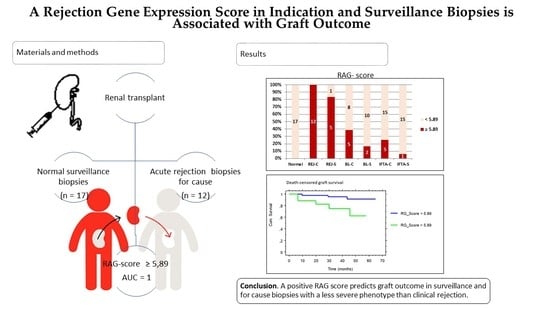
A Rejection Gene Expression Score in Indication and Surveillance Biopsies Is Associated with Graft Outcome
Abstract
:1. Introduction
2. Results
2.1. Patients
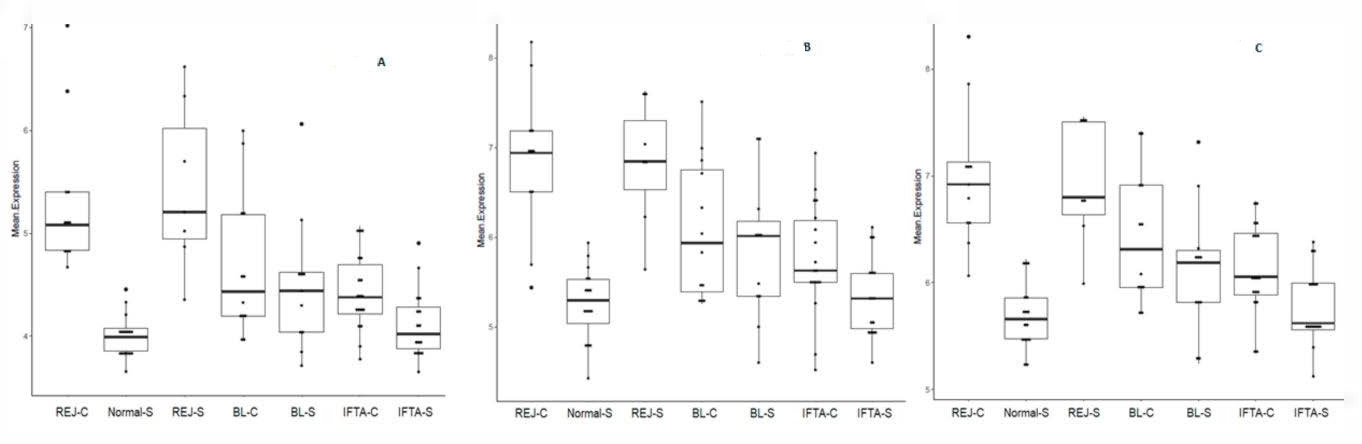
2.2. Principal Component Analysis Using Gene Transcripts
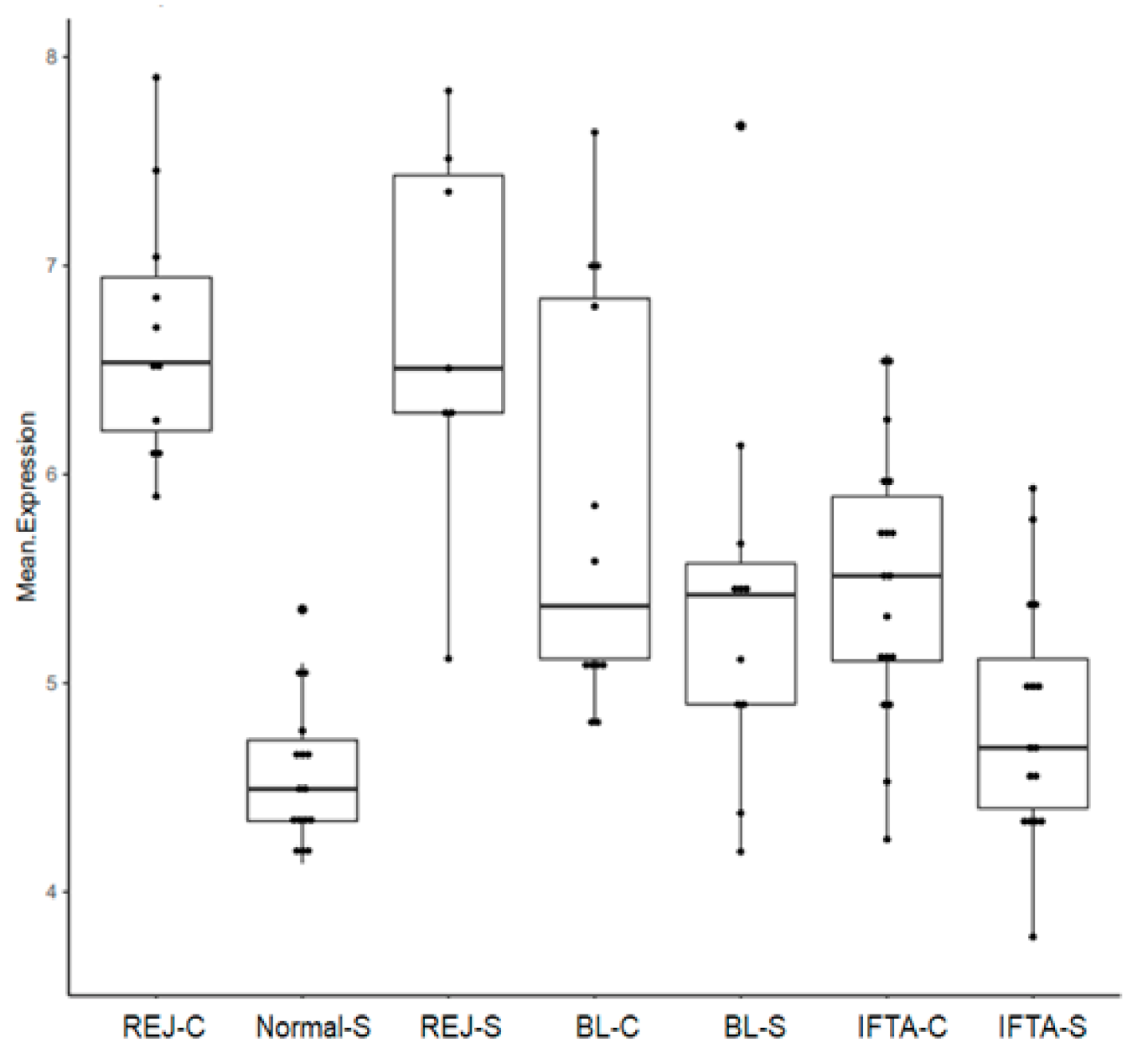
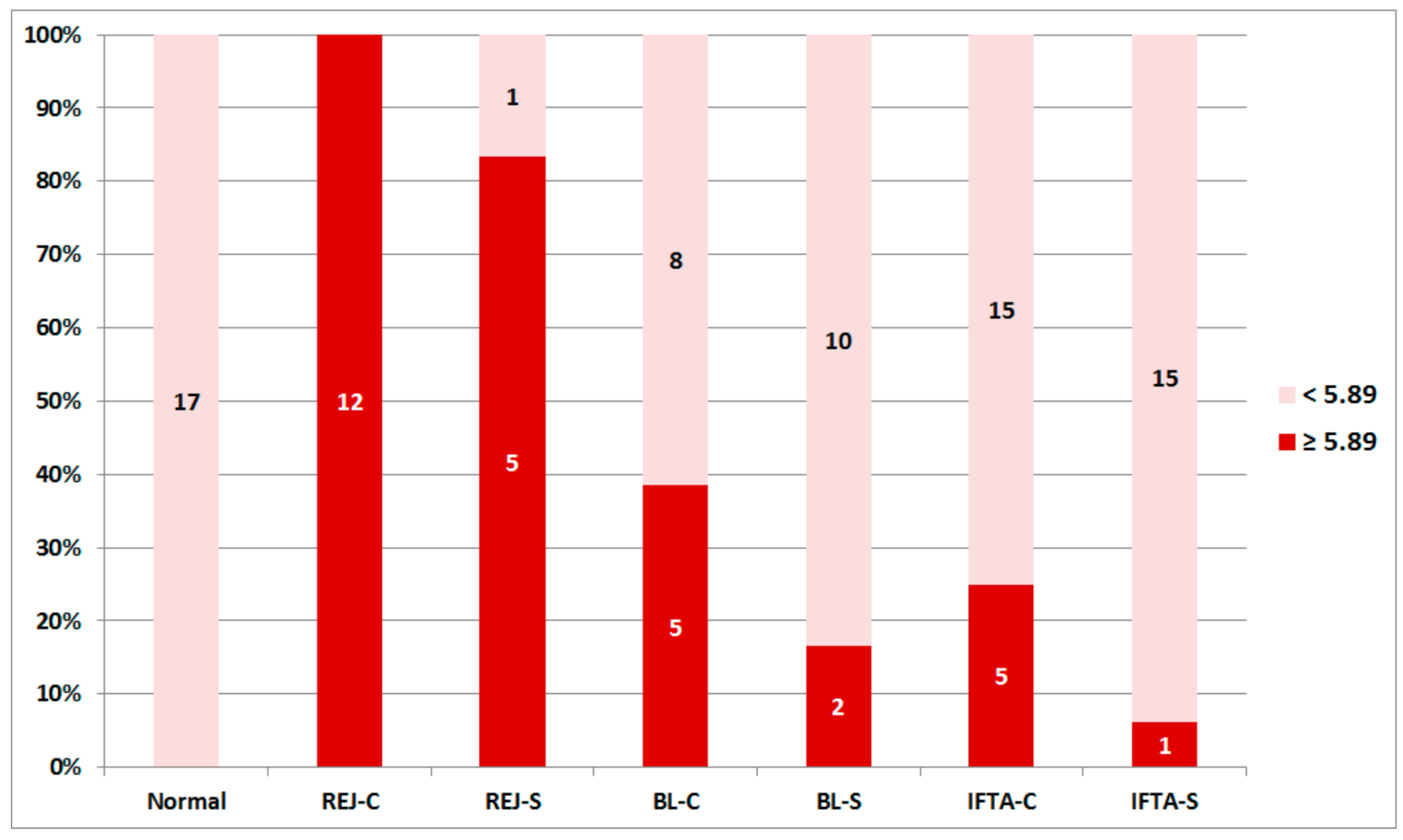
2.3. Rejection-Associated Gene Score (RAG Score)
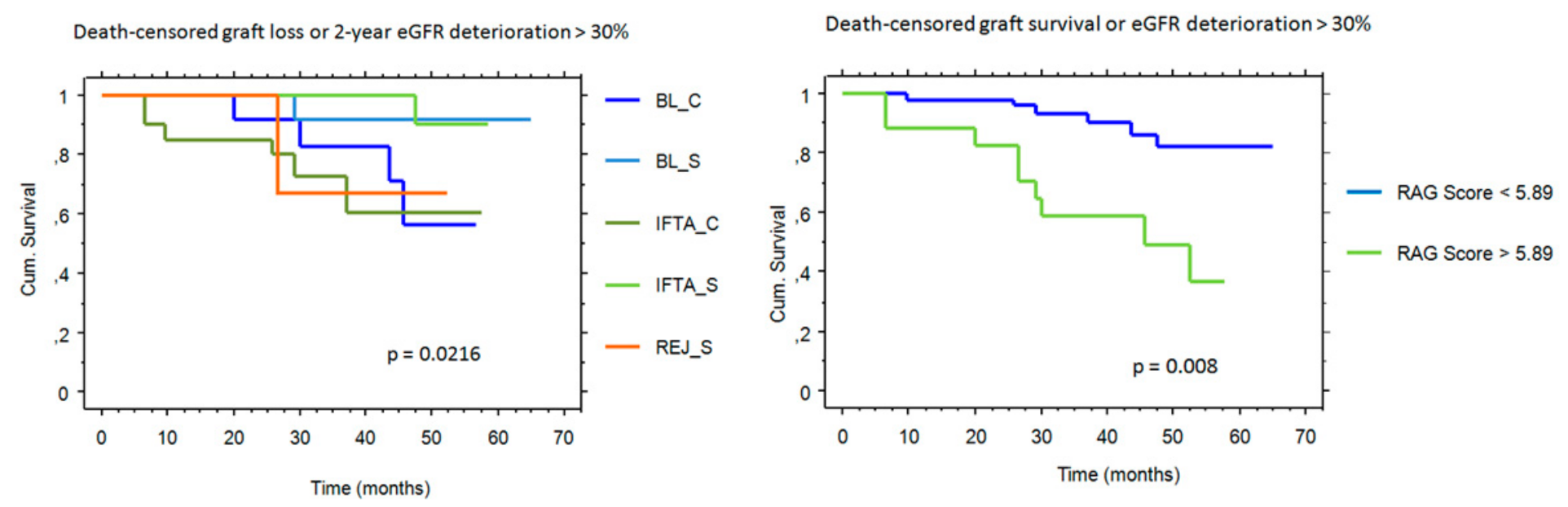
2.4. RAG Score and Outcome
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Biopsies
4.3. RNA Extraction and Microarray Hybridization
4.4. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IFTA | Interstitial fibrosis and tubular atrophy |
| BL-C | Borderline changes in biopsies for cause |
| BL-S | Borderline changes in surveillance biopsies |
| IFTA-C | Interstitial fibrosis and tubular atrophy in biopsies for cause |
| IFTA-S | Interstitial fibrosis and tubular atrophy in surveillance biopsies |
| REJ-C | Rejection in biopsies for cause |
| REJ-S | Rejection in surveillance biopsies |
| RAG | Rejection-associated gene score |
References
- Mueller, T.F.; Einecke, G.; Reeve, J.; Sis, B.; Mengel, M.; Jhangri, G.S.; Bunnag, S.; Cruz, J.; Wishart, D.S.; Meng, C.; et al. Microarray Analysis of Rejection in Human Kidney Transplants Using Pathogenesis-Based Transcript Sets. Arab. Archaeol. Epigr. 2007, 7, 2712–2722. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Pereira, A.B.; Chang, J.; Matas, A.; Picton, M.; De Freitas, D.; Bromberg, J.; Seron, D.; Sellarés, J.; Einecke, G.; et al. Potential Impact of Microarray Diagnosis of T Cell-Mediated Rejection in Kidney Transplants: The INTERCOM Study. Arab. Archaeol. Epigr. 2013, 13, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Pereira, A.B.; Chang, J.; Matas, A.; Picton, M.; De Freitas, D.; Bromberg, J.; Seron, D.; Sellarés, J.; Einecke, G.; et al. Microarray Diagnosis of Antibody-Mediated Rejection in Kidney Transplant Biopsies: An International Prospective Study (INTERCOM). Arab. Archaeol. Epigr. 