On the Edge of Dispensability, the Chloroplast ndh Genes
Abstract
:1. Introduction
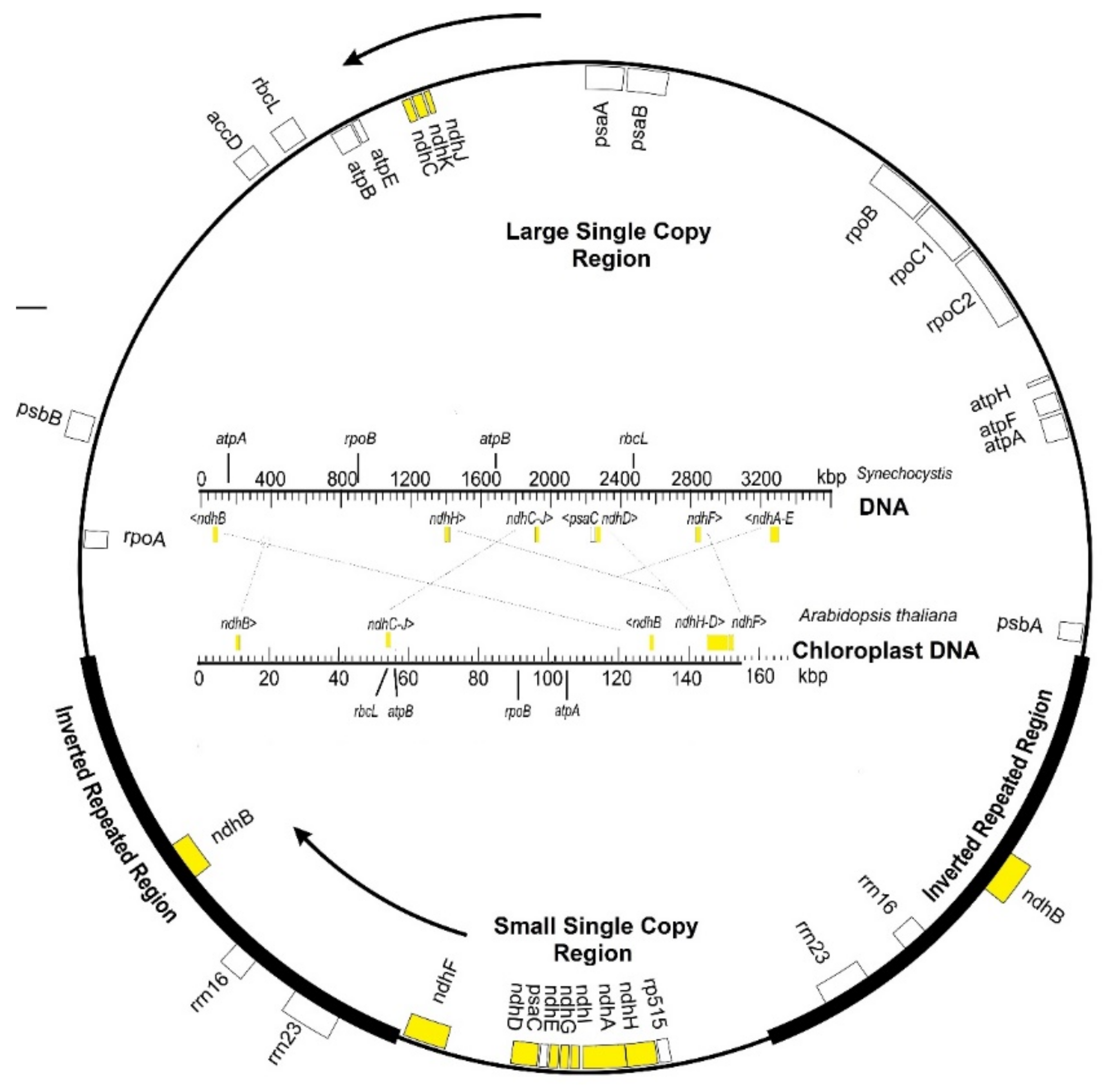
2. Chloroplast ndh Genes
3. Functional Role of the Thylakoid Ndh Complex
4. Dispensing with the Role of the ndh Genes
5. Plant Death or Species Extinction, a ndh Dilemma?
6. Concluding Remarks and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohyama, K.; Fukuzawa, H.; Kochi, T.; Shira, H.; Sano, T.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z.; Aota, S.; et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashida, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome. Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Martin, W.; Herrmann, R.G. Gene transfer from organelles to nucleus: How much, what happens, and why? Plant Physiol. 1998, 118, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Martin, W.; Rujan, T.; Richly, E.; Hansen, A.; Cornelsen, S.; Lins, T.; Leister, D.; Stoebe, B.; Hasegawa, M.; Penny, D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 2002, 99, 12246–12251. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, K.H.; Morden, C.W.; Palmer, J.D. Function and evolution of a minimal plastid genome from a non-photosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 1992, 89, 10648–10652. [Google Scholar] [CrossRef] [Green Version]
- Haberhausen, G.; Zetsche, K. Functional loss of all ndh genes in an otherwise relatively unaltered plastid genome of the holoparasitic flowering plant Cuscuta reflexa. Plant Mol. Biol. 1994, 24, 217–222. [Google Scholar] [CrossRef]
- Funk, H.T.; Berg, S.; Krupinska, K.; Maier, U.G.; Krause, K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007, 7, 45. [Google Scholar] [CrossRef] [Green Version]
- Sazanov, L.; Burrows, P.A.; Nixon, P.J. The plastid ndh genes code for an NADH-specific dehydrogenase: Purification and characterization of a mitochondrial-like complex I from pea thylakoid membranes. Proc. Natl. Acad. Sci. USA 1998, 95, 1319–1324. [Google Scholar] [CrossRef] [Green Version]
- Casano, L.M.; Zapata, J.M.; Martín, M.; Sabater, B. Chlororespiration and poising of cyclic electron transport: Plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. J. Biol. Chem. 2000, 275, 942–948. [Google Scholar] [CrossRef] [Green Version]
- Martín, M.; Sabater, B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 2010, 48, 636–645. [Google Scholar] [CrossRef]
- Martín, M.; Casano, L.M.; Sabater, B. Identification of the product of ndhA gene as a thylakoid protein synthesized in response to photooxidative treatment. Plant Cell Physiol. 1996, 37, 293–298. [Google Scholar] [CrossRef] [Green Version]
- Martín, M.; Casano, L.M.; Zapata, J.M.; Guéra, A.; del Campo, E.M.; Schmitz-Linneweber, C.; Maier, R.M.; Sabater, B. Role of thylakoid Ndh complex and peroxidase in the protection against photo-oxidative stress: Fluorescence and enzyme activities in wild-type and ndhF-deficient tobacco. Physiol. Plant. 2004, 122, 443–452. [Google Scholar] [CrossRef]
- Friedrich, T.; Scheide, D. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000, 479, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Freyer, R.; López, C.; Maier, R.M.; Martín, M.; Sabater, B.; Kossel, H. Editing of chloroplast ndhB encoded transcript shows divergence between closely related members of the grass family (poaceae). Plant Mol. Biol. 1995, 29, 679–684. [Google Scholar] [CrossRef]
