CX-5461 Inhibits Pancreatic Ductal Adenocarcinoma Cell Growth, Migration and Induces DNA Damage
Abstract
:1. Introduction
2. Results
2.1. CX-5461 Inhibits Pancreatic Cancer Cell Growth in Nanomolar Range
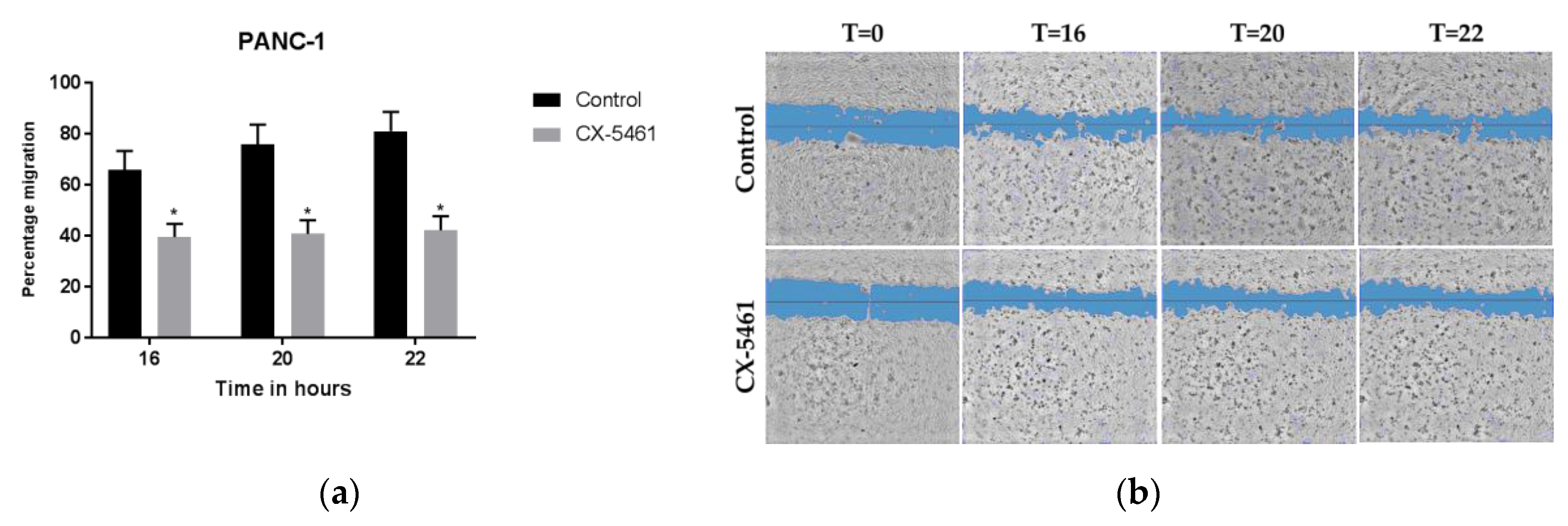
2.2. CX-5461 Inhibits PANC-1 Cell Migration
2.3. CX-5461 Induces mRNA Expression of EMT Markers
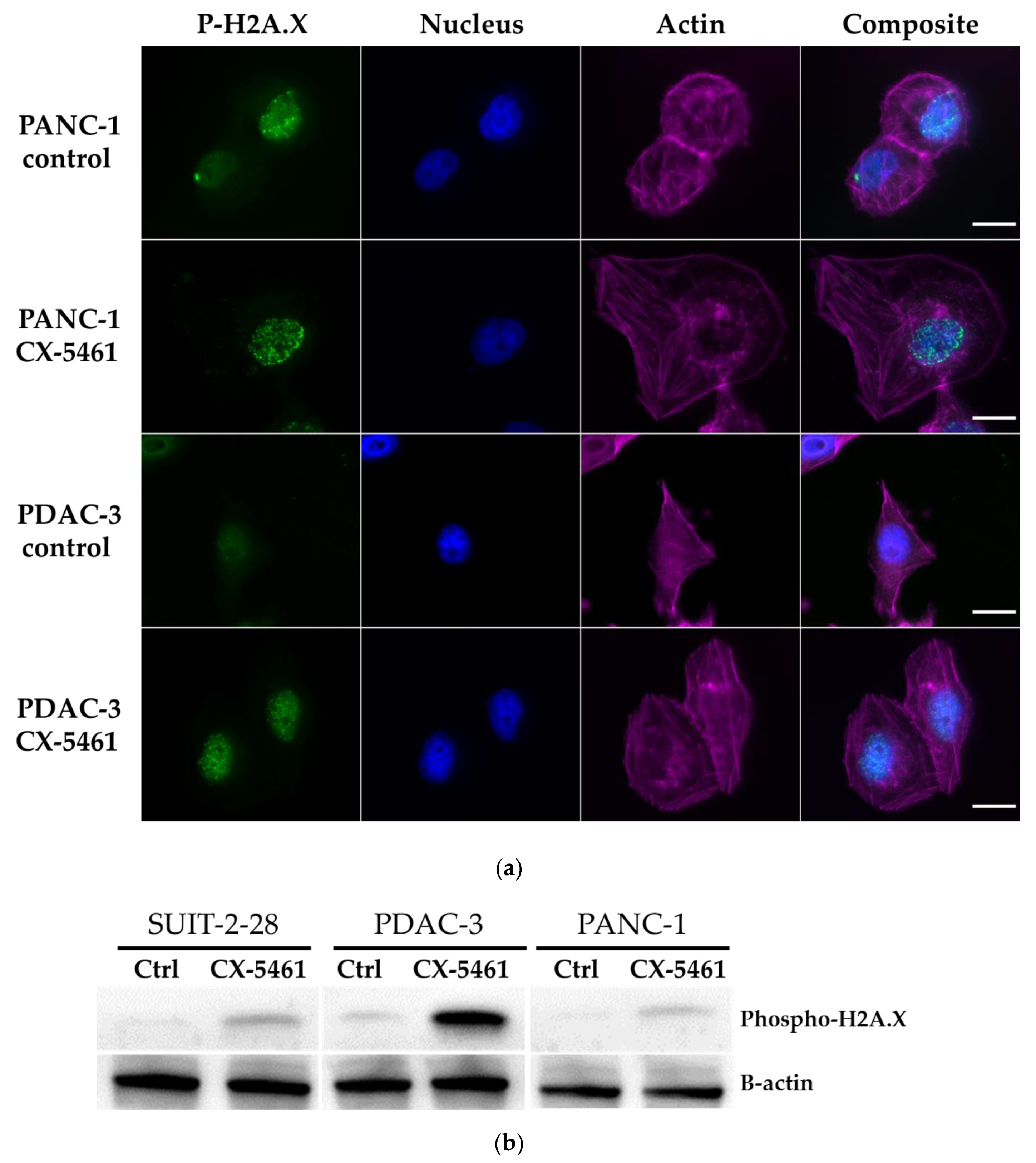
2.4. DNA Damage Induced by CX-5461
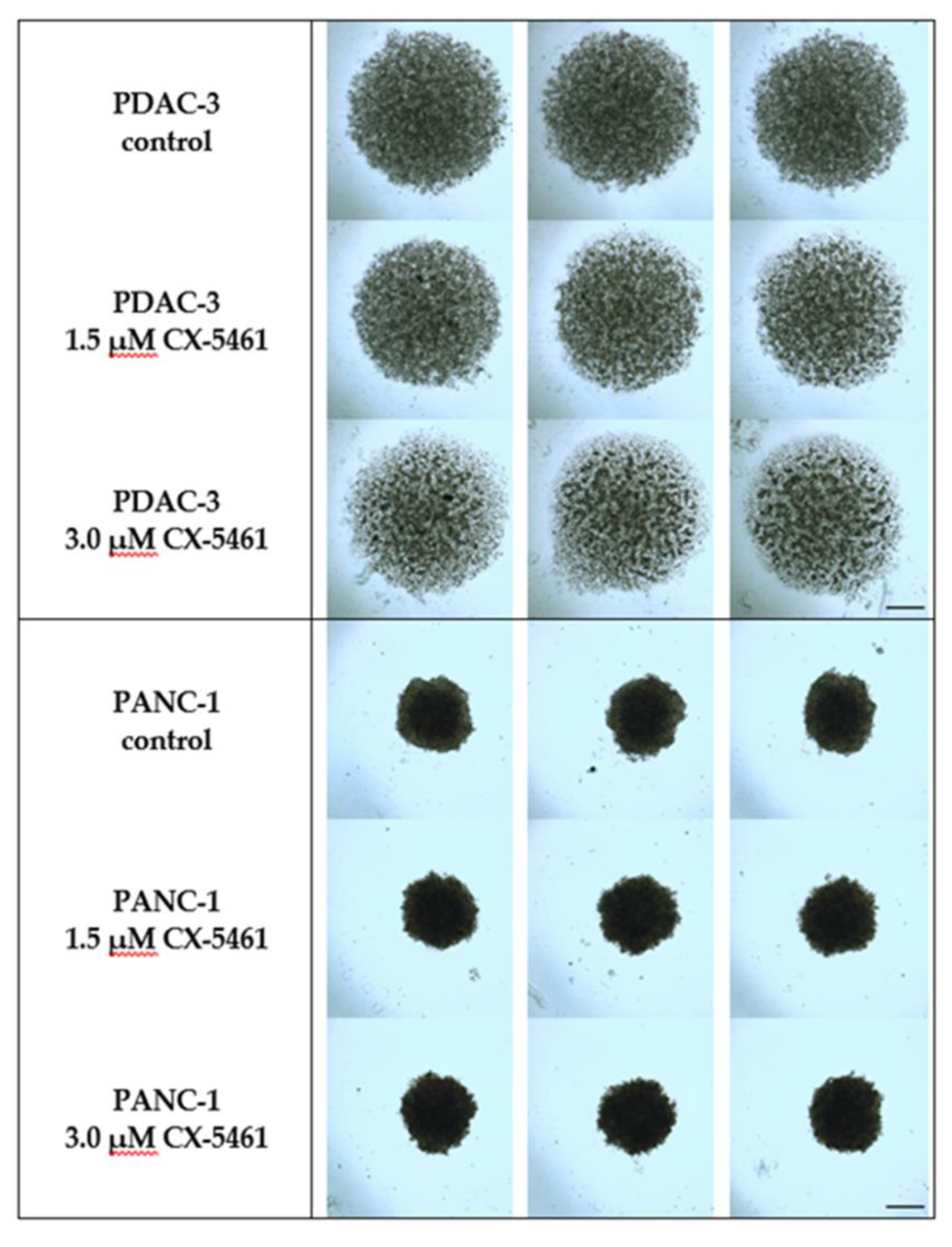
2.5. The Effect of CX-5461 in 3D Spheroid Models
2.6. Combination of CX-5461 and Gemcitabine Leads to An Antagonistic Effect
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Chemicals
4.3. Sulforhodamine B (SRB) Assay
4.4. Migration Assay
4.5. qRT-PCR
4.6. Immunofluoresent Staining and Imaging
4.7. Western Blot
4.8. Spheroid Formation
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019 (US statistics). CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsolic, I.; Jurada, D.; Pullen, N.; Oren, M.; Eliopoulos, A.G.; Volarevic, S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin. Cancer Biol. 2016, 37–38, 36–50. [Google Scholar] [CrossRef] [PubMed]
- El Hassouni, B.; Sarkisjan, D.; Vos, J.C.; Giovannetti, E.; Peters, G.J. Targeting the ribosome biogenesis key molecule fibrillarin to avoid chemoresistance. Curr. Med. Chem. 2019, 26, 6020–6032. [Google Scholar] [CrossRef]
