Investigating the Association Between Citrus Huanglongbing and Chlorophyll Content Using Hyperspectral Detection
Abstract
1. Introduction
- (1)
- The relationships between chlorophyll content and hyperspectral responses in healthy and HLB-infected citrus leaves will be compared and analyzed;
- (2)
- Optimal feature selection methods will be explored, spectral band differences between healthy and HLB-infected leaves will be compared, the predictive performance of feature bands will be evaluated, and optimal chlorophyll content estimation models for healthy and HLB-infected leaves will be established;
- (3)
- The coupling mechanism between HLB infection and changes in chlorophyll content in leaves will be investigated via a comparison of LCC estimation models for healthy and HLB-infected leaves.
2. Materials and Methods
2.1. Study Area
2.2. Hyperspectral Data Acquisition and Spectral Preprocessing
2.3. Chlorophyll Acquisition and TR-PCDR Detection
2.4. Exploring the Correlations Between HLB Citrus Leaves and Chlorophyll Contents Using Hyperspectral Data
2.4.1. Comparative Analysis of the Chlorophyll and Hyperspectral Responses for Healthy and HLB-Infected Citrus Leaves
2.4.2. Comparative Analysis of the Feature Bands Between Healthy and HLB-Infected Leaves
2.4.3. Comparison of Models for Estimating Chlorophyll Contents in Healthy and HLB-Infected Citrus Leaves
3. Results and Analysis
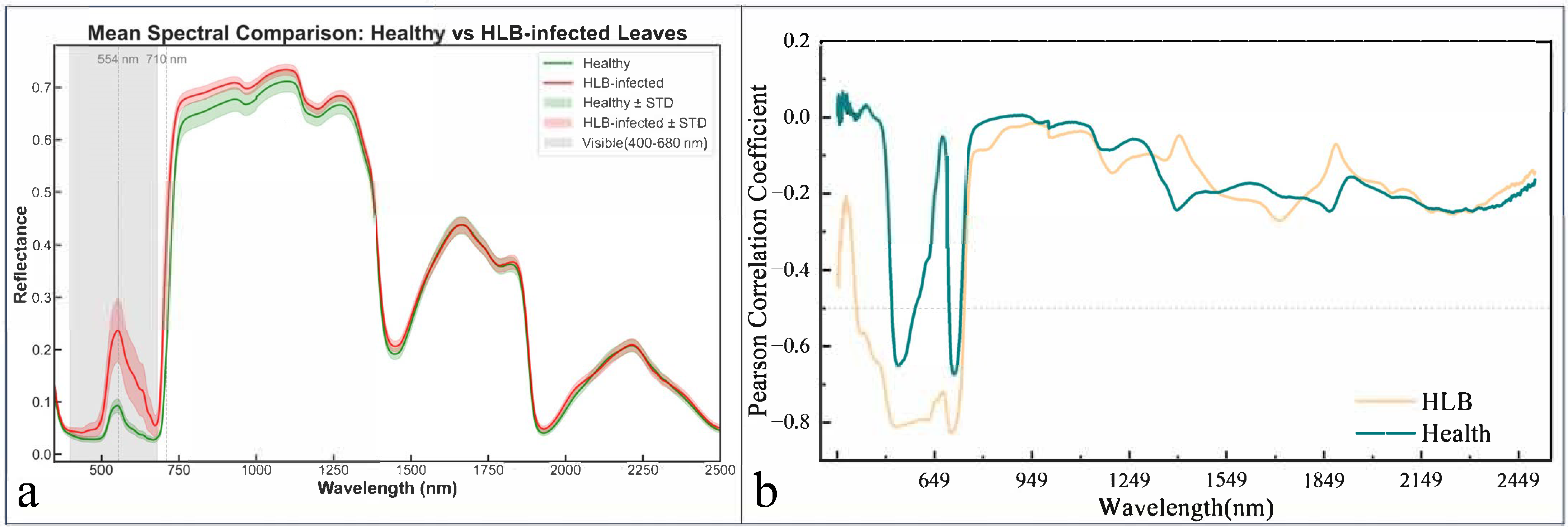
3.1. Comparative Analysis of the Chlorophyll and Hyperspectral Responses for Healthy and HLB-Infected Mianju Citrus Leaves
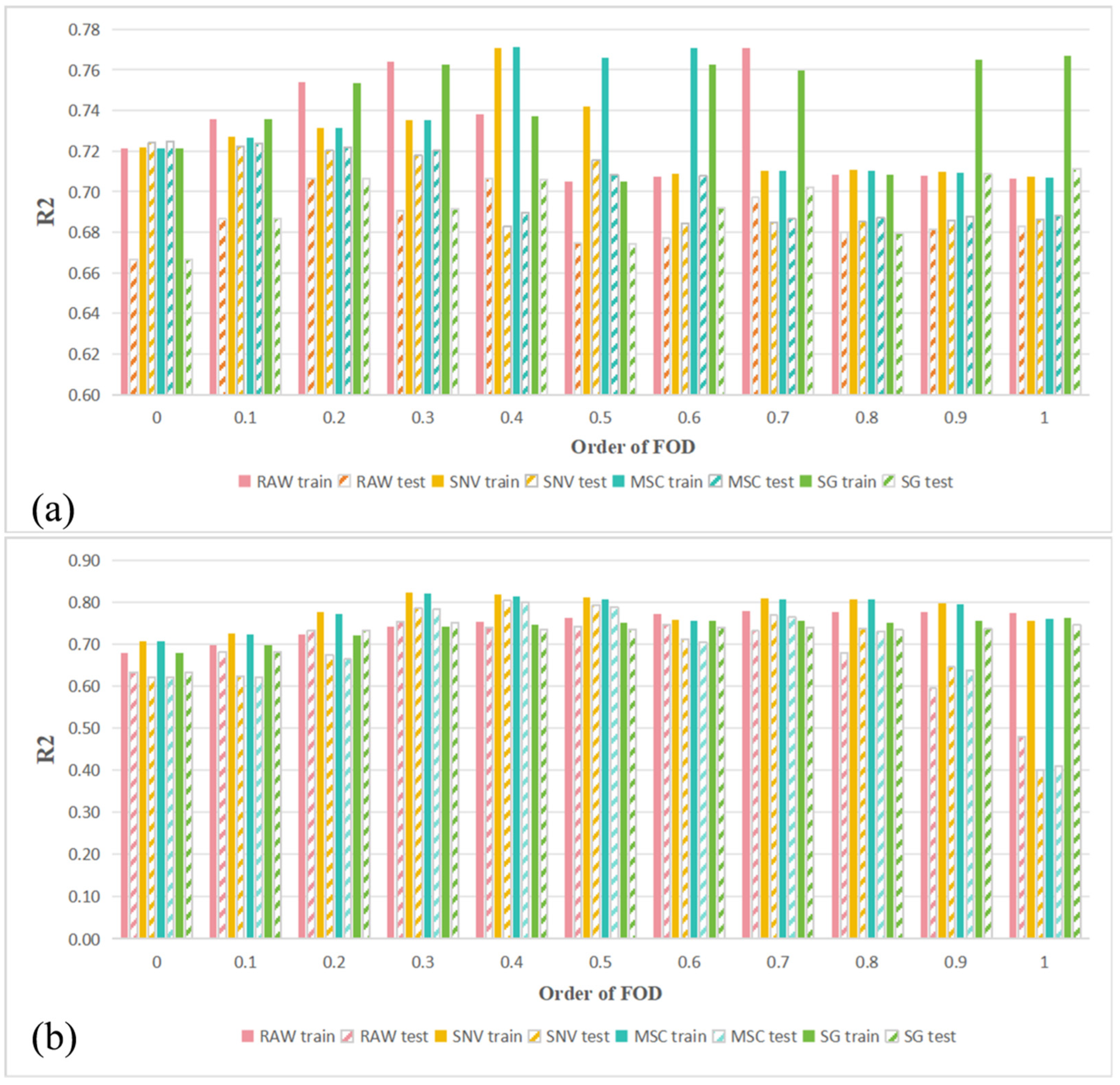
3.2. Comparative Analysis of Combinations of Spectral Preprocessing for Healthy and HLB-Infected Mianju Citrus Leaves
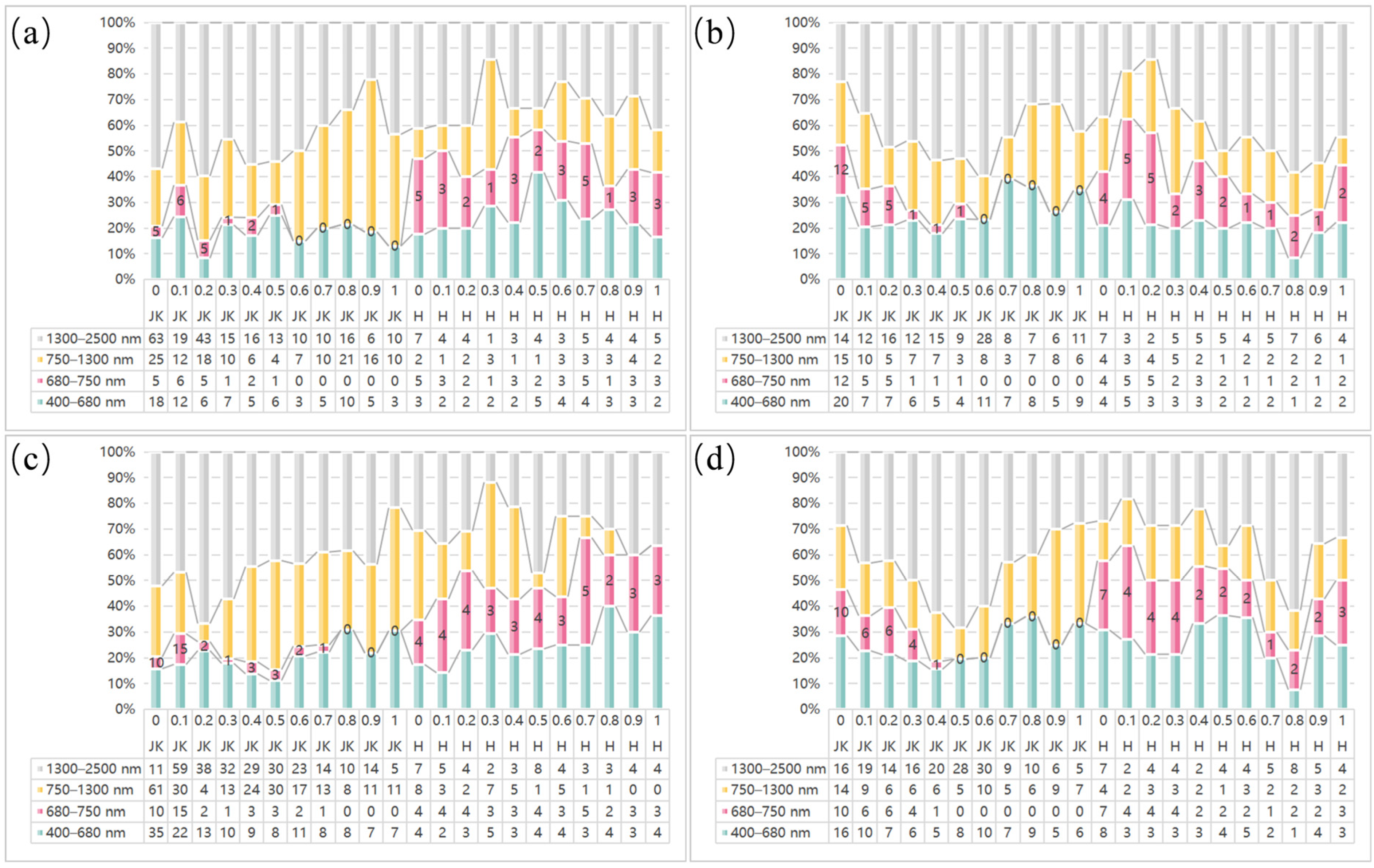
3.3. Comparative Analysis of the LASSO Feature Bands for Healthy and HLB-Infected Mianju Citrus Leaves
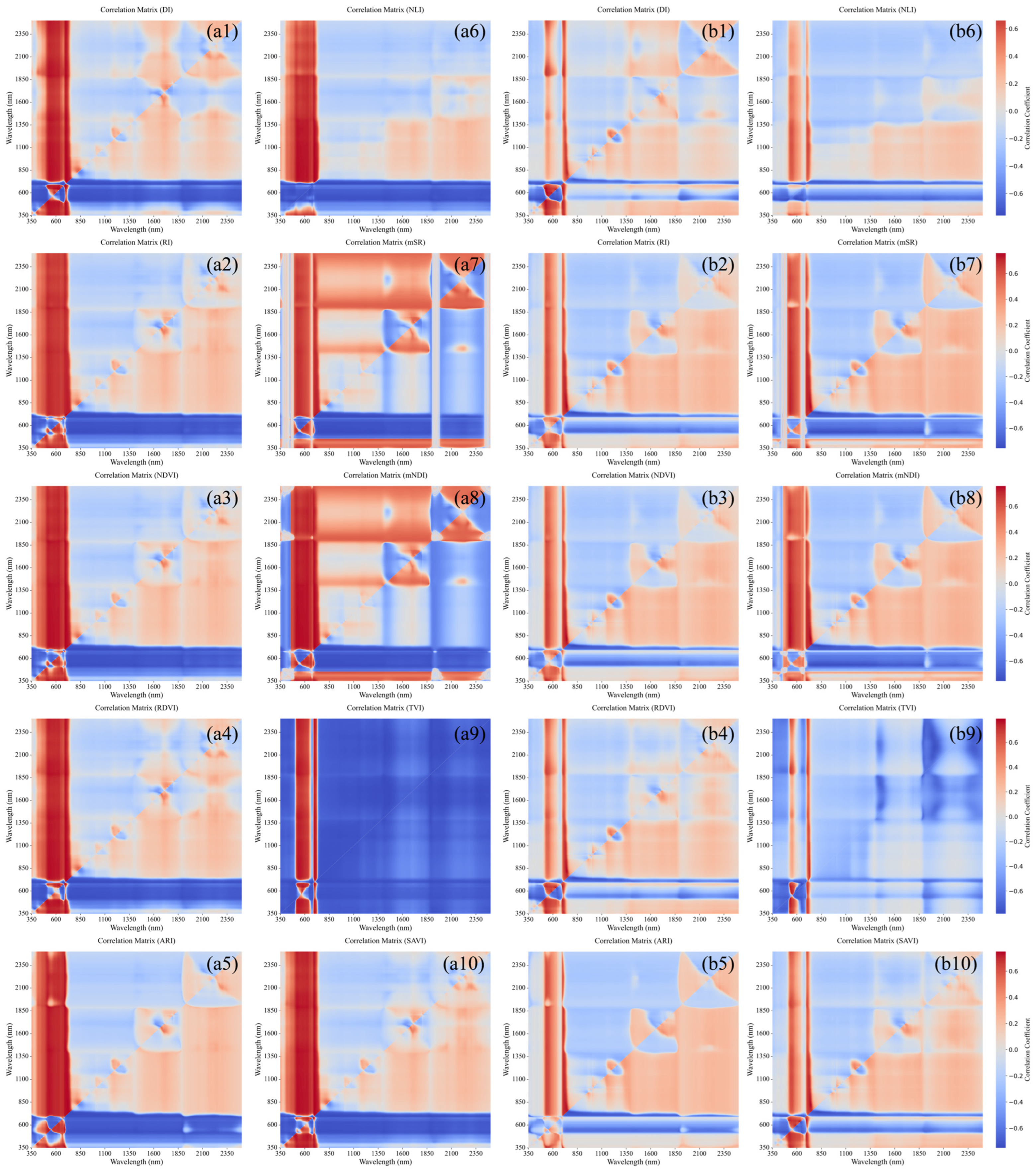