2013, 13, 2865–2874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatri, P.; Roedder, S.; Kimura, N.; De Vusser, K.; Morgan, A.A.; Gong, Y.; Fischbein, M.P.; Robbins, R.C.; Naesens, M.; Butte, A.J.; et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J. Exp. Med. 2013, 210, 2205–2221. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.; Worschech, A.; Marincola, F.M. The immunologic constant of rejection. Trends Immunol. 2008, 29, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Modena, B.D.; Kurian, S.M.; Gaber, L.W.; Waalen, J.; I Su, A.; Gelbart, T.; Mondala, T.S.; Head, S.R.; Papp, S.; Heilman, R.; et al. Gene Expression in Biopsies of Acute Rejection and Interstitial Fibrosis/Tubular Atrophy Reveals Highly Shared Mechanisms That Correlate with Worse Long-Term Outcomes. Arab. Archaeol. Epigr. 2016, 16, 1982–1998. [Google Scholar] [CrossRef] [Green Version]
- Moreso, F.; Ibernon, M.; Goma, M.; Carrera, M.; Fulladosa, X.; Hueso, M.; Gil-Vernet, S.; Cruzado, J.M.; Torras, J.; Grinyó, J.M.; et al. Subclinical Rejection Associated with Chronic Allograft Nephropathy in Protocol Biopsies as a Risk Factor for Late Graft Loss. Arab. Archaeol. Epigr. 2006, 6, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Scholten, E.M.; Rowshani, A.T.; Cremers, S.; Bemelman, F.J.; Eikmans, M.; Van Kan, E.; Mallat, M.J.; Florquin, S.; Surachno, J.; Berge, I.J.T.; et al. Untreated Rejection in 6-Month Protocol Biopsies Is Not Associated with Fibrosis in Serial Biopsies or with Loss of Graft Function. J. Am. Soc. Nephrol. 2006, 17, 2622–2632. [Google Scholar] [CrossRef] [Green Version]
- Loupy, A.; Vernerey, D.; Tinel, C.; Aubert, O.; Van Huyen, J.-P.D.; Rabant, M.; Verine, J.; Nochy, D.; Empana, J.-P.; Martinez, F.; et al. Subclinical Rejection Phenotypes at 1 Year Post-Transplant and Outcome of Kidney Allografts. J. Am. Soc. Nephrol. 2015, 26, 1721–1731. [Google Scholar] [CrossRef] [Green Version]
- Kurian, S.M.; Velazquez, E.; Thompson, R.; Whisenant, T.; Rose, S.; Riley, N.; Harrison, F.; Gelbart, T.; Friedewald, J.; Charette, J.; et al. Orthogonal Comparison of Molecular Signatures of Kidney Transplants with Subclinical and Clinical Acute Rejection: Equivalent Performance is Agnostic to Both Technology and Platform. Arab. Archaeol. Epigr. 2017, 17, 2103–2116. [Google Scholar] [CrossRef]