- Bywater, M.J.; Poortinga, G.; Sanij, E.; Hein, N.; Peck, A.; Cullinane, C.; Wall, M.; Cluse, L.; Drygin, D.; Anderes, K.; et al. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell 2012, 22, 51–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef] [Green Version]
- Penzo, M.; Montanaro, L.; Treré, D.; Derenzini, M. The Ribosome Biogenesis—Cancer Connection. Cells 2019, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.; Yu, M.; Jia, J.K.; Li, W.X.; Tian, Y.W.; Xue, H.Z. Possible molecular markers for the diagnosis of pancreatic ductal adenocarcinoma. Med. Sci. Monitor 2018, 24, 2368–2376. [Google Scholar] [CrossRef] [Green Version]
- Khot, A.; Brajanovski, N.; Cameron, D.P.; Hein, N.; Maclachlan, K.H.; Sanij, E.; Lim, J.; Soong, J.; Link, E.; Blombery, P.; et al. First-in-Human RNA Polymerase I Transcription Inhibitor CX-5461 in Patients with Advanced Hematologic Cancers: Results of a Phase I Dose-Escalation Study. Cancer Discov. 2019, 9, 1036–1049. [Google Scholar] [CrossRef] [Green Version]
- Hald, Ø.H.; Olsen, L.; Gallo-Oller, G.; Elfman, L.H.M.; Løkke, C.; Kogner, P.; Sveinbjörnsson, B.; Flægstad, T.; Johnsen, J.I.; Einvik, C. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene 2019, 38, 2800–2813. [Google Scholar] [CrossRef]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Dos Santos, N.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- Ismael, M.; Webb, R.; Ajaz, M.; Kirkby, K.J.; Coley, H.M. The Targeting of RNA Polymerase I Transcription Using CX-5461 in Combination with Radiation Enhances Tumour Cell Killing Effects in Human Solid Cancers. Cancers 2019, 11, 1429. [Google Scholar] [CrossRef] [Green Version]