3.4. Comparative Analysis of the Optimal Spectral Indices for Healthy and HLB-Infected Mianju Citrus Leaves
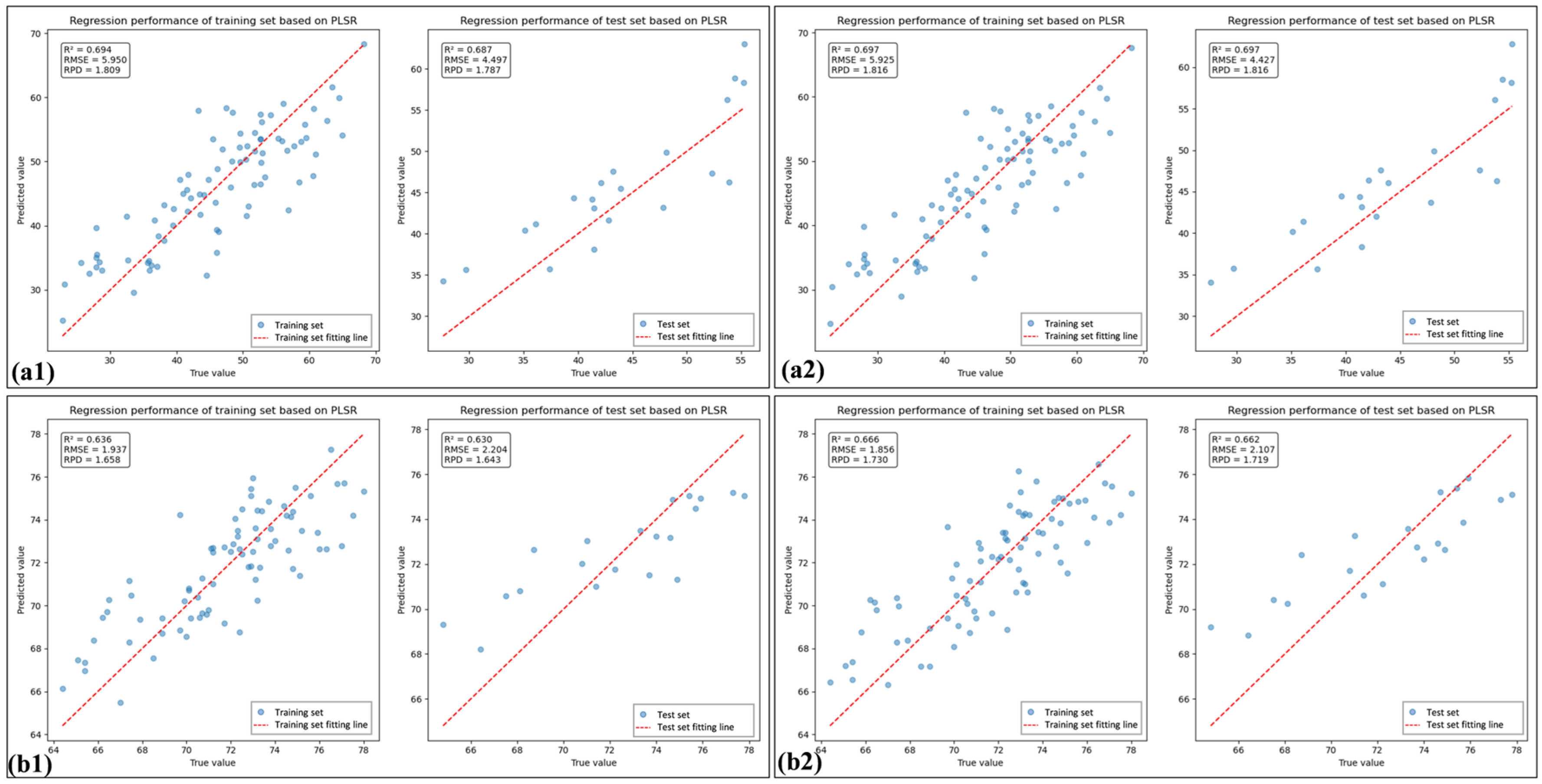
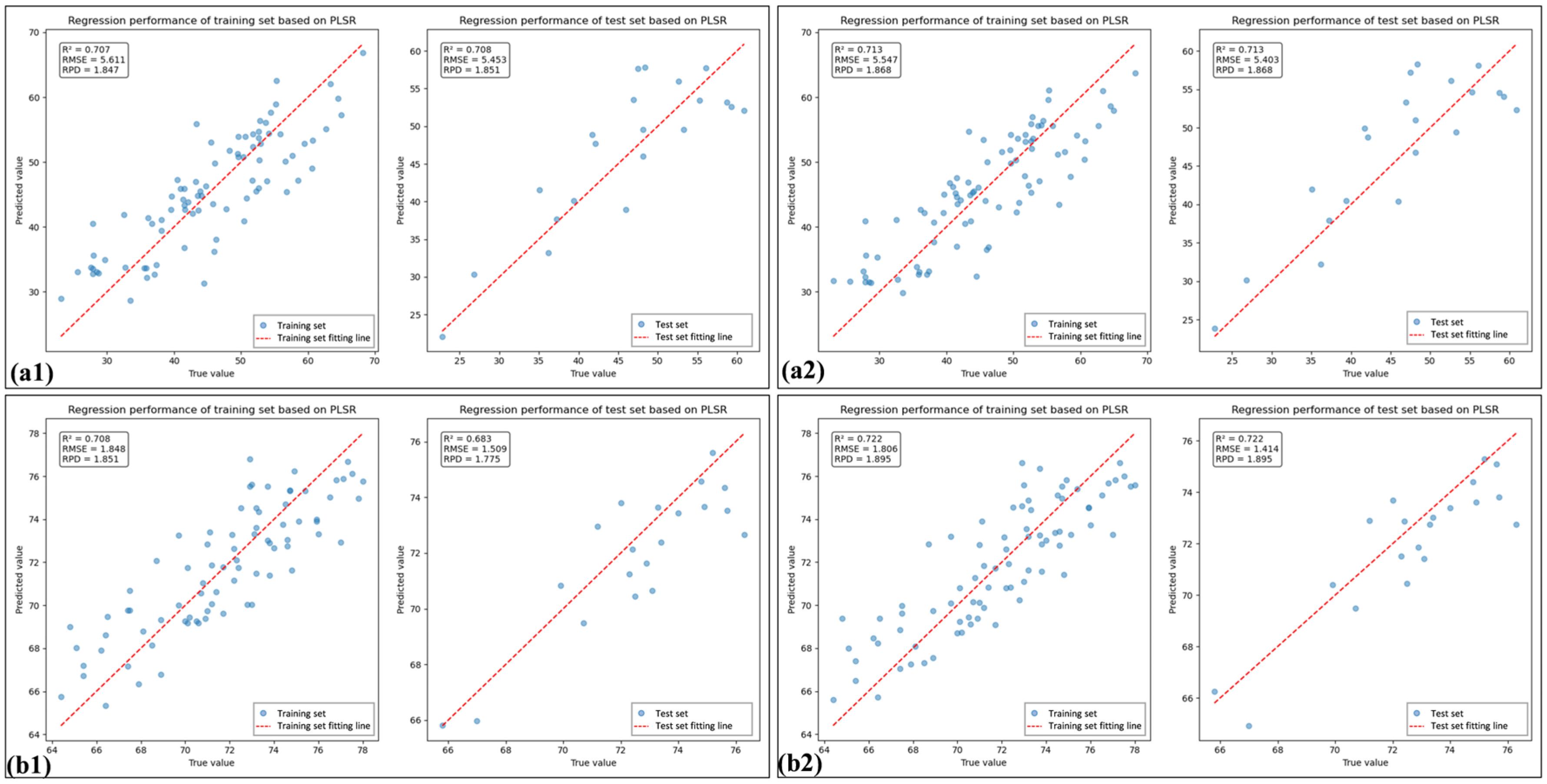
3.5. Comparison of the Chlorophyll Estimation Models for Healthy and HLB-Infected Mianju Citrus Leaves
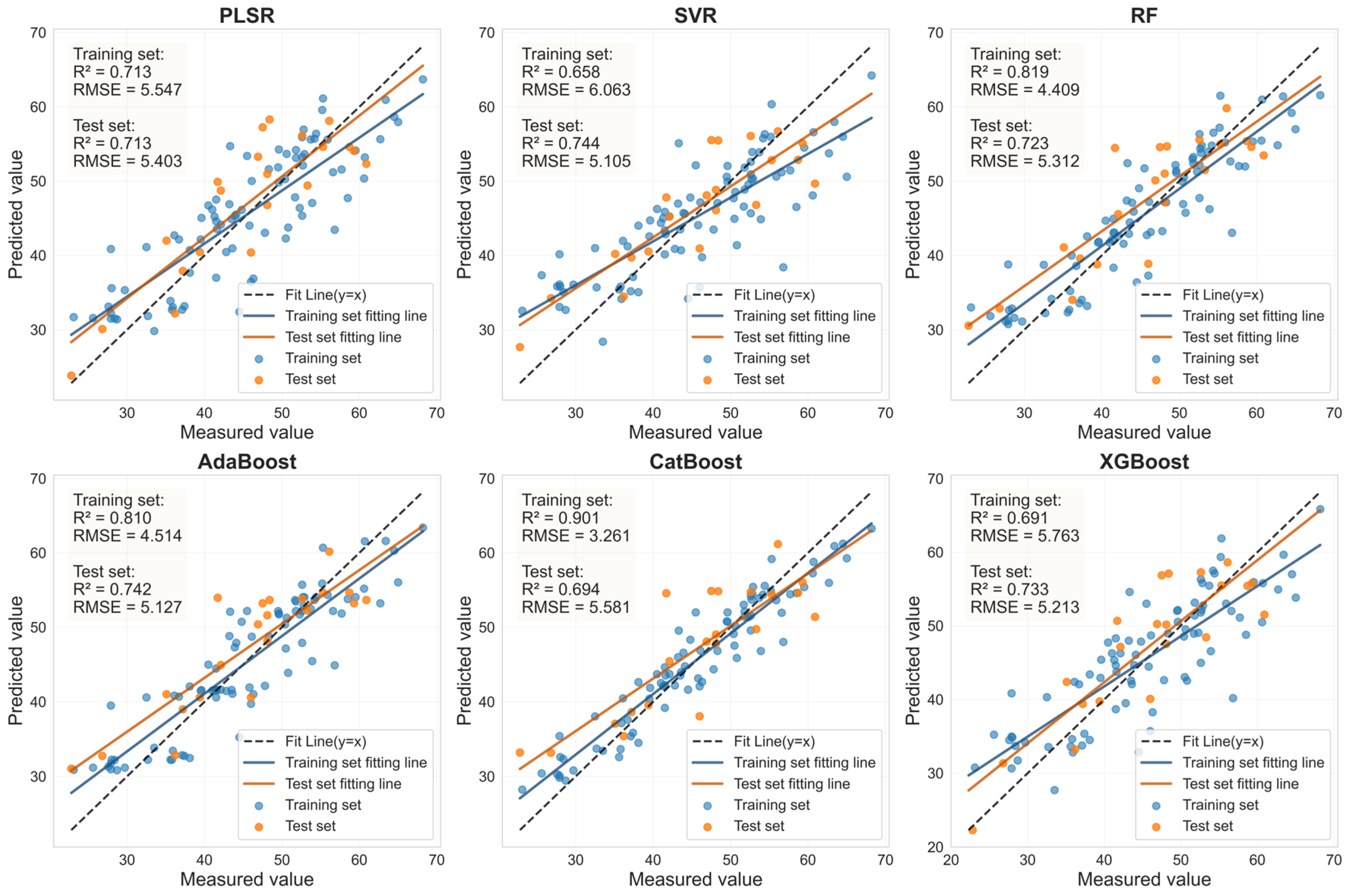
3.6. Estimation of LCC Using LASSO-Coupled Machine Learning Models on an External Dataset
4. Discussion
4.1. Effects of HLB Infection on the Spectral Characteristics of Citrus Leaves
4.2. Linear Regression Analysis for Chlorophyll Estimation Using Feature Bands of Healthy and HLB-Infected Leaves
4.3. Future Perspectives in Chlorophyll Estimation for HLB-Infected Citrus
5. Conclusions
- (1)
- HLB-infected leaves exhibited significant spectral differences in the visible and red-edge regions, with average reflectance at 554 nm and 710 nm being 0.144 and 0.209 higher, respectively, compared to healthy leaves. The infected leaves also showed a distinct blue shift in the red-edge position.