- De Freitas, D.G.; Sellarés, J.; Mengel, M.; Chang, J.; Hidalgo, L.G.; Famulski, K.S.; Sis, B.; Einecke, G.; Halloran, P.F. The Nature of Biopsies with “Borderline Rejection” and Prospects for Eliminating This Category. Arab. Archaeol. Epigr. 2011, 12, 191–201. [Google Scholar] [CrossRef]
- Solez, K.; Axelsen, R.A.; Benediktsson, H.; Burdick, J.F.; Cohen, A.H.; Colvin, R.B.; Croker, B.P.; Droz, D.; Dunnill, M.S.; Halloran, P.F.; et al. International standardization of criteria for the histologic diagnosis of renal allograft rejection: The Banff working classification of kidney transplant pathology. Kidney Int. 1993, 44, 411–422. [Google Scholar] [CrossRef] [Green Version]
- Becker, J.U.; Chang, A.; Nickeleit, V.; Randhawa, P.; Roufosse, C. Banff Borderline Changes Suspicious for Acute T Cell–Mediated Rejection: Where Do We Stand? Arab. Archaeol. Epigr. 2016, 16, 2654–2660. [Google Scholar] [CrossRef] [Green Version]
- McRae, M.; Bouchard-Boivin, F.; Béland, S.; Noël, R.; Côté, I.; Lapointe, I.; Lesage, J.; Latulippe, E.; Riopel, J.; Santoriello, D.; et al. Impact of the Current versus the Previous Diagnostic Threshold on the Outcome of Patients with Borderline Changes Suspicious for T Cell–mediated Rejection Diagnosed on Indication Biopsies. Transplantation 2018, 102, 2120–2125. [Google Scholar] [CrossRef] [Green Version]
- Loupy, A.; Haas, M.; Roufosse, C.; Naesens, M.; Adam, B.; Afrouzian, M.; Akalin, E.; Alachkar, N.; Bagnasco, S.; Becker, J.U.; et al. The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell– and antibody-mediated rejection. Arab. Archaeol. Epigr. 2020, 20, 2318–2331. [Google Scholar] [CrossRef]
- Nankivell, B.J.; Agrawal, N.; Sharma, A.; Taverniti, A.; P’Ng, C.H.; Shingde, M.; Wong, G.; Chapman, J.R. The clinical and pathological significance of borderline T cell–mediated rejection. Arab. Archaeol. Epigr. 2019, 19, 1452–1463. [Google Scholar] [CrossRef]