- Maier, R.M.; Neckermann, K.; Igloi, G.L.; Kossel, H. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995, 251, 614–628. [Google Scholar] [CrossRef]
- Hiratsuka, J.; Shimada, H.; Whittier, R.; Ishibashi, T.; Sakamoto, M.; Mori, M.; Kondo, C.; Honji, Y.; Sun, C.-R.; Meng, B.-Y.; et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989, 217, 185–194. [Google Scholar] [CrossRef]
- Martín, M.; Funk, H.T.; Serrot, P.H.; Poltnigg, P.; Sabater, B. Functional characterization of the thylakoid Ndh complex phosphorylation by site-directed mutations in the ndhF gene. Biochim. Biophys. Acta BBA Bioenerg. 2009, 1787, 920–928. [Google Scholar] [CrossRef] [Green Version]
- Martínez, P.; López, C.; Roldán, M.; Sabater, B.; Martín, M. Plastid DNA of five ecotypes of Arabidopsis thaliana: Sequence of ndhG gene and maternal inheritance. Plant Sci. 1997, 123, 113–122. [Google Scholar] [CrossRef]
- Berger, S.; Ellersiek, U.; Westhoff, P.; Steinmuller, K. Studies on the expression of NDH-H, a subunit of the NAD(P)H-plastoquinone-oxidoreductase of higher plant chloroplasts. Planta 1993, 190, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Burrows, P.A.; Sazanov, L.A.; Svab, Z.; Maliga, P.; Nixon, P.J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998, 17, 868–871. [Google Scholar] [CrossRef]
- Barrett, C.F.; Freudenstein, J.V.; Li, J.; Mayfield-Jones, D.R.; Perez, L.; Santos, C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 2014, 31, 3095–3112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Hou, B.-W.; Niu, Z.-T.; Liu, W.; Xue, Q.-Y.; Ding, X.-Y. Comparative chloroplast genomes of photosynthetic orchids: Insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS ONE 2014, 9, e99016. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Chen, J.J.; Huang, Y.T.; Chan, M.T.; Daniell, H.; Chang, W.J.; Hsu, C.T.; Liao, D.C.; Wu, F.H.; Lin, S.Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 9040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakasugi, T.; Tsudzuki, J.; Ito, S.; Nakashima, K.; Tsudzuki, T.; Sugiura, M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 1994, 91, 9794–9798. [Google Scholar] [CrossRef] [Green Version]
- Braukmann, T.W.A.; Kuzmina, M.; Stefanovíc, S. Loss of all ndh genes in Gnetales and conifers: Extent and evolutionary significance for seed plant phylogeny. Curr. Genet. 2009, 55, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.S.; Wang, Y.N.; Liu, S.M.; Chaw, S.M. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 Cp Protein-coding genes of Gnetum parvifolium: Insights into CpDNA evolution and phylogeny of extant seed plants. Mol. Biol. Evol. 2007, 24, 1366–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirao, T.; Watanabe, A.; Kurita, M.; Kondo, T.; Takata, K. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: Diversified genomic structure of coniferous species. BMC Plant Biol. 2008, 8, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrot, P.H.; Sabater, B.; Martín, M. Activity, polypeptide and gene identification of thylakoid Ndh complex in trees: Potential physiological relevance of fluorescence assays. Physiol. Plant. 2012, 146, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Blazier, J.C.; Guisinger, M.M.; Jansen, R.K. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol. Biol. 2011, 76, 263–272. [Google Scholar] [CrossRef]
- Braukmann, T.W.A.; Stefanovíc, S. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol. Biol. 2012, 79, 5–20. [Google Scholar] [CrossRef]