- (2)
- Comparative analysis of the characteristic bands selected by LASSO and spectral indices revealed that the proportion of characteristic bands for HLB-infected leaves located in the NIR region was markedly greater than that for healthy leaves. Specifically, more than 66% of the bands in the SI_CB characteristic band for HLB-infected leaves were in the NIR region.
- (3)
- The modeling results based on six machine learning algorithms indicated that the feature bands selected by LASSO outperformed those derived from spectral indices for predicting citrus LCC. Among the different models, PLSR yielded the best performance, achieving an Rv2 of 0.956 for healthy leaves and an Rv2 of 0.816 for HLB-infected leaves, which confirms its high estimation accuracy and robustness.
- (4)
- According to the statistical table of LCC in sweet citrus leaves, the average LCC of healthy leaves in November was approximately 1.57 times that of HLB-infected leaves. By comparing the LCC estimation models, this study confirmed that the optimal model for healthy leaves outperformed that for HLB-infected leaves. Furthermore, quantitative relationships between the characteristic bands and chlorophyll content were established for both healthy and HLB-infected leaves.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, M.; Liu, S.; Ou, Y.; Zhang, P. An efficient and lightweight citrus leaf disease detection model based on YOLOv8. Acta Agric. Zhejiangensis 2025, 37, 2198–2208. [Google Scholar]
- Zheng, G.; Yu, M. The achievements, problems and suggestions of the development of citrus export trade in China. S. China Fruits 2024, 53, 234–242. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, L.; Yu, G.; Liu, G. Impact of Huanglongbing on Citrus Orchards: A Spatiotemporal Study in Xunwu County, Jiangxi Province. Agriculture 2024, 14, 55. [Google Scholar] [CrossRef]
- Chen, X.; Wu, Y.; Lin, J.; Mao, J.; Wu, Z.; Huang, Z. Research Progress in Pathogenic Mechanism and Control Technologies of Citrus Huanglongbing. Guangxi Sci. 2025, 32, 245–254. [Google Scholar] [CrossRef]
- Deng, X.; Huang, Z.; Zheng, Z.; Lan, Y.; Dai, F. Field detection and classification of citrus Huanglongbing based on hyperspectral reflectance. Comput. Electron. Agric. 2019, 167, 105006. [Google Scholar] [CrossRef]
- Boakye, D.A.; Alferez, F. Fruit Yield in Sweet Orange Trees under Huanglongbing (HLB) Conditions Is Influenced by Reproductive Phenological Characteristics of the Scion-Rootstock Combination. Agriculture 2022, 12, 1750. [Google Scholar] [CrossRef]
- Chen, S.; Shen, Y.; Xie, P.; Lu, X.; He, Y.; Cen, H. In-situ diagnosis of citrus Huanglongbing using sun-induced chlorophyll fluorescence. Trans. Chin. Soc. Agric. Eng. 2022, 38, 324–332. [Google Scholar]
- Shi, Y.; Zeng, J.; Xue, J.; Jia, S.; Wang, M.; Guo, S. Effects of Candidatus Liberibacter asiaticus on assimilate accumulation and mineral nutrient transport in citrus leaves. J. Plant Nutr. Fertil. 2023, 29, 949–960. [Google Scholar] [CrossRef]
- Qin, H.; Liu, T.; Kong, L.; Yu, X.; Wang, X.; Huang, M. Rapid Detection of Citrus Huanglongbing Based on Extraction of Characteristic Wavelength of Visible Spectrum and Classification Algorithm. Spectrosc. Spectr. Anal. 2024, 44, 1518–1525. [Google Scholar]
- Weng, H.; Liu, Y.; Captoline, I.; Li, X.; Ye, D.; Wu, R. Citrus Huanglongbing detection based on polyphasic chlorophyll a fluorescence coupled with machine learning and model transfer in two citrus cultivars. Comput. Electron. Agric. 2021, 187, 106289. [Google Scholar] [CrossRef]
- Li, D.; Hu, Q.; Ruan, S.; Liu, J.; Zhang, J.; Hu, C.; Liu, Y.; Dian, Y.; Zhou, J. Utilizing Hyperspectral Reflectance and Machine Learning Algorithms for Non-Destructive Estimation of Chlorophyll Content in Citrus Leaves. Remote Sens. 2023, 15, 4934. [Google Scholar] [CrossRef]
- Li, X.; Wei, Z.; Peng, F.; Liu, J.; Han, G. Estimating the distribution of chlorophyll content in CYVCV infected lemon leaf using hyperspectral imaging. Comput. Electron. Agric. 2022, 198, 107036. [Google Scholar] [CrossRef]
- Li, J.; Sheng, Y.; Wang, W.; Liu, J.; Li, X. Estimating Wheat Chlorophyll Content Using a Multi-Source Deep Feature Neural Network. Agriculture 2025, 15, 1624. [Google Scholar] [CrossRef]
- Ding, S.; Jing, J.; Dou, S.; Zhai, M.; Zhang, W. Citrus Canopy SPAD Prediction under Bordeaux Solution Coverage Based on Texture- and Spectral-Information Fusion. Agriculture 2023, 13, 1701. [Google Scholar] [CrossRef]
- Ali, A.; Imran, M.; Ali, A.; Khan, M.A. Evaluating Sentinel-2 red edge through hyperspectral profiles for monitoring LAI & chlorophyll content of Kinnow Mandarin orchards. Remote Sens. Appl. Soc. Environ. 2022, 26, 100719. [Google Scholar] [CrossRef]