- Dale, L.; Brennan, C.; Batal, I.; Morris, H.; Jain, N.G.; Valeri, A.; Husain, S.A.; King, K.; Tsapepas, D.; Cohen, D.; et al. Treatment of borderline infiltrates with minimal inflammation in kidney transplant recipients has no effect on allograft or patient outcomes. Clin. Transplant. 2020, 34, e14019. [Google Scholar] [CrossRef]
- Seifert, M.E.; Yanik, M.V.; Feig, D.I.; Hauptfeld-Dolejsek, V.; Mroczek-Musulman, E.C.; Kelly, D.R.; Rosenblum, F.; Mannon, R.B. Subclinical inflammation phenotypes and long-term outcomes after pediatric kidney transplantation. Arab. Archaeol. Epigr. 2018, 18, 2189–2199. [Google Scholar] [CrossRef] [PubMed]
- Mengel, M.; Chang, J.; Kayser, D.; Gwinner, W.; Schwarz, A.; Einecke, G.; Broecker, V.; Famulski, K.; De Freitas, D.G.; Guembes-Hidalgo, L.; et al. The Molecular Phenotype of 6-Week Protocol Biopsies from Human Renal Allografts: Reflections of Prior Injury but Not Future Course. Arab. Archaeol. Epigr. 2010, 11, 708–718. [Google Scholar] [CrossRef]
- Boor, P.; Floege, J. Renal Allograft Fibrosis: Biology and Therapeutic Targets. Arab. Archaeol. Epigr. 2015, 15, 863–886. [Google Scholar] [CrossRef]
- Matas, A.J.; Helgeson, E.S.; Gaston, R.; Cosio, F.; Mannon, R.B.; Kasiske, B.L.; Hunsicker, L.; Gourishankar, S.; Rush, D.N.; Cecka, J.M.; et al. Inflammation in areas of fibrosis: The DeKAF prospective cohort. Arab. Archaeol. Epigr. 2020, 20, 2509–2521. [Google Scholar] [CrossRef] [PubMed]
- Haas, M.; Loupy, A.; Lefaucheur, C.; Roufosse, C.; Glotz, D.; Seron, D.; Nankivell, B.J.; Halloran, P.F.; Colvin, R.B.; Akalin, E.; et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell–mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Arab. Archaeol. Epigr. 2018, 18, 293–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venner, J.; Famulski, K.S.; Badr, D.; Hidalgo, L.G.; Chang, J.; Halloran, P.F. Molecular Landscape of T Cell-Mediated Rejection in Human Kidney Transplants: Prominence of CTLA4 and PD Ligands. Arab. Archaeol. Epigr. 2014, 14, 2565–2576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rush, D.N.; Nickerson, P.; Gough, J.; McKenna, R.; Grimm, P.; Cheang, M.; Trpkov, K.; Solez, K.; Jeffery, J. Beneficial effects of treatment of early subclinical rejection: A randomized study. J. Am. Soc. Nephrol. 1998, 9, 2129–2134. [Google Scholar] [PubMed]