- Peredo, E.L.; King, U.M.; Les, D.H. The plastid genome of Najas flexilis: Adaptation to submersed environments is accompanied by the complete loss of the NDH Complex in an Aquatic Angiosperm. PLoS ONE 2013, 8, e68591. [Google Scholar] [CrossRef] [Green Version]
- Ruhlman, T.A.; Chang, W.J.; Chen, J.J.; Huang, Y.T.; Chan, M.T.; Zhang, J.; Liao, D.C.; Blazier, J.C.; Jin, X.; Shih, M.C.; et al. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 2015, 15, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, W.-L.; Wang, R.-N.; Zhang, N.-Y.; Fan, W.-B.; Fang, M.-F.; Li, Z.-H. Molecular evolution of chloroplast genomes of orchid species: Insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, S.-S.; Fu, P.-C.; Zhou, X.-J.; Cheng, Y.-W.; Zhang, F.-Q.; Chen, S.-L.; Gao, Q.-B. The complete plastome sequences of seven species in Gentiana sect. Kudoa (Gentianaceae): Insights into plastid gene loss and molecular evolution. Front. Plant Sci. 2018, 9, 493. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Wang, J.; Jia, Y.; Li, W.; Fusheng Xu, F.; Wang, X. Comparative chloroplast genome analyses of species in Gentiana section Cruciata (Gentianaceae) and the development of authentication markers. Int. J. Mol. Sci. 2018, 19, 1962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mower, J.P.; Guo, W.; Partha, R.; Fan, W.; Levsen, N.; Wolff, K.; Jacqueline, M.; Nugent, J.M.; Pabón-Mora, N.; González, F. Plastomes from tribe Plantagineae (Plantaginaceae) reveal infrageneric structural synapormorphies and localized hypermutation for Plantago and functional loss of ndh genes from Littorella. Mol. Phylogenet. Evol. 2021, 162, 107217. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.G.; Barrett, C.F.; Gomez, M.S.; Lam, V.K.Y.; Henriquez, C.L.; Les, D.H.; Davis, J.I.; Cuenca, A.; Petersen, G.; Seberg, O.; et al. Plastid phylogenomics and molecular evolution of Alismatales. Cladistics 2016, 32, 160–178. [Google Scholar] [CrossRef]
- Maurya, S.; Darshetkar, A.M.; Datar, M.N.; Tamhankar, S.; Li, P.; Choudhary, R.K. Plastome data provide insights into intra and interspecific diversity and ndh gene loss in Capparis (Capparaceae). Phytotaxa 2020, 432, 206–220. [Google Scholar] [CrossRef]
- Fajardo, D.; Senalik, D.; Ames, M.; Zhu, H.; Steffan, S.A.; Harbut, R.; Polashock, J.; Vorsa, N.; Gillespie, E.; Kron, K.; et al. Complete Plastid Genome Sequence of Vaccinium macrocarpon: Structure, Gene Content, and Rearrangements Revealed by Next Generation Sequencing. Tree Gen. Genom. 2013, 9, 489–498. [Google Scholar] [CrossRef]
- Szczecinska, M.; Gomolinska, A.; Szkudlarz, P.; Sawick, J. Plastid and nuclear genomic resources of a relict and endangered plant species: Chamaedaphne calyculata (L.) Moench (Ericaceae). Turk. J. Bot. 2014, 38, 1229–1238. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Copetti, D.; Búrquez, A.; Bustamante, E.; Charboneau, J.L.M.; Luis, E.; Eguiarte, L.E.; Kumar, S.; Lee, H.O.; Lee, J.; et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): Loss of the ndh gene suite and inverted repeat. Am. J. Bot. 2015, 102, 1115–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabater, B.; Martín, M.; Schmitz-Linneweber, C.; Maier, R.M. Is clustering of plastid RNA editing sites a consequence of transitory loss of gene function?—Implications for past environmental and evolutionary events in plants. Perspect. Plant Ecol. Evol. Syst. 2002, 5, 81–90. [Google Scholar] [CrossRef]