- Daemen, A.; Peterson, D.; Sahu, N.; McCord, R.; Du, X.; Liu, B.; Kowanetz, K.; Hong, R.; Moffat, J.; Gao, M.; et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. USA 2015, 112, E4410–E4417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannetti, E.; van der Borden, C.L.; Frampton, A.E.; Ali, A.; Firuzi, O.; Peters, G.J. Never let it go: Stopping key mechanisms underlying metastasis to fight pancreatic cancer. Semin. Cancer Biol. 2017, 44, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Prakash, V.; Carson, B.B.; Feenstra, J.M.; Dass, R.A.; Sekyrova, P.; Hoshino, A.; Petersen, J.; Guo, Y.; Parks, M.M.; Kurylo, C.M.; et al. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019, 10, 2110. [Google Scholar] [CrossRef] [PubMed]
- Bergman, A.M.; Eijk, P.P.; Ruiz Van Haperen, V.W.T.; Smid, K.; Veerman, G.; Hubeek, I.; Van Den Ijssel, P.; Ylstra, B.; Peters, G.J. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 2005, 65, 9510–9516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelison, R.; Dobbin, Z.C.; Katre, A.A.; Jeong, D.H.; Zhang, Y.; Chen, D.; Petrova, Y.; Llaneza, D.C.; Steg, A.D.; Parsons, L.; et al. Targeting RNA-polymerase I in both chemosensitive and chemoresistant populations in