- Rush, D.; Arlen, D.; Boucher, A.; Busque, S.; Cockfield, S.M.; Girardin, C.; Knoll, G.; Lachance, J.-G.; Landsberg, D.; Shapiro, J.; et al. Lack of Benefit of Early Protocol Biopsies in Renal Transplant Patients Receiving TAC and MMF: A Randomized Study. Arab. Archaeol. Epigr. 2007, 7, 2538–2545. [Google Scholar] [CrossRef]
- Lipman, M.L.; Shen, Y.; Jeffery, J.R.; Gough, J.; McKenna, R.M.; Grimm, P.C.; Rush, D.N. Immune-activation gene expression in clinically stable renal allograft biopsies: Molecular evidence for subclinical rejection1,2. Transplantation 1998, 66, 1673–1681. [Google Scholar] [CrossRef]
- Choi, B.S.; Shin, M.J.; Shin, S.J.; Kim, Y.S.; Choi, Y.J.; Kim, Y.-S.; Moon, I.S.; Kim, S.Y.; Koh, Y.B.; Bang, B.K.; et al. Clinical Significance of an Early Protocol Biopsy in Living-Donor Renal Transplantation: Ten-Year Experience at a Single Center. Arab. Archaeol. Epigr. 2005, 5, 1354–1360. [Google Scholar] [CrossRef]
- Nankivell, B.J.; Borrows, R.J.; Fung, C.L.-S.; O’Connell, P.J.; Allen, R.D.M.; Chapman, J.R. Natural History, Risk Factors, and Impact of Subclinical Rejection in Kidney Transplantation. Transplantation 2004, 78, 242–249. [Google Scholar] [CrossRef]
- García-Carro, C.; Dörje, C.; Åsberg, A.; Midtvedt, K.; Scott, H.; Reinholt, F.P.; Holdaas, H.; Seron, D.; Reisæter, A.V. Inflammation in Early Kidney Allograft Surveillance Biopsies with and without Associated Tubulointerstitial Chronic Damage as a Predictor of Fibrosis Progression and Development of de novo Donor Specific Antibodies. Transplantation 2017, 101, 1410–1415. [Google Scholar] [CrossRef] [Green Version]
- Wiebe, C.; Gibson, I.W.; Blydt-Hansen, T.D.; Karpinski, M.; Ho, J.; Storsley, L.; Goldberg, A.; Birk, P.E.; Rush, D.N.; Nickerson, P.W. Evolution and Clinical Pathologic Correlations of de novo Donor-Specific HLA Antibody Post Kidney Transplant. Arab. Archaeol. Epigr. 2012, 12, 1157–1167. [Google Scholar] [CrossRef]
