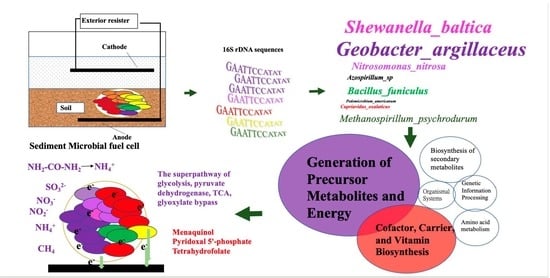
Functional Prediction of Microbial Communities in Sediment Microbial Fuel Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sediment MFCs and Sequence Processing
2.2. Functional Prediction of Microbial Communities
2.3. Statistical Analysis
3. Results
3.1. Functional Feature Prediction
3.2. Functional Features Related to Power Generation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slate, A.J.; Whitehead, K.A.; Brownson, D.A.; Banks, C.E. Microbial fuel cells: An overview of current technology. Renew. Sustain. Energy Rev. 2019, 101, 22. [Google Scholar] [CrossRef]
- Logan, B.E.; Rossi, R.; Ragab, A.; Saikaly, P.E. Electroactive microorganisms in bioelectrochemical systems. Nat. Rev. Genet. 2019, 17, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Logan, B.E. Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Genet. 2009, 7, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Dong, H.; Reguera, G.; Beyenal, H.; Haluk, B.; Liu, J.; Yu, H.-Q.; Fredrickson, J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Genet. 2016, 14, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Bond, D.R.; Holmes, D.E.; Tender, L.M.; Lovley, D.R. Electrode-Reducing Microorganisms That Harvest Energy from Marine Sediments. Science 2002, 295, 483–485. [Google Scholar] [CrossRef]
- Holmes, D.E.; Bond, D.R.; O’Neil, R.A.; Reimers, C.E.; Tender, L.R.; Lovley, D.R. Microbial Communities Associated with Electrodes Harvesting Electricity from a Variety of Aquatic Sediments. Microb. Ecol. 2004, 48, 178–190. [Google Scholar] [CrossRef]
- Kouzuma, A.; Kasai, T.; Nakagawa, G.; Yamamuro, A.; Abe, T.; Watanabe, K. Comparative Metagenomics of Anode-Associated Microbiomes Developed in Rice Paddy-Field Microbial Fuel Cells. PLoS ONE 2013, 8, e77443. [Google Scholar] [CrossRef]
- Yamamuro, A.; Kouzuma, A.; Abe, T.; Watanabe, K. Metagenomic Analyses Reveal the Involvement of Syntrophic Consortia in Methanol/Electricity Conversion in Microbial Fuel Cells. PLoS ONE 2014, 9, e98425. [Google Scholar] [CrossRef]
- Kondaveeti, S.; Mohanakrishna, G.; Lee, J.-K.; Kalia, V.C. Methane as a Substrate for Energy Generation Using Microbial Fuel Cells. Indian J. Microbiol. 2019, 59, 121–124. [Google Scholar] [CrossRef]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidropoulos, D.; Spear, J.R.; Caporaso, G.; Blekhman, R.; Knight, R.; et al. BugBase predicts organism-level microbiome phenotypes. bioRxiv 2017. Preprint. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.; Liu, D.; Wang, S.-H.; Lin, C.-H. Dynamic Changes in Soil Microbial Communities with Glucose Enrichment in Sediment Microbial Fuel Cells. Indian J. Microbiol. 2021, 61, 497–505. [Google Scholar] [CrossRef]
- Liu, D.; Kuo, J.; Wang, S.-H.; Puah, K.M.; Lin, C.-H. Comparative Microbial Communities of Anode Associated Soils in Sediment Microbial Fuel Cells of Rice Field and Drainage Ditch Soils. Pol. J. Environ. Stud. 2021, 31, 179–188. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 9 August 2021).