epithelial ovarian cancer. Clin. Cancer Res. 2017, 23, 6529–6540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caparello, C.; Meijer, L.L.; Garajova, I.; Falcone, A.; Le Large, T.Y.; Funel, N.; Kazemier, G.; Peters, G.J.; Vasile, E.; Giovannetti, E. FOLFIRINOX and translational studies: Towards personalized therapy in pancreatic cancer. World J. Gastroenterol. 2016, 22, 6987. [Google Scholar] [CrossRef]
- Reni, M.; Cereda, S.; Milella, M.; Novarino, A.; Passardi, A.; Mambrini, A.; Di Lucca, G.; Aprile, G.; Belli, C.; Danova, M.; et al. Maintenance sunitinib or observation in metastatic pancreatic adenocarcinoma: A phase II randomised trial. Eur. J. Cancer 2013, 49, 3609–3615. [Google Scholar] [CrossRef]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.-O.; Hochhauser, D.; Arnold, D.; Oh, D.-Y.; et al. Maintenance Olaparib for Germline BRCA -Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef]
- Buchholz, M.; Biebl, A.; Neeße, A.; Wagner, M.; Iwamura, T.; Leder, G.; Adler, G.; Gress, T.M. SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo. Cancer Res. 2003, 63, 4945–4951. [Google Scholar]
- Funel, N.; Giovannetti, E.; Del Chiaro, M.; Mey, V.; Pollina, L.E.; Nannizzi, S.; Boggi, U.; Ricciardi, S.; Del Tacca, M.; Bevilacqua, G.; et al. Laser microdissection and primary cell cultures improve pharmacogenetic analysis in pancreatic adenocarcinoma. Lab. Investig. 