- Hruba, P.; Brabcova, I.; Gueler, F.; Krejcik, Z.; Stranecky, V.; Svobodová, E.; Maluskova, J.; Gwinner, W.; Honsova, E.; Lodererova, A.; et al. Molecular diagnostics identifies risks for graft dysfunction despite borderline histologic changes. Kidney Int. 2015, 88, 785–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeve, J.; Einecke, G.; Mengel, M.; Sis, B.; Kayser, N.; Kaplan, B.; Halloran, P.F. Diagnosing Rejection in Renal Transplants: A Comparison of Molecular- and Histopathology-Based Approaches. Arab. Archaeol. Epigr. 2009, 9, 1802–1810. [Google Scholar] [CrossRef] [PubMed]
- Cosio, F.G.; Grande, J.P.; Wadei, H.; Larson, T.S.; Griffin, M.D.; Stegall, M.D. Predicting Subsequent Decline in Kidney Allograft Function from Early Surveillance Biopsies. Arab. Archaeol. Epigr. 2005, 5, 2464–2472. [Google Scholar] [CrossRef] [PubMed]
- Park, W.D.; Griffin, M.D.; Cornell, L.D.; Cosio, F.G.; Stegall, M.D. Fibrosis with Inflammation at One Year Predicts Transplant Functional Decline. J. Am. Soc. Nephrol. 2010, 21, 1987–1997. [Google Scholar] [CrossRef] [Green Version]
- Toki, D.; Zhang, W.; Hor, K.L.M.; Liuwantara, D.; Alexander, S.I.; Yi, Z.; Sharma, R.; Chapman, J.R.; Nankivell, B.J.; Murphy, B.; et al. The Role of Macrophages in the Development of Human Renal Allograft Fibrosis in the First Year after Transplantation. Arab. Archaeol. Epigr. 2014, 14, 2126–2136. [Google Scholar] [CrossRef]
- Maluf, D.G.; Mas, V.R.; Archer, K.J.; Yanek, K.; Gibney, E.M.; King, A.L.; Cotterell, A.; Fisher, R.A.; Posner, M.P. Molecular Pathways Involved in Loss of Kidney Graft Function with Tubular Atrophy and Interstitial Fibrosis. Mol. Med. 2008, 14, 276–285. [Google Scholar] [CrossRef]
- Mengel, M.; Reeve, J.; Bunnag, S.; Einecke, G.; Sis, B.; Mueller, T.; Kaplan, B.; Halloran, P.F. Molecular Correlates of Scarring in Kidney Transplants: The Emergence of Mast Cell Transcripts. Arab. Archaeol. Epigr. 2008, 9, 169–178. [Google Scholar] [CrossRef]
- Madill-Thomsen, K.; Perkowska-Ptasinska, A.; Böhmig, G.A.; Eskandary, F.; Einecke, G.; Gupta, G.; Halloran, P.F. The MMDx-Kidney study group Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Arab. Archaeol. Epigr. 2020, 20, 1341–1350. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]

| Available Biopsies | Included Biopsies | |||