- Ansar, A.; Muhammad, I. Evaluating the potential of red edge position (REP) of hyperspectral remote sensing data for real time estimation of LAI & chlorophyll content of kinnow mandarin (Citrus reticulata) fruit orchards. Sci. Hortic. 2020, 267, 109326. [Google Scholar] [CrossRef]
- Xiao, B.; Li, S.; Dou, S.; He, H.; Fu, B.; Zhang, T.; Sun, W.; Yang, Y.; Xiong, Y.; Shi, J.; et al. Comparison of leaf chlorophyll content retrieval performance of citrus using FOD and CWT methods with field-based full-spectrum hyperspectral reflectance data. Comput. Electron. Agric. 2024, 217, 108559. [Google Scholar] [CrossRef]
- Peng, Z.; Guan, L.; Liao, Y.; Lian, S. Estimating Total Leaf Chlorophyll Content of Gannan Navel Orange Leaves Using Hyperspectral Data Based on Partial Least Squares Regression. IEEE Access 2019, 7, 155540–155551. [Google Scholar] [CrossRef]
- Yue, X.; Quan, D.; Hong, T.; Wang, J.; Qu, X.; Gan, H. Non-destructive hyperspectral measurement model of chlorophyll content for citrus leaves. Trans. Chin. Soc. Agric. Eng. 2015, 31, 294–302. [Google Scholar]
- Shen, J.; Xiong, Y.; Xu, Y.; Liu, H.; Cheng, D. Study on hyperspectral remote sensing estimation Models of pigment contents of pomelo leaves under different stresses. Guangdong Agric. Sci. 2018, 45, 118–129. [Google Scholar] [CrossRef]
- Ali, A.; Imran, M. Remotely sensed real-time quantification of biophysical and biochemical traits of Citrus (Citrus sinensis L.) fruit orchards—A review. Sci. Hortic. 2021, 282, 110024. [Google Scholar] [CrossRef]
- Wang, K.; Guo, D.; Zhang, Y.; Deng, L.; Xie, R.; Lv, Q.; Yi, S.; Zheng, Y.; Ma, Y.; He, S. Detection of Huanglongbing (citrus greening) based on hyperspectral image analysis and PCR. Front. Agric. Sci. Eng. 2019, 6, 172. [Google Scholar] [CrossRef]
- Chen, S.; Zhai, L.; Zhou, Y.; Xie, J.; Shao, Y.; Wang, W.; Li, H.; He, Y.; Cen, H. Early diagnosis and mechanistic understanding of citrus Huanglongbing via sun-induced chlorophyll fluorescence. Comput. Electron. Agric. 2023, 215, 108357. [Google Scholar] [CrossRef]
- Wan, L.; Zhou, W.; He, Y.; Wanger, T.C.; Cen, H. Combining transfer learning and hyperspectral reflectance analysis to assess leaf nitrogen concentration across different plant species datasets. Remote Sens. Environ. 2022, 269, 112826. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, T.; Gu, X.; Chen, D.; Wang, C.; Wu, W.; Zhu, Q.; Zhao, C. Hyperspectral remote sensing for tobacco quality estimation, yield prediction, and stress detection: A review of applications and methods. Front. Plant Sci. 2023, 14, 1073346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ru, G.; Zhao, Z.; Wang, D. Hyperspectral Prediction Models of Chlorophyll Content in Paulownia Leaves under Drought Stress. Sensors 2024, 24, 6309. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, H.; Qiao, X.; Yan, X.; Feng, M.; Xiao, L.; Song, X.; Zhang, M.; Shafiq, F.; Yang, W.; et al. Hyperspectral estimation of canopy chlorophyll of winter wheat by using the optimized vegetation indices. Comput. Electron. Agric. 2022, 193, 106654. [Google Scholar] [CrossRef]
- Zhang, Y.; Chang, Q.; Chen, Y.; Liu, Y.; Jiang, D.; Zhang, Z. Hyperspectral Estimation of Chlorophyll Content in Apple Tree Leaf Based on Feature Band Selection and the CatBoost Model. Agronomy 2023, 13, 2075. [Google Scholar] [CrossRef]
- Li, L.; Wu, Y.; Wang, H.; He, J.; Wang, Q.; Xu, J.; Xia, Y.; Yuan, W.; Chen, S.; Tao, L.; et al. Prediction Model of Flavonoids Content in Ancient Tree Sun−Dried Green Tea under Abiotic Stress Based on LASSO−Cox. Agriculture 2024, 14, 296. [Google Scholar] [CrossRef]
- Zhan, B.; Li, P.; Li, M.; Luo, W.; Zhang, H. Detection of Soluble Solids Content (SSC) in Pears Using Near-Infrared Spectroscopy Combined with LASSO–GWF–PLS Model. Agriculture 2023, 13, 1491. [Google Scholar] [CrossRef]
- Jiang, J.; Zhou, G.; Ji, H.; Pan, R.; Yang, L.; Zhao, S.; Fang, Z.; Liu, X.; Ma, Y.; Zhu, X.; et al. Advanced hyper-spectral feature engineering for real-time prediction of tea quality in support of high-grade cultivation. Comput. Electron. Agric. 2025, 236, 110485. [Google Scholar] [CrossRef]
- Laha, S.K. A sparse approach to transfer function estimation via Least Absolute Shrinkage and Selection Operator. IFAC J. Syst. Control 2025, 31, 100299. [Google Scholar] [CrossRef]
- De, X.; Li, H.; Zhang, J.; Li, N.; Wan, H.; Ma, Y. Prediction of the Calorific Value and Moisture Content of Caragana korshinskii Fuel Using Hyperspectral Imaging Technology and Various Stoichiometric Methods. Agriculture 2025, 15, 1557. [Google Scholar] [CrossRef]
- Xiang, S.; Xu, Z.; Zhang, Y.; Zhang, Q.; Zhou, X.; Yu, H.; Li, B.; Li, Y. Construction and Application of ReliefF-RFE Feature Selection Algorithm for Hyperspectral Image Classification. Spectrosc. Spectr. Anal. 2022, 42, 3283–3290. [Google Scholar] [CrossRef]