2008, 88, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Keepers, Y.P.; Pizao, P.E.; Peters, G.J.; van Ark-Otte, J.; Winograd, B.; Pinedo, H.M. Comparison of the sulforhodamine B protein and tetrazolium (MTT) assays for in vitro chemosensitivity testing. Eur. J. Cancer Clin. Oncol. 1991, 27, 897–900. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |
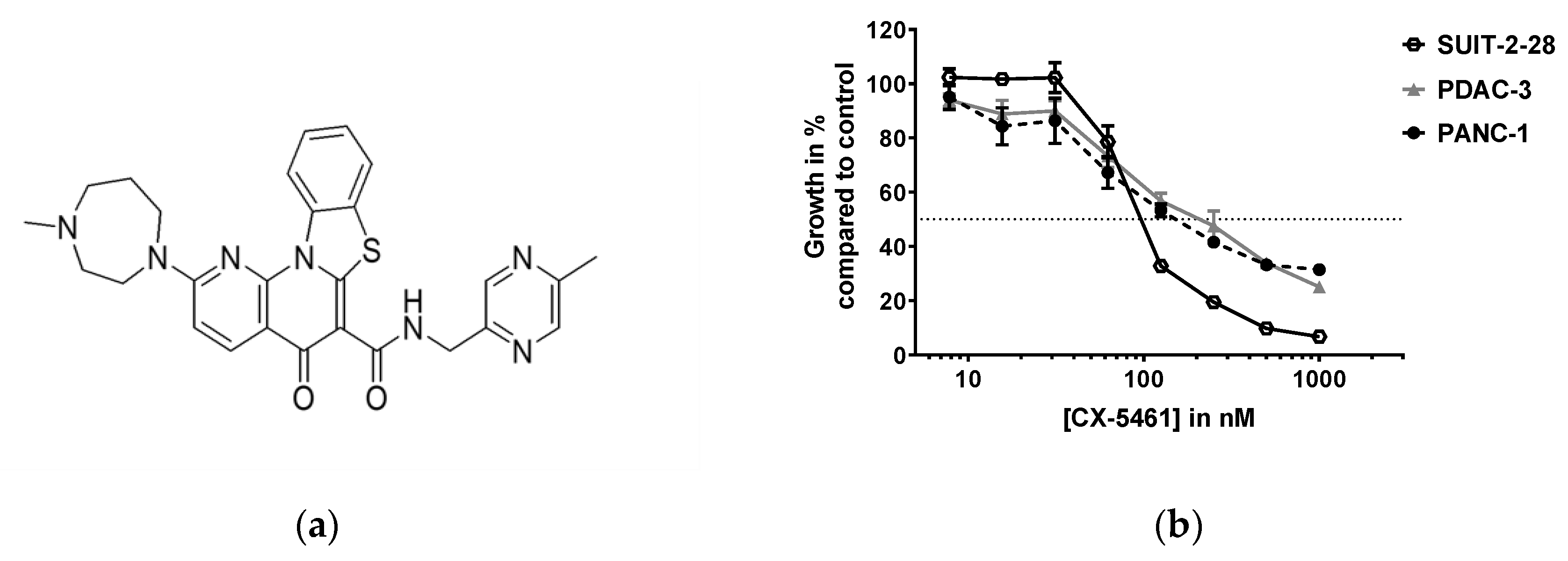

| Cell Line | Average IC50 (nM) ± SEM | Average Doubling Time in Hours ± SEM |
|---|---|---|
| SUIT-2-28 | 77 ± 14 | 16.4 ± 0.20 |
| PDAC-3 | 114 ± 30 | 18.8 ± 1.36 |
| PANC-1 | 199 ± 26 | 28.4 ± 1.91 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Hassouni, B.; Mantini, G.; Immordino, B.; Peters, G.J.; Giovannetti, E. CX-5461 Inhibits Pancreatic Ductal Adenocarcinoma Cell Growth, Migration and Induces DNA Damage. Molecules 2019, 24, 4445. https://doi.org/10.3390/molecules24244445
El Hassouni B, Mantini G, Immordino B, Peters GJ, Giovannetti E. CX-5461 Inhibits Pancreatic Ductal Adenocarcinoma Cell Growth, Migration and Induces DNA Damage. Molecules. 2019; 24(24):4445. https://doi.org/10.3390/molecules24244445
Chicago/Turabian StyleEl Hassouni, Btissame, Giulia Mantini, Benoît Immordino, Godefridus J. Peters, and Elisa Giovannetti. 2019. "CX-5461 Inhibits Pancreatic Ductal Adenocarcinoma Cell Growth, Migration and Induces DNA Damage" Molecules 24, no. 24: 4445. https://doi.org/10.3390/molecules24244445