- Marín, D.; Martín, M.; Serrot, P.; Sabater, B. Thermodynamic balance of photosynthesis and transpiration at increasing CO2 concentrations and rapid light fluctuations. BioSystems 2014, 116, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Martín, M.; Marín, D.; Serrot, P.H.; Sabater, B. Evolutionary reversion of editing sites of ndh genes suggests their origin in the Permian-Triassic before the increase of atmospheric CO2. Front. Ecol. Evol. 2015, 3, 81. [Google Scholar] [CrossRef] [Green Version]
- Zhe, M.; Zhang, L.; Liu, F.; Huang, Y.; Fan, W.; Yang, J.; Zhu, A. Plastid RNA editing reduction accompanied with genetic variations in Cymbidium, a genus with diverse lifestyle modes. Plant Divers. 2021, in press. [Google Scholar] [CrossRef]
- Ni, Z.; Ye, Y.; Bai, T.; Xu, M.; Xu, L.A. Complete chloroplast genome of Pinus massoniana (Pinaceae): Gene rearrangements, loss of ndh genes, and short inverted repeats contraction, expansion. Molecules 2017, 22, 1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeb, U.; Dong, W.-L.; Zhang, T.-T.; Wang, R.-N.; Shahzad, K.; Ma, X.-F.; Li, Z.-H. Comparative plastid genomics of Pinus species: Insights into sequence variations and phylogenetic relationships. J. Syst. Evol. 2020, 58, 118–132. [Google Scholar] [CrossRef]
- Sokołowska, J.; Fuchs, H.; Celínski, K. New insight into taxonomy of European mountain pines, Pinus mugo complex, based on complete chloroplast genomes sequencing. Plants 2021, 10, 1331. [Google Scholar] [CrossRef]
- Casano, L.M.; Lascano, H.R.; Martín, M.; Sabater, B. Topology of the plastid Ndh complex and its NDH-F subunit in thylakoid membranes. Biochem. J. 2004, 382, 145–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lennon, A.M.; Prommeenate, P.; Nixon, P.J. Location, expression, and orientation of the putative chlororespiratory enzymes, Ndh, and IMMUTANS, in higher plant plastids. Planta 2003, 218, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Cuello, J.; Quiles, M.J.; Albacete, M.E.; Sabater, B. Properties of a large complex of NADH dehydrogenase from barley leaves. Plant Cell Physiol. 1995, 36, 265–271. [Google Scholar] [CrossRef]
- Corneille, S.; Courmac, L.; Guedeney, G.; Havaux, M.; Peltier, G. Reduction of the plastoquinone pool by exogenous NADH and NADPH in higher plant chloroplasts. Characterization of a NAD(P)H-plastoquinone oxidoreductase activity. Biochim. Biophys. Acta BBA Bioenerg. 1998, 1363, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Díaz, M.; de Haro, V.; Muñoz, R.; Quiles, M.J. Chlororespiration is involved in the adaptation of Brassica plants to heat and high light intensity. Plant Cell Environ. 2007, 30, 1578–1585. [Google Scholar] [CrossRef]
- Joët, T.; Cournac, L.; Peltier, G.; Havaux, M. Cyclic electron flow around Photosystem I in C3 plants. In vivo control by the redox state of chloroplast and involvement of the NADH-dehydrogenase complex. Plant Physiol. 2002, 128, 760–769. [Google Scholar] [CrossRef]