- Najafabadi, M.Y.; Earl, H.J.; Tulpan, D.; Sulik, J.; Eskandari, M. Application of Machine Learning Algorithms in Plant Breeding: Predicting Yield From Hyperspectral Reflectance in Soybean. Front. Plant Sci. 2021, 11, 624273. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhang, C.; Bian, X.; An, J.; Wang, Y.; Ou, X.; Kamruzzaman, M. Improved prediction of vitamin C and reducing sugar content in sweetpotatoes using hyperspectral imaging and LARS-enhanced LASSO variable selection. J. Food Compos. Anal. 2024, 132, 106350. [Google Scholar] [CrossRef]
- Liu, S.; Yu, H.; Zhang, J.; Zhou, H.; Kong, L.; Zhang, L.; Dang, J.; Sui, Y. Study on Inversion Model of Chlorophyll Content in Soybean Leaf Based on Optimal Spectral Indices. Spectrosc. Spectr. Anal. 2021, 41, 1912–1919. [Google Scholar]
- Chen, Q.; Chang, Q.; Guo, S.; Zhang, Y. Estimation of Chlorophyll Content in Winter Wheat Based on Red Edge Characteristics and Continuous Wavelet Transform. J. Triticeae Crops 2022, 42, 883–891. [Google Scholar]
- Liu, X.; Li, Z.; Xiang, Y.; Tang, Z.; Huang, X.; Shi, H.; Sun, T.; Yang, W.; Cui, S.; Chen, G.; et al. Estimation of Winter Wheat Chlorophyll Content Based on Wavelet Transform and the Optimal Spectral Index. Agronomy 2024, 14, 1309. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Liu, M.; Wang, C.; Xia, X. A hyperspectral index sensitive to subtle changes in the canopy chlorophyll content under arsenic stress. Int. J. Appl. Earth Obs. Geoinf. 2015, 36, 41–53. [Google Scholar] [CrossRef]
- Ye, X.; Kitaya, M.; Abe, S.; Sheng, F.; Zhang, S. A New Small-Size Camera with Built-In Specific-Wavelength LED Lighting for Evaluating Chlorophyll Status of Fruit Trees. Sensors 2023, 23, 4636. [Google Scholar] [CrossRef]
- Lou, B.; Bai, X.; Bai, Y.; Deng, C.; RoyChowdhury, M.; Chen, C.; Song, Y. Detection and Molecular Characterization of a 16SrII-A* Phytoplasma in Grapefruit (Citrus paradisi) with Huanglongbing-like Symptoms in China. J. Phytopathol. 2014, 162, 387–395. [Google Scholar] [CrossRef]
- Ignatov, K.B.; Barsova, E.V.; Fradkov, A.F.; Blagodatskikh, K.A.; Kramarova, T.V.; Kramarov, V.M. A strong strand displacement activity of thermostable DNA polymerase markedly improves the results of DNA amplification. BioTechniques 2014, 57, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Lou, B.; Song, Y.; RoyChowdhury, M.; Deng, C.; Niu, Y.; Fan, Q.; Tang, Y.; Zhou, C. Development of a Tandem Repeat-Based Polymerase Chain Displacement Reaction Method for Highly Sensitive Detection of ‘Candidatus Liberibacter asiaticus’. Phytopathology 2018, 108, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, X.; Zhang, N.; Guo, H.; Wu, H.; Wang, X. Optimizing the estimation of cotton leaf SPAD and LAI values via UAV multispectral imagery and LASSO regression. Smart Agric. Technol. 2025, 12, 101098. [Google Scholar] [CrossRef]
- Daniel, A.S.; John, A.G. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Tuerxun, N.; Naibi, S.; Zheng, J.; Wang, R.; Wang, L.; Lu, B.; Yu, D. Accurate estimation of Jujube leaf chlorophyll content using optimized spectral indices and machine learning methods integrating geospatial information. Ecol. Inform. 2025, 85, 102980. [Google Scholar] [CrossRef]
- Shi, H.; Guo, J.; An, J.; Tang, Z.; Wang, X.; Li, W.; Zhao, X.; Jin, L.; Xiang, Y.; Li, Z.; et al. Estimation of Chlorophyll Content in Soybean Crop at Different Growth Stages Based on Optimal Spectral Index. Agronomy 2023, 13, 663. [Google Scholar] [CrossRef]
- Song, Z.; Liu, Y.; Yu, J.; Guo, Y.; Jiang, D.; Zhang, Y.; Guo, Z.; Chang, Q. Estimation of Chlorophyll Content in Apple Leaves Infected with Mosaic Disease by Combining Spectral and Textural Information Using Hyperspectral Images. Remote Sens. 2024, 16, 2190. [Google Scholar] [CrossRef]
- Du, M.; Pang, S.; Dong, L.; Mo, K.; Hou, M.; Wang, S.; Zou, X. Research Progress on the Molecular Mechanisms of Interactions Between Candidatus Liberibacter and Citrus. Acta Hortic. Sin. 2024, 51, 1623–1638. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, H.; Xu, H.; Rao, Y.; Jiang, X.; Sun, X. Visual discrimination of citrus HLB based on image features. Vib. Spectrosc. 2019, 102, 103–111. [Google Scholar] [CrossRef]
- Weng, H.; Lv, J.; Cen, H.; He, M.; Zeng, Y.; Hua, S.; Li, H.; Meng, Y.; Fang, H.; He, Y. Hyperspectral reflectance imaging combined with carbohydrate metabolism analysis for diagnosis of citrus Huanglongbing in different seasons and cultivars. Sens. Actuators B Chem. 2018, 275, 50–60. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, J.; Yan, K.; Lu, X.; Li, W.; Tian, H.; Wang, L.; Deng, J.; Lan, Y. Advances and Developments in Monitoring and Inversion of the Biochemical Information of Crop Nutrients Based on Hyperspectral Technology. Agronomy 2023, 13, 2163. [Google Scholar] [CrossRef]