|---|---|---|---|---|
| For Cause | Surveillance | For Cause | Surveillance | |
| Normal | 0 | 39 | 0 | 17 |
| Borderline changes | 19 | 18 | 13 | 12 |
| IFTA | 35 | 32 | 20 | 16 |
| TCMR | 6 | 5 | 6 a | 5 c |
| ABMR | 6 | 1 | 6 b | 1 d |
| Total | 66 | 95 | 45 | 51 |
| Variable | Normal | REJ-C | REJ-S | BL-C | BL-S | IFTA-C | IFTA-S | p-Value |
|---|---|---|---|---|---|---|---|---|
| N | 17 | 12 | 6 | 13 | 12 | 20 | 16 | |
| Age (y) | 54 ± 13 | 43 ± 12 | 53 ± 21 | 59 ± 11 b | 57 ± 13 b,c | 46 ± 15 c | 52 ± 17 | 0.060 |
| Sex (m/f) | 12/5 | 6/6 | 4/2 | 8/5 | 9/3 | 16/4 | 10/6 | 0.610 |
| 1st Tx/Re-Tx | 17/0 | 8/4 | 6/0 | 13/0 | 12/0 | 18/2 | 12/4 | 0.002 |
| Donor age (y) | 56 ± 15 | 44 ± 16 | 59 ± 15 | 59 ± 14 | 53 ± 16 | 58 ± 11 | 57 ± 16 | 0.226 |
| HLA (A + B + DR) mm | 3.8 ± 1.0 | 3.7 ± 1.4 | 4.0 ± 1.9 | 3.9 ± 1.0 | 3.8 ± 0.7 | 3.5 ± 1.3 | 3.7 ± 1.1 | 0.942 |
| DGF (no/yes) | 15/2 | 10/1 | 6/0 | 5/8 | 10/2 | 14/1 | 13/3 | 0.010 |
| Rejection (no/yes) | 17/0 | 8/4 | 4/2 | 12/1 | 9/3 | 19/1 | 14/2 | 0.082 |
| Induction (ATG/IL2RAb) | 4/13 | 7/5 | 2/4 | 4/15 | 7/9 | 4/14 | 7/9 | 0.295 |
| Immunosuppression TAC + MMF + S/other | 16/1 | 10/2 | 4/2 | 9/4 | 12/0 | 14/6 | 16/0 | 0.036 |
| Time biopsy (m) | 6 ± 5 | 51 ± 57 a | 6 ± 6 b | 25 ± 27 | 7 ± 4 b,d | 77 ± 78 a,c,e | 7 ± 5 b,f | <0.001 |
| Creatinine (mg/dL) | 1.3 ± 0.3 | 2.0 ± 0.6 a | 1.4 ± 0.2 | 2.5 ± 0.8 a,b,c | 1.3 ± 0.3 b,d | 2.1 ± 0.7 a,c,e | 1.4 ± 0.3 b,d,f | <0.001 |
| UPCR (g/g) | 0.4 ± 0.3 | 1.7 ± 1.5 a | 0.3 ± 0.1 b | 1.1 ± 1.2 a,b,c | 0.3 ± 0.2 b,d | 1.3 ± 0.9 a,c,e | 0.3 ± 0.3 b,d,f | <0.001 |
| DSA (no/yes) | 17/0 | 8/4 | 5/1 | 13/0 | 12/0 | 20/0 | 16/0 | <0.001 |
| Variable | Normal | REJ-C | REJ-S | BL-C | BL-S | IFTA-C | IFTA-S |
|---|---|---|---|---|---|---|---|
| N | 17 | 12 | 6 | 13 | 12 | 20 | 16 |
| Glomeruli (N) | 16 ± 8 | 16 ± 6 | 17 ± 5 | 17 ± 6 | 21 ± 11 | 17 ± 8 | 23 ± 12 |
| Gs (%) | 7 ± 7 | 22 ± 23 | 12 ± 16 | 22 ± 15 | 12 ± 14 | 31 ± 24 | 10 ± 6 |
| g | 0.1 ± 0.3 | 1.4 ± 1.1 | 0.8 ± 1.3 | 0.2 ± 0.4 | 0.4 ± 0.8 | 0.3 ± 0.6 | 0.2 ± 0.4 |
| i | 0.1 ± 0.2 | 1.3 ± 1.1 | 1.5 ± 0.5 | 1.2 ± 0.7 | 0.7 ± 0.6 | 0.1 ± 0.3 | 0.1 ± 0.2 |