- Rumeau, D.; Peltier, G.; Cournac, L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007, 30, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.; Marín, D.; Serrot, P.H.; Sabater, B. The rise of the photosynthetic rate when light intensity increases is delayed in ndh gene-defective tobacco at high but not at low CO2 concentrations. Front. Plant Sci. 2015, 6, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heber, U.; Walker, D. Concerning a dual function of coupled cyclic electron transport in leaves. Plant Physiol. 1992, 100, 1621–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casano, L.M.; Martín, M.; Sabater, B. Sensitivity of superoxide dismutase transcript levels and activities to oxidative stress is lower in mature-senescent than in young barley leaves. Plant Physiol. 1994, 106, 1033–1039. [Google Scholar] [CrossRef] [Green Version]
- Casano, L.M.; Martín, M.; Zapata, J.M.; Sabater, B. Leaf age- and paraquat concentration-dependent effects on the levels of enzymes protecting against photooxidative stress. Plant Sci. 1999, 149, 13–22. [Google Scholar] [CrossRef]
- Abarca, D.; Martín, M.; Sabater, B. Differential leaf stress responses in young and senescent plants. Physiol. Plant. 2001, 113, 409–415. [Google Scholar] [CrossRef]
- Abarca, D.; Roldán, M.; Martín, M.; Sabater, B. Arabidopsis thaliana ecotype Cvi shows an increased tolerance to photo-oxidative stress and contains a new chloroplastic copper/zinc superoxide dismutase isoenzyme. J. Exp. Bot. 2001, 52, 1417–1425. [Google Scholar] [CrossRef] [Green Version]
- Zapata, J.M.; Guera, A.; Esteban-Carrasco, A.; Martín, M.; Sabater, B. Chloroplasts regulate leaf senescence: Delayed senescence in transgenic ndhF-defective tobacco. Cell Death Differ. 2005, 12, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Sabater, B.; Martín, M. Chloroplast control of leaf senescence. In Plastid Development in Leaves during Growth and Senescence. Advances in Photosynthesis and Respiration; Biswal, B., Krupinska, K., Biswal, U., Eds.; Springer Science+Business Media: Dordrecht, The Netherlands, 2013; Volume 36, pp. 529–550. [Google Scholar] [CrossRef]
- Sabater, B.; Martín, M. Hypothesis: Increase of the ratio singlet oxygen plus superoxide radical to hydrogen peroxide changes stress defense response to programmed leaf death. Front. Plant Sci. 2013, 4, 479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabater, B.; Martín, M. Control of leaf cell death by reactive oxygen species. In Programmed Cell Death in Plants and Animals; Rice, J., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2016; Chapter 3; pp. 75–93. ISBN 978-1-63484-505-2. [Google Scholar]
- Yamamoto, H.; Peng, L.; Fukao, Y.; Shikanai, T. A Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex of Arabidopsis. Plant Cell 2011, 23, 1480–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurisu, G.; Zhang, H.; Smith, J.L.; Cramer, W.A. Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science 2003, 302, 1009–1014. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.R.; Diaz, Y.C.; Penha, H.A.; Pinheiro, D.G.; Fernandes, C.C.; Miranda, V.F.; Michael, T.P.; Varani, A.M. The Chloroplast Genome of Utricularia reniformis Sheds Light on the Evolution of the ndh Gene Complex of Terrestrial Carnivorous Plants from the Lentibulariaceae Family. PLoS ONE 2016, 11, e0165176. [Google Scholar] [CrossRef]
- Gruzdev, E.V.; Kadnikov, V.V.; Beletsky, A.V.; Kochieva, E.Z.; Mardanov, A.V.; Skryabin, K.G.; Ravin, N.V. Plastid genomes of carnivorous plants Drosera rotundifolia and Nepenthes × ventrata reveal evolutionary patterns resembling those observed in parasitic plants. Int. J. Mol. Sci. 2019, 20, 4107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horváth, E.M.; Peter, S.O.; Joët, T.; Rumeau, D.; Cournac, L.; Horváth, G.V.; Kavanagh, T.A.; Schäfer, C.; Peltier, G.; Medgyesy, P. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 2000, 123, 1337–1349. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Geng, Q.; Du, Y.; Yang, X.; Zhai, H. Induction of cyclic electron flow around photosystem I during heat stress in grape leaves. Plant Sci. 2017, 256, 65–71. [Google Scholar] [CrossRef]