- Yan, K.; Song, X.; Yang, J.; Xiao, J.; Xu, X.; Guo, J.; Zhu, H.; Lan, Y.; Zhang, Y. Citrus huanglongbing detection: A hyperspectral data-driven model integrating feature band selection with machine learning algorithms. Crop Prot. 2025, 188, 107008. [Google Scholar] [CrossRef]
- Dong, R.; Shiraiwa, A.; Ichinose, K.; Pawasut, A.; Sreechun, K.; Mensin, S.; Hayashi, T. Hyperspectral Imaging and Machine Learning for Huanglongbing Detection on Leaf-Symptoms. Plants 2025, 14, 451. [Google Scholar] [CrossRef]
- Tang, Y.; Jin, Y.; Wu, L.; Yan, D.; Zhang, X.; Li, F. Comparison of Methods for Estimating Chlorophyll Content in Maize Leaves by Red Edge Position. J. Maize Sci. 2024, 32, 46–54. [Google Scholar] [CrossRef]
- Kabir, M.; Unal, F.; Akinci, T.C.; Martinez-Morales, A.A.; Ekici, S. Revealing GLCM Metric Variations across a Plant Disease Dataset: A Comprehensive Examination and Future Prospects for Enhanced Deep Learning Applications. Electronics 2024, 13, 2299. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, Y.; Zhao, Z.; Xie, M.; Hou, D. Estimation of Relative Chlorophyll Content in Spring Wheat Based on Multi-Temporal UAV Remote Sensing. Agronomy 2023, 13, 211. [Google Scholar] [CrossRef]
- Kasim, N.; Shi, Q.; Wang, J.; Sawut, R.; Nurmemet, I.; Isak, G. Estimation of spring wheat chlorophyll content based on hyperspectral features and PLSR model. Trans. Chin. Soc. Agric. Eng. 2017, 33, 208–216. [Google Scholar] [CrossRef]

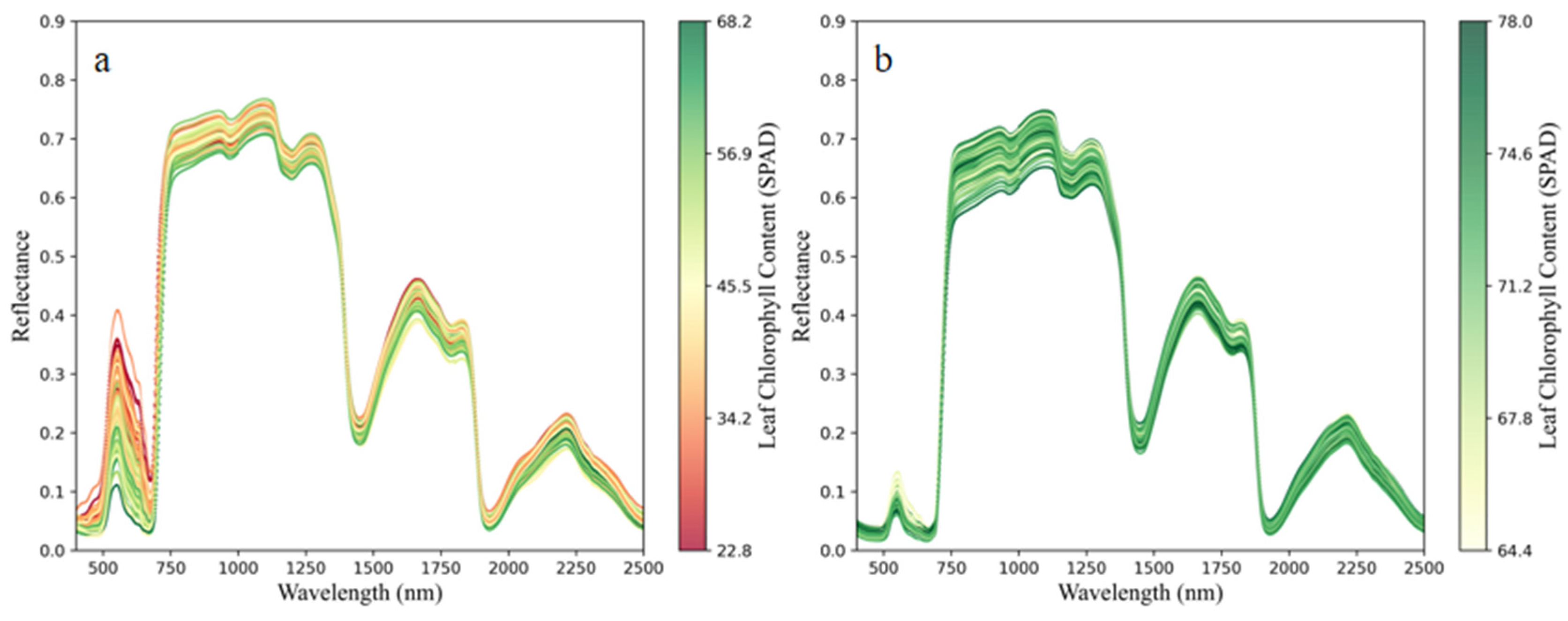



| Health Status | Sample Size | Max | Mean | Min | Standard Deviation | Coefficient of Variation |
|---|---|---|---|---|---|---|
| Healthy | 100 | 78 | 71.9 | 64.4 | 3.322 | 0.046 |
| HLB-infected | 105 | 68.2 | 45.6 | 22.8 | 10.359 | 0.227 |
| Spectral Index | Formula | Reference |
|---|---|---|
| Ratio index (RI) | Ri/Rj | [47] |
| Difference index (DI) | Ri − Rj | [47] |
| Normalized difference vegetation index (NDVI) | Ri − Rj/Ri + Rj | [47] |
| Renormalized difference vegetation index (RDVI) | (Ri − Rj)/SQRT(Ri + Rj) | [47] |
| Anth reflectance index (ARI) | (Rj − Rj)/(Rj × Rj) | [47] |
| Nonlinear vegetation index (NLI) | (R2i − Rj)/(R2i + Rj) | [47] |
| Modified simple ratio (mSR) | Ri − R445/Rj − R445 | [46] |
| Modified normalized difference index (mNDI) | Ri − Rj/Ri + Rj − 2R445 | [46] |
| Triangular vegetation index (TVI) | 0.5 × (120 × (Ri − R500)) − 200 × (Rj − R500) | [46] |
| Soil–adjusted vegetation index (SAVI) | [48] |
| Leaf Condition | Feature Selection | Pretreatment | FOD Order | Band Number | Rc2 | RMSEc | Rv2 | RMSEv |
|---|---|---|---|---|---|---|---|---|
| Health | LASSO | RAW | 1 | 23 | 0.95 | 0.76 | 0.94 | 0.81 |
| SNV | 1 | 28 | 0.92 | 0.93 | 0.90 | 1.00 | ||
| SG | 0.8 | 34 | 0.92 | 0.94 | 0.93 | 0.88 | ||
| MSC | 0.9 | 27 | 0.92 | 0.91 | 0.91 | 0.94 | ||
| RFE | RAW | 0.8 | 30 | 0.85 | 1.27 | 0.81 | 1.40 | |
| SNV | 0.9 | 30 | 0.86 | 1.22 | 0.84 | 1.30 | ||
| SG | 0.5 | 30 | 0.78 | 1.54 | 0.79 | 1.46 | ||
| MSC | 0.8 | 30 | 0.84 | 1.29 | 0.85 | 1.26 | ||
| CARS | RAW | 0.9 | 143 | 0.98 | 0.45 | 0.89 | 1.06 | |
| SNV | 0.6 | 190 | 0.91 | 0.97 | 0.90 | 1.04 | ||
| SG | 0.7 | 123 | 0.94 | 0.77 | 0.88 | 1.13 | ||
| MSC | 1 | 144 | 0.92 | 0.94 | 0.85 | 1.25 | ||
| HLB | LASSO | RAW | 0.9 | 14 | 0.82 | 4.34 | 0.82 | 4.70 |
| SNV | 1 | 9 | 0.80 | 4.51 | 0.81 | 4.75 | ||
| SG | 0.5 | 17 | 0.77 | 4.92 | 0.75 | 5.41 | ||
| MSC | 1 | 12 | 0.81 | 4.44 | 0.78 | 5.00 | ||
| RFE | RAW | 0.5 | 30 | 0.75 | 5.08 | 0.75 | 5.42 | |
| SNV | 0.8 | 30 | 0.74 | 5.17 | 0.74 | 5.48 | ||
| SG | 0.1 | 30 | 0.75 | 5.11 | 0.73 | 5.63 | ||
| MSC | 0.7 | 30 | 0.75 | 5.12 | 0.74 | 5.44 | ||
| CARS | RAW | 0.1 | 81 | 0.68 | 5.79 | 0.68 | 6.07 | |
| SNV | 0.1 | 144 | 0.74 | 5.23 | 0.73 | 5.61 | ||
| SG | 0.4 | 60 | 0.76 | 5.03 | 0.65 | 6.34 | ||
| MSC | 0.2 | 61 | 0.75 | 5.07 | 0.71 | 5.79 |
| Spectral Index | HLB | Healthy | ||
|---|---|---|---|---|
| Band Combination | Correlation Coefficient | Band Combination | Correlation Coefficient | |
| RI | (713, 712) | 0.826 | (820, 739) | 0.747 |
| DI | (830, 703) | 0.841 | (643, 529) | 0.754 |
| NDVI | (830, 699) | 0.833 | (819, 741) | 0.747 |
| RDVI | (714, 713) | 0.838 | (636, 631) | 0.742 |
| ARI | (1896, 689) | 0.795 | (751, 750) | 0.736 |
| NLI | (842, 699) | 0.835 | (352, 531) | 0.659 |
| mSR | (714, 713) | 0.825 | (814, 739) | 0.757 |
| mNDI | (830, 699) | 0.835 | (814, 739) | 0.757 |
| TVI | (842, 702) | 0.835 | (530, 537) | 0.791 |
| SAVI | (818, 700) | 0.835 | (636, 631) | 0.751 |
| Sample | Feature Combination Name | Feature Variable | Rc2 | RMSEc | Rv2 | RMSEv |
|---|---|---|---|---|---|---|
