transl. Med. 2025, 23, 1065. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Sequence (5′–3′) |
|---|---|
| TaPDS Frw | TTTGCTCCAGCAGAGGAATGG |
| TaPDS Frw T7pol | TAATACGACTCACTATAGGTTTGCTCCAGCAGAGGAATGG |
| TaPDS Rev | AAACCCTTCGATCGGTGATCG |
| TaPDS Rev T7pol | TAATACGACTCACTATAGGAAACCCTTCGATCGGTGATCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rito, M.; Caeiro, S.; Rosa, P.; Azevedo, C.; Correia, S. Populus Callus Cell Lines: A Novel Source of Extracellular Vesicles with Nanocarrier Potential. Curr. Issues Mol. Biol. 2025, 47, 1015. https://doi.org/10.3390/cimb47121015
Rito M, Caeiro S, Rosa P, Azevedo C, Correia S. Populus Callus Cell Lines: A Novel Source of Extracellular Vesicles with Nanocarrier Potential. Current Issues in Molecular Biology. 2025; 47(12):1015. https://doi.org/10.3390/cimb47121015
Chicago/Turabian StyleRito, Miguel, Sandra Caeiro, Pedro Rosa, Cristina Azevedo, and Sandra Correia. 2025. "Populus Callus Cell Lines: A Novel Source of Extracellular Vesicles with Nanocarrier Potential" Current Issues in Molecular Biology 47, no. 12: 1015. https://doi.org/10.3390/cimb47121015
APA StyleRito, M., Caeiro, S., Rosa, P., Azevedo, C., & Correia, S. (2025). Populus Callus Cell Lines: A Novel Source of Extracellular Vesicles with Nanocarrier Potential. Current Issues in Molecular Biology, 47(12), 1015. https://doi.org/10.3390/cimb47121015