- Strand, D.D.; Fisher, N.; Kramer, D.M. The higher plant plastid NAD(P)H dehydrogenase-like complex (NDH) is a high efficiency proton pump that increases ATP production by cyclic electron flow. J. Biol. Chem. 2017, 292, 11850–11860. [Google Scholar] [CrossRef] [Green Version]
- Paredes, M.; Quiles, M.J. Stimulation of chlororespiration by drought under heat and high illumination in Rosa meillandina. J. Plant Physiol. 2013, 170, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.V.; Quiles, M.J. Involvement of chlororespiration in chilling stress in the tropical species Spathiphyllum wallisü. Plant Cell Environ. 2015, 38, 525–533. [Google Scholar] [CrossRef]
- Li, Q.; Yao, Z.J.; Mi, H. Alleviation of photoinhibition by co-ordination of chlororespiration and cyclic electron flow mediated by NDH under heat stressed condition in tobacco. Front. Plant Sci. 2016, 7, 285. [Google Scholar] [CrossRef] [Green Version]
- Alafari, H.A.; Abd-Elgawadab, M.E. Differential expression gene/protein contribute to heat stress-responsive in Tetraena propinqua in Saudi Arabia. Saudi J. Biol. Sci. 2021, 28, 5017–5027. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, Q.S.; Zhao, W.; Liu, Z.; Ma, M.Y.; Zhong, M.Y.; Wang, M.X.; Xu, B. Chlororespiration serves as photoprotection for the photo-inactivated oxygen-evolving complex in Zostera marina, a marine angiosperm. Plant Cell Physiol. 2020, 61, 1517–1529. [Google Scholar] [CrossRef]
- Zhao, J.; Yu, W.; Zhang, L.; Liu, J. Chlororespiration protects the photosynthetic apparatus against photoinhibition by alleviating inhibition of photodamaged-PSII repair in Haematococcus pluvialis at the green motile stage. Algal Res. 2021, 54, 102140. [Google Scholar] [CrossRef]
- Sun, Y.; Deng, T.; Zhang, A.; Moore, M.J.; Landis, J.B.; Lin, N.; Zhang, H.; Zhang, X.; Huang, J.; Zhang, X.; et al. Genome sequencing of the endangered Kingdonia uniflora (Circaeasteraceae, Ranunculales) reveals potential mechanisms of evolutionary specialization. iScience 2020, 23, 101124. [Google Scholar] [CrossRef]
- Folk, R.A.; Sewnath, N.; Xiang, C.-L.; Sinn, B.T.; Guralnick, R.P. Degradation of key photosynthetic genes in the critically endangered semi-aquatic flowering plant Saniculiphyllum guangxiense (Saxifragaceae). BMC Plant Biol. 2020, 20, 324. [Google Scholar] [CrossRef]
- Su, Y.; Huang, L.; Wang, Z.; Wang, T. Comparative chloroplast genomics between the invasive weed Mikania micrantha and its indigenous congener Mikania cordata: Structure variation, identification of highly divergent regions, divergence time estimation, and phylogenetic analysis. Mol. Phylogenet. Evol. 2018, 126, 181–195. [Google Scholar] [CrossRef]