| t | 0 | 1.2 ± 0.9 | 1.8 ± 0.7 | 1.0 ± 0 | 1.0 ± 0.6 | 0.2 ± 0.4 | 0.1 ± 0.3 |
| v | 0 | 0.3 ± 0.6 | 0.5 ± 0.5 | 0 | 0 | 0 | 0 |
| ah | 0.2 ± 0.4 | 1.4 ± 1.2 | 0 | 1.0 ± 0.9 | 0.7 ± 0.-6 | 1.3 ± 1.2 | 0.6 ± 0.6 |
| cg | 0 | 0.7 ± 0.9 | 0 | 0 | 0.2 ± 0.6 | 0.2 ± 0.5 | 0 |
| ci | 0.1 ± 0.3 | 1.2 ± 0.9 | 0.8 ± 1.0 | 1.3 ± 0.6 | 0.9 ± 0.8 | 1.8 ± 0.8 | 1.3 ± 0.5 |
| ct | 0.6 ± 0.5 | 1.1 ± 0.8 | 1.0 ± 0.6 | 1.2 ± 0.4 | 1.0 ± 0.4 | 1.6 ± 0.9 | 1.1 ± 0.3 |
| cv | 0.4 ± 0.7 | 0.8 ± 0.8 | 0.3 ± 0.5 | 1.0 ± 0.8 | 0.4 ± 0.5 | 1.3 ± 1.2 | 0.9 ± 0.8 |
| mm | 0.1 ± 0.2 | 0.5 ± 0.7 | 0 | 0.1 ± 0.3 | 0 | 0.2 ± 0.7 | 0 |
| ptc | 0.1 ± 0.2 | 1.4 ± 0.8 | 0.7 ± 1.2 | 0.6 ± 0.8 | 0.4 ± 0.8 | 0.5 ± 0.8 | 0.1 ± 0.2 |
| C4d | 0 | 0.3 ± 0.5 | 0 | 0 | 0 | 0 | 0 |
| i-IFTA | 0.4 ± 0.9 | 2.1 ± 1.3 | 2.2 ± 1.3 | 2.1 ± 0.8 | 1.3 ± 1.1 | 2.2 ± 1.0 | 1.8 ± 0.9 |
| t-IFTA | 0.2 ± 0.4 | 0.9 ± 0.6 | 0.8 ± 0.4 | 0.7 ± 0.7 | 0.6 ± 0.7 | 0.7 ± 0.6 | 0.7 ± 0.5 |
| i-total | 0.1 ± 0.2 | 1.3 ± 0.6 | 1.7 ± 0.6 | 1.5 ± 0.7 | 0.8 ± 0.6 | 0.7 ± 0.5 | 0.4 ± 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chamoun, B.; Caraben, A.; Torres, I.B.; Sellares, J.; Jiménez, R.; Toapanta, N.; Cidraque, I.; Gabaldon, A.; Perelló, M.; Gonzalo, R.; et al. A Rejection Gene Expression Score in Indication and Surveillance Biopsies Is Associated with Graft Outcome. Int. J. Mol. Sci. 2020, 21, 8237. https://doi.org/10.3390/ijms21218237
Chamoun B, Caraben A, Torres IB, Sellares J, Jiménez R, Toapanta N, Cidraque I, Gabaldon A, Perelló M, Gonzalo R, et al. A Rejection Gene Expression Score in Indication and Surveillance Biopsies Is Associated with Graft Outcome. International Journal of Molecular Sciences. 2020; 21(21):8237. https://doi.org/10.3390/ijms21218237
Chicago/Turabian StyleChamoun, Betty, Anna Caraben, Irina B. Torres, Joana Sellares, Raquel Jiménez, Néstor Toapanta, Ignacio Cidraque, Alejandra Gabaldon, Manel Perelló, Ricardo Gonzalo, and et al. 2020. "A Rejection Gene Expression Score in Indication and Surveillance Biopsies Is Associated with Graft Outcome" International Journal of Molecular Sciences 21, no. 21: 8237. https://doi.org/10.3390/ijms21218237