| HLB | SI | RI, DI, NDVI, RDVI, ARI, NLI, mSR, mNDI, TVI, SAVI | 0.694 | 5.950 | 0.687 | 4.497 |
| SI_PLS | DI, NDVI, RDVI, NLI, mSR, mNDI, TVI, SAVI | 0.697 | 5.925 | 0.697 | 4.427 | |
| SI_CB | 689, 699, 700, 702, 703, 712, 713, 714, 818, 830, 842, 1896 | 0.707 | 5.611 | 0.708 | 5.453 | |
| SI_CB_PLS | 702, 703, 713, 714, 1896 | 0.713 | 5.547 | 0.713 | 5.403 | |
| Health | SI | RI, DI, NDVI, RDVI, ARI, NLI, mSR, mNDI, TVI, SAVI | 0.636 | 1.937 | 0.630 | 2.204 |
| SI_PLS | DI, ARI, NLI, mSR, TVI, SAVI | 0.666 | 1.856 | 0.662 | 2.107 | |
| SI_CB | 352,5 29, 530, 531, 537, 631, 636, 643, 739, 741, 750, 751, 814, 819, 820 | 0.708 | 1.848 | 0.683 | 1.509 | |
| SI_CB_PLS | 352, 529, 643, 739, 741, 750, 814, 819, 820 | 0.722 | 1.895 | 0.722 | 1.414 |
| Sample | Feature Variable | Wavelength/nm |
|---|---|---|
| HLB | RAW-FOD0.9-LASSO | 404, 409, 511, 720, 722, 725, 818, 840, 1077, 1245, 2319, 2320, 2239, 2399 |
| SI_CB_PLS | 702, 703, 713, 714, 1896 | |
| Health | RAW-FOD1.0-LASSO | 438, 558, 593, 751, 823, 863, 887, 899, 923, 953, 965, 1070, 1076, 1742, 1777, 1906, 2125, 2283, 2322, 2412, 2423, 2424, 2483 |
| SI_CB_PLS | 352, 529, 643, 739, 741, 750, 814, 819, 820 |
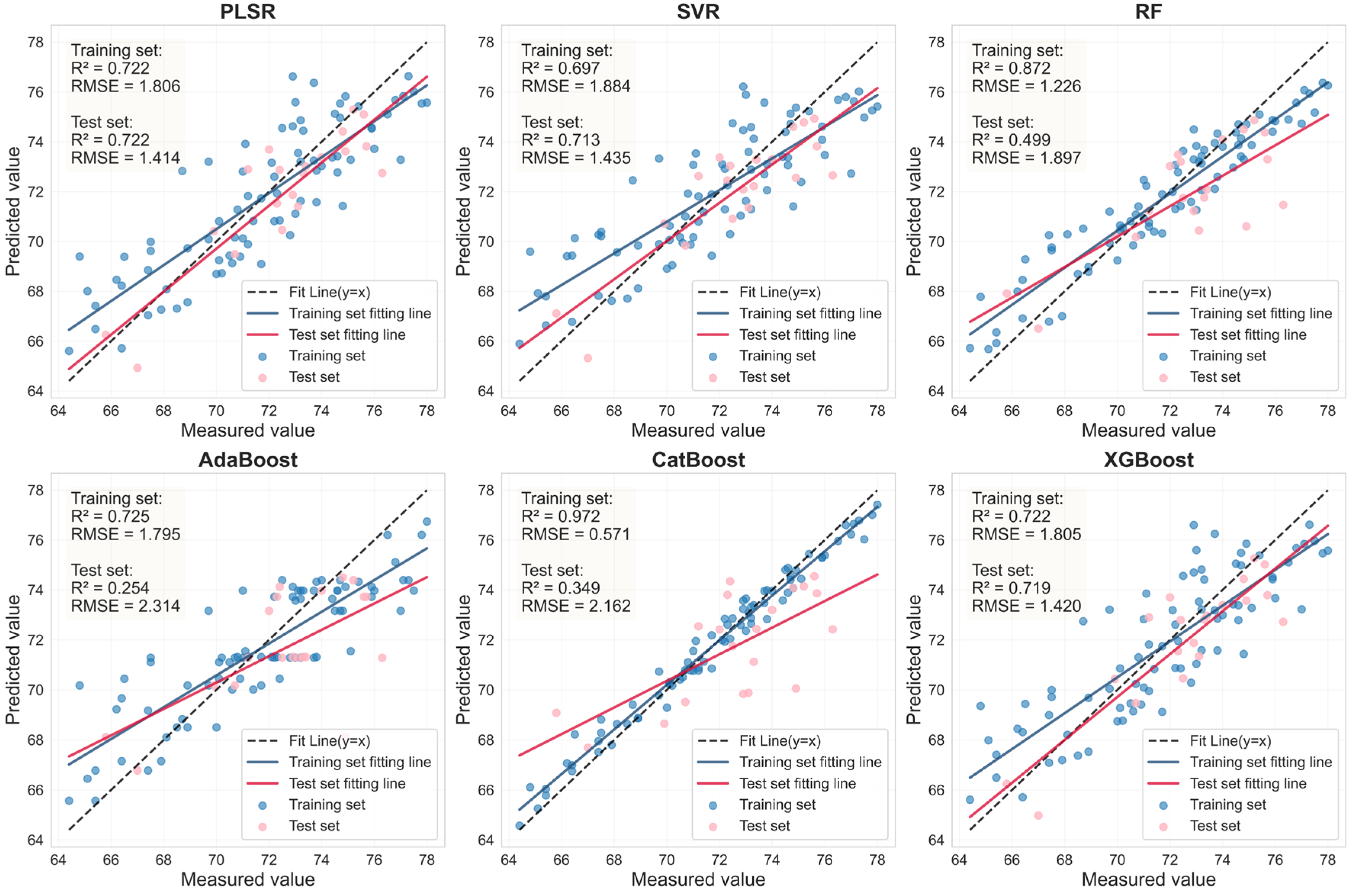
| Sample | Feature Variable | Model | Number | Rc2 | RMSEc | Rv2 | RMSEv | RPD |
|---|---|---|---|---|---|---|---|---|
| Health | RAW- FOD1.0- LASSO | PLSR | 23 | 0.956 | 0.683 | 0.956 | 0.675 | 4.767 |
| SVR | 23 | 0.958 | 0.664 | 0.921 | 0.907 | 3.548 | ||
| RF | 23 | 0.926 | 0.885 | 0.599 | 2.038 | 1.578 | ||
| AdaBoost | 23 | 0.947 | 0.753 | 0.587 | 2.068 | 1.556 | ||
| CatBoost | 23 | 1.000 | 0.000 | 0.595 | 2.046 | 1.572 | ||
| XGBoost | 23 | 0.961 | 0.644 | 0.940 | 0.790 | 4.074 | ||
| SI_CB _PLS | PLSR | 9 | 0.722 | 1.895 | 0.722 | 1.414 | 1.895 | |
| SVR | 9 | 0.697 | 1.884 | 0.713 | 1.435 | 1.867 | ||
| RF | 9 | 0.872 | 1.226 | 0.499 | 1.897 | 1.412 | ||
| AdaBoost | 9 | 0.725 | 1.795 | 0.254 | 2.314 | 1.158 | ||
| CatBoost | 9 | 0.972 | 0.571 | 0.349 | 2.162 | 1.239 | ||
| XGBoost | 9 | 0.722 | 1.805 | 0.719 | 1.420 | 1.886 | ||
| HLB | RAW- FOD0.9- LASSO | PLSR | 14 | 0.817 | 4.360 | 0.816 | 4.614 | 2.331 |
| SVR | 14 | 0.820 | 4.322 | 0.766 | 5.198 | 2.069 | ||
| RF | 14 | 0.947 | 2.337 | 0.792 | 4.908 | 2.191 | ||
| AdaBoost | 14 | 0.946 | 2.355 | 0.807 | 4.723 | 2.277 | ||
| CatBoost | 14 | 0.999 | 0.285 | 0.737 | 5.514 | 1.950 | ||
| XGBoost | 14 | 0.818 | 4.341 | 0.801 | 4.793 | 2.244 | ||
| SI_CB _PLS | PLSR | 5 | 0.713 | 5.547 | 0.713 | 5.403 | 1.868 | |
| SVR | 5 | 0.658 | 6.063 | 0.744 | 5.105 | 1.977 | ||
| RF | 5 | 0.819 | 4.409 | 0.723 | 5.312 | 1.900 | ||
| AdaBoost | 5 | 0.810 | 4.514 | 0.742 | 5.127 | 1.968 | ||
| CatBoost | 5 | 0.901 | 3.261 | 0.694 | 5.581 | 1.808 | ||
| XGBoost | 5 | 0.691 | 5.763 | 0.733 | 5.213 | 1.936 |
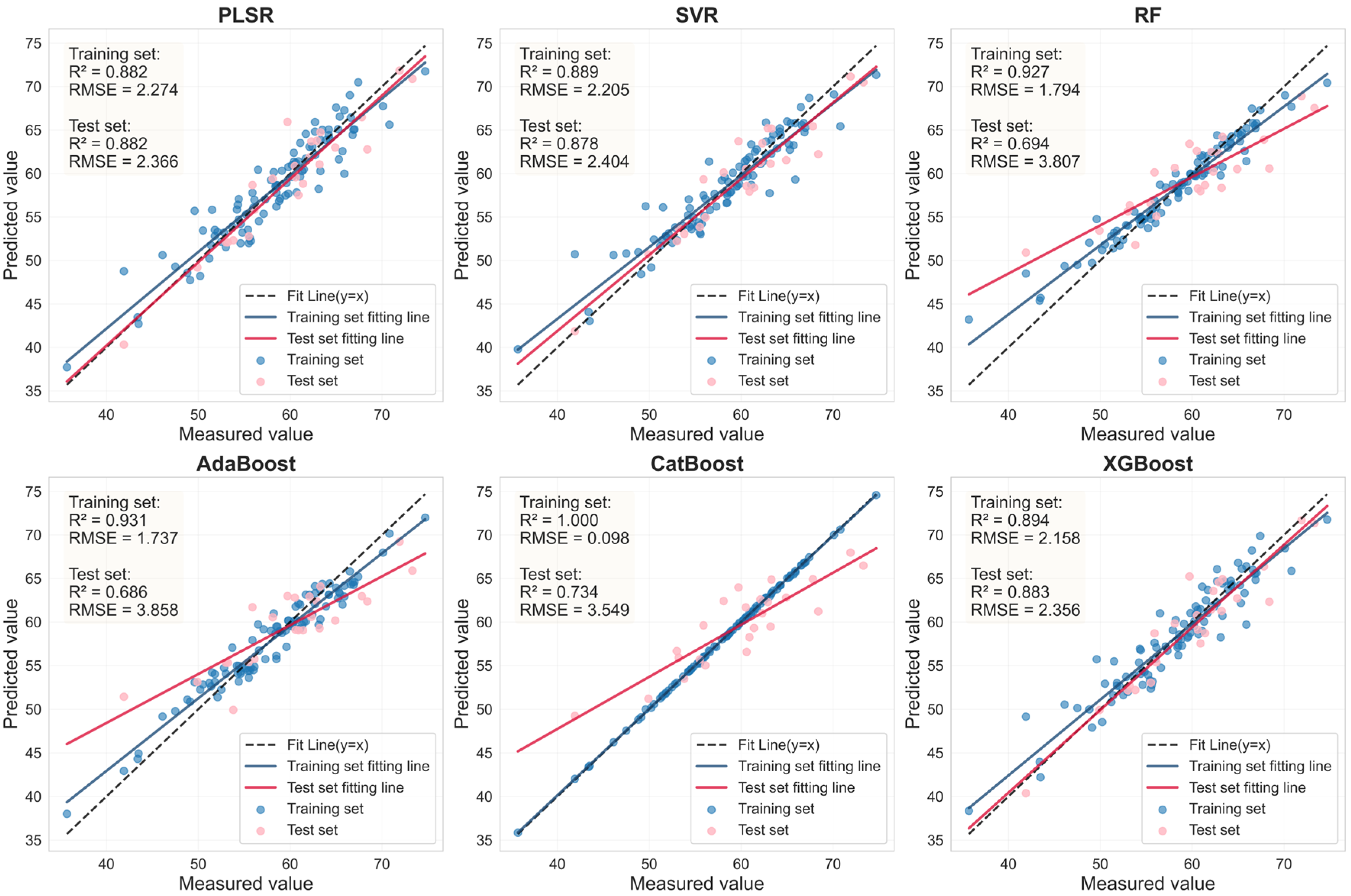
| Sample | Feature Variable | Num | Model | Rc2 | RMSEc | Rv2 | RMSEv | RPD |
|---|---|---|---|---|---|---|---|---|
| Health | RAW- FOD1.0- LASSO | 44 | PLSR | 0.882 | 2.274 | 0.882 | 2.366 | 2.908 |
| SVR | 0.889 | 2.205 | 0.878 | 2.404 | 2.862 | |||
| RF | 0.927 | 1.794 | 0.694 | 3.807 | 1.807 | |||
| AdaBoost | 0.931 | 1.737 | 0.686 | 3.858 | 1.783 | |||
| CatBoost | 1.000 | 0.098 | 0.734 | 3.549 | 1.939 | |||
| XGBoost | 0.894 | 2.158 | 0.883 | 2.356 | 2.920 | |||
| HLB | RAW- FOD0.9- LASSO | 34 | PLSR | 0.734 | 4.063 | 0.730 | 3.583 | 1.923 |
| SVR | 0.728 | 4.110 | 0.722 | 3.636 | 1.895 | |||
| RF | 0.878 | 2.756 | 0.473 | 5.001 | 1.378 | |||
| AdaBoost | 0.869 | 2.852 | 0.493 | 4.907 | 1.404 | |||
| CatBoost | 1.000 | 0.167 | 0.575 | 4.494 | 1.533 | |||
| XGBoost | 0.734 | 4.066 | 0.707 | 3.729 | 1.848 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, S.; Li, M.; Qi, X.; Hou, Y.; Yuan, S.; Song, Y.; Mei, Z.; Qi, G. Investigating the Association Between Citrus Huanglongbing and Chlorophyll Content Using Hyperspectral Detection. Sensors 2025, 25, 7292. https://doi.org/10.3390/s25237292
Dou S, Li M, Qi X, Hou Y, Yuan S, Song Y, Mei Z, Qi G. Investigating the Association Between Citrus Huanglongbing and Chlorophyll Content Using Hyperspectral Detection. Sensors. 2025; 25(23):7292. https://doi.org/10.3390/s25237292
Chicago/Turabian StyleDou, Shiqing, Minglan Li, Xiangqian Qi, Yichang Hou, Shixin Yuan, Yaqin Song, Zhengmin Mei, and Genhong Qi. 2025. "Investigating the Association Between Citrus Huanglongbing and Chlorophyll Content Using Hyperspectral Detection" Sensors 25, no. 23: 7292. https://doi.org/10.3390/s25237292
APA StyleDou, S., Li, M., Qi, X., Hou, Y., Yuan, S., Song, Y., Mei, Z., & Qi, G. (2025). Investigating the Association Between Citrus Huanglongbing and Chlorophyll Content Using Hyperspectral Detection. Sensors, 25(23), 7292. https://doi.org/10.3390/s25237292