- Scobeyeva, V.A.; Artyushin, I.V.; Krinitsina, A.A.; Nikitin, P.A.; Antipin, M.I.; Kuptsov, S.V.; Belenikin, M.S.; Omelchenko, D.O.; Logacheva, M.D.; Konorov, E.A.; et al. Gene loss, pseudogenization in plastomes of genus Allium (Amaryllidaceae), and putative selection for adaptation to environmental conditions. Front. Genet. 2021, 12, 674783. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, Z.; Sablok, G.; Daskalova, E.; Zahmanova, G.; Apostolova, E.; Yahubyan, G.; Baev, V. Chloroplast genome analysis of resurrection tertiary relict Haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 2017, 8, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, D.-F.; Yu, Y.; Deng, Y.-Q.; Li, J.; Liu, H.-Y.; Dong Zhou, S.-D.; He, X.-J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.-L.; Fan, W.-B.; Wang, N.; Dong, P.-B.; Zhang, T.-T.; Yue, M.; Li, Z.-H. Evolutionary analysis of plastid genomes of seven Lonicera L. species: Implications for sequence divergence and phylogenetic relationships. Int. J. Mol. Sci. 2018, 19, 4039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Zhong, S.; Zhang, M.; Liang, Y.; Gong, G.; Chang, X.; Tan, F.; Yang, H.; Qiu, X.; Luo, L.; et al. Potential role of photosynthesis in the regulation of reactive oxygen species and defence responses to Blumeria graminis f. sp. tritici in wheat. Int. J. Mol. Sci. 2020, 21, 5767. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 2007, 51, 941–954. [Google Scholar] [CrossRef]
- Reape, T.J.; Molony, E.M.; McCabe, P.F. Programmed cell death in plants: Distinguishing between different modes. J. Exp. Bot. 2008, 59, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Doyle, S.M.; Diamond, M.; McCabe, P.F. Chloroplast and reactive oxygen species involvement in apoptotic-like programmed cell death in Arabidopsis suspension cultures. J. Exp. Bot. 2010, 61, 473–482. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.S.; Braun, D.M. Genetic analyses of cell death in maize (Zea mays, poaceae) leaves reveal a distinct pathway operating in the camouflage1 mutant. Am. J. Bot. 2010, 97, 357–364. [Google Scholar] [CrossRef]
- Nilo, R.; Saffie, C.; Lilley, K.; Baeza-Yates, R.; Cambiazo, V.; Campos-Vargas, R.; González, M.; Meisel, L.A.; Retamales, J.; Silva, H.; et al. Proteomic analysis of peach fruit mesocarp softening and chilling injury using difference gel electrophoresis (DIGE). BMC Genom. 2010, 11, 43. [Google Scholar] [CrossRef] [Green Version]
- Mayta, M.L.; Hajirezaei, M.-R.; Carrillo, N.; Lodeyro, A.F. Leaf senescence: The chloroplast connection comes of age. Plants 2019, 8, 495. [Google Scholar] [CrossRef] [Green Version]
- McCoy, S.R.; Kuehl, J.V.; Boore, J.L.; Raubeson, L.A. The complete plastid genome sequence of Welwitschia mirabilis: An unusually compact plastome with accelerated divergence rates. BMC Evol. Biol. 2008, 8, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gechev, T.S.; Van Breusegem, F.; Stone, J.M.; Denev, J.; Laloi, C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 2006, 28, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Mubarakshina, M.M.; Ivanov, B.N.; Naydov, I.A.; Hillier, W.; Badger, M.R.; Krieger-Liszkay, A. Production and diffusion of chloroplastic H2O2 and its implication to signalling. J. Exp. Bot. 2010, 61, 3577–3587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiciarz, M.; Gubernator, B.; Kruk, J.; Niewiadomska, E. Enhanced chloroplastic generation of H2O2 in stress resistant Thellungiella salsuginea in comparison to Arabidopsis thaliana. Physiol. Plant. 2015, 153, 467–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowler, C.; Van Montagu, M.; Inzé, D. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Kurepa, J.; Hérouart, D.; Van Montagu, M.; Inzé, D. Differential expression of CuZn- and Fe-superoxide dismutase genes of tobacco during development, oxidative stress and hormonal treatments. Plant Cell Physiol. 1997, 38, 463–470. [Google Scholar] [CrossRef] [Green Version]