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011, 40, D109–D114. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2017, 46, D633–D639. [Google Scholar] [CrossRef]
- Markowitz, V.M.; Chen, I.-M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Jacob, B.; Huang, J.; Williams, P.; et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2011, 40, D115–D122. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Djemiel, C.; Maron, P.-A.; Terrat, S.; Dequiedt, S.; Cottin, A.; Ranjard, L. Inferring microbiota functions from taxonomic genes: A review. Gigascience 2022, 11, giab090. [Google Scholar] [CrossRef]
- Mukherjee, A.; Chettri, B.; Langpoklakpam, J.S.; Basak, P.; Prasad, A.; Mukherjee, A.K.; Bhattacharyya, M.; Singh, A.K.; Chattopadhyay, D. Bioinformatic Approaches Including Predictive Metagenomic Profiling Reveal Characteristics of Bacterial Response to Petroleum Hydrocarbon Contamination in Diverse Environments. Sci. Rep. 2017, 7, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, J.; Sengupta, A.; Grewal, P.; Dick, W.A. Functional Predictions of Microbial Communities in Soil as Affected by Long-term Tillage Practices. Agric. Environ. Lett. 2017, 2, 170031. [Google Scholar] [CrossRef]
- Sengupta, A.; Hariharan, J.; Grewal, P.S.; Dick, W.A. Bacterial community dissimilarity in soils is driven by long-term land-use practices. Agrosyst. Geosci. Environ. 2020, 3, e20031. [Google Scholar] [CrossRef]
- Dube, J.P.; Valverde, A.; Steyn, J.M.; Cowan, D.A.; van der Waals, J.E. Differences in Bacterial Diversity, Composition and Function due to Long-Term Agriculture in Soils in the Eastern Free State of South Africa. Diversity 2019, 11, 61. [Google Scholar] [CrossRef]
- Lüneberg, K.; Schneider, D.; Siebe, C.; Daniel, R. Drylands soil bacterial community is affected by land use change and different irrigation practices in the Mezquital Valley, Mexico. Sci. Rep. 2018, 8, 1413. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.E.; Vannette, R.L.; Igwe, A.; Blundell, R.; Casteel, C.L.; Gaudin, A.C.M. Effects of Agricultural Management on Rhizosphere Microbial Structure and Function in Processing Tomato Plants. Appl. Environ. Microbiol. 2019, 85, 16. [Google Scholar] [CrossRef]
- Gumiere, T.; Gumiere, S.; Matteau, J.-P.; Constant, P.; Létourneau, G.; Rousseau, A.N. Soil Bacterial Community Associated with High Potato Production and Minimal Water Use. Front. Environ. Sci. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Luo, O.; Kong, G.; Wang, B.; Li, X.; Li, E.; Li, J.; Liu, F.; Xu, M. Deciphering the Anode-Enhanced Azo Dye Degradation in Anaerobic Baffled Reactors Integrating with Microbial Fuel Cells. Front. Microbiol. 2018, 9, 2117. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, H.; Yang, Y.-L.; Yang, X.-L.; Wu, Y.; Zhang, S.; Song, H.-L. Effects of graphite and Mn ore media on electro-active bacteria enrichment and fate of antibiotic and corresponding resistance gene in up flow microbial fuel cell constructed wetland. Water Res. 2019, 165, 114988. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cai, Y.; Gu, Z.; Yang, Y.-L.; Zhang, S.; Yang, X.-L.; Song, H.-L. Accumulation of sulfonamide resistance genes and bacterial community function prediction in microbial fuel cell-constructed wetland treating pharmaceutical wastewater. Chemosphere 2020, 248, 126014. [Google Scholar] [CrossRef]
- Zhu, K.; Xu, Y.; Yang, X.; Fu, W.; Dang, W.; Yuan, J.; Wang, Z. Sludge Derived Carbon Modified Anode in Microbial Fuel Cell for Performance Improvement and Microbial Community Dynamics. Membranes 2022, 12, 120. [Google Scholar] [CrossRef]
- Holmkvist, L.; Ferdelman, T.G.; Jørgensen, B.B. A cryptic sulfur cycle driven by iron in the methane zone of marine sediment (Aarhus Bay, Denmark). Geochim. Cosmochim. Acta 2011, 75, 3581–3599. [Google Scholar] [CrossRef]
- Aoyagi, T.; Kimura, M.; Yamada, N.; Navarro, R.R.; Itoh, H.; Ogata, A.; Sakoda, A.; Katayama, Y.; Takasaki, M.; Hori, T. Dynamic transition of chemolithotrophic sulfur-oxidizing bacteria in response to amendment with nitrate in deposited marine sediments. Front. Microbiol. 2015, 6, 426. [Google Scholar] [CrossRef]
- Vuillemin, A.; Horn, F.; Friese, A.; Winkel, M.; Alawi, M.; Wagner, D.; Henny, C.; Orsi, W.D.; Crowe, S.A.; Kallmeyer, J. Metabolic potential of microbial communities from ferruginous sediments. Environ. Microbiol. 2018, 20, 4297–4313. [Google Scholar] [CrossRef] [PubMed]
- Kraft, B.; Strous, M.; Tegetmeyer, H.E. Microbial nitrate respiration—Genes, enzymes and environmental distribution. J. Biotechnol. 2011, 155, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ward, B.B.; Sigman, D.M. Global Nitrogen Cycle: Critical Enzymes, Organisms, and Processes for Nitrogen Budgets and Dynamics. Chem. Rev. 2020, 120, 5308–5351. [Google Scholar] [CrossRef]
- Oshiki, M.; Araki, M.; Hirakata, Y.; Hatamoto, M.; Yamaguchi, T.; Araki, N. Ureolytic Prokaryotes in Soil: Community Abundance and Diversity