- Prochazkova, D.; Sairam, R.K.; Srivastava, G.C.; Singh, D.V. Oxidative stress and antioxidant activity as the basis of senescence in maize. Plant Sci. 2001, 161, 765–771. [Google Scholar] [CrossRef]
- Ohe, M.; Rapolu, M.; Mieda, T.; Miyagawa, Y.; Yabuta, Y.; Yoshimura, K.; Shigeoka, S. Decline of leaf photooxidative stress tolerance with age in tobacco. Plant Sci. 2005, 168, 1487–1493. [Google Scholar] [CrossRef]
- Orr, W.C.; Sohal, R.S. Extension of lifespan by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science 1994, 263, 1128–1130. [Google Scholar] [CrossRef]
- Lascano, H.R.; Casano, L.M.; Martín, M.; Sabater, B. The activity of the chloroplastic Ndh complex is regulated by phosphorylation of the NDH-F subunit. Plant Physiol. 2003, 132, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Nashilevitz, S.; Melamed-Bessudo, C.; Izkovich, Y.; Rogachev, I.; Osorio, S.; Itkin, M.; Adato, A.; Pankratov, J.; Hirschberg, J.; Femie, A.R.; et al. An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. Plant. Cell 2010, 22, 1977–1997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieger-Liszkay, A.; Trosch, M.; Krupinska, K. Generation of reactive oxygen species in thylakoids from senescing flag leaves of the barley varieties Lomerit and Carina. Planta 2015, 241, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Kane, D.J.; Sarafian, T.A.; Anton, R.; Hahn, H.; Gralla, E.B.; Valentine, J.S.; Örd, T.; Bredesen, D.E. Bcl-2 inhibition of neural death: Decreased generation of reactive oxygen species. Science 1993, 262, 1274–1277. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef]
- Yu, Z.-H.; Perdue, T.D.; Heimer, Y.M.; Jones, A.M. Mitochondrial involvement in tracheary element programmed cell death. Cell Death Differ. 2002, 9, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef]
- Ruiz-Pesini, E.; Mishmar, D.; Brandon, M.; Procaccio, V.; Wallace, D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 2004, 303, 223–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Ndh Gene | Encoded Polypeptide | Homologous Polypeptides in Respiratory Complex I (References) |
|---|---|---|
| ndhA | NDH-A | ND1/NuoH/FpoH, EchB, NQ08 [11,13] |
| ndhB | NDH-B | ND2/NuoN/FpoO, NQO14 [13,14,15] |
| ndhC | NDH-C | ND3/NuoA/FpoA, NQ07 [14,15,16] |
| ndhD | NDH-D | ND4/NuoM/FpoM, NQ013 [13] |
| ndhE | NDH-E | ND4L/NuoK/FpoK, NQ011 [13,15,16] |
| ndhF | NDH-F | ND5/NuoL/FpoL, NQ012 [13,17] |
| ndhG | NDH-G | ND6/NuoJ/FpoJ, NQ010 [13,18] |
| ndhH | NDH-H | 49(IP)/NuoD/FpoD, EchE, NQ05 [13,19,20] |
| ndhI | NDH-I | TYKY/NuoI/FpoI, EchF, NQ09 [13,20] |
| ndhJ | NDH-J | 30(IP)/NuoC/FpoC, EchD, NQ04 [13,20] |
| ndhK | NDH-K | PSTT/NuoB/FpoB, EchC, NQ06 [13,20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabater, B. On the Edge of Dispensability, the Chloroplast ndh Genes. Int. J. Mol. Sci. 2021, 22, 12505. https://doi.org/10.3390/ijms222212505
Sabater B. On the Edge of Dispensability, the Chloroplast ndh Genes. International Journal of Molecular Sciences. 2021; 22(22):12505. https://doi.org/10.3390/ijms222212505
Chicago/Turabian StyleSabater, Bartolomé. 2021. "On the Edge of Dispensability, the Chloroplast ndh Genes" International Journal of Molecular Sciences 22, no. 22: 12505. https://doi.org/10.3390/ijms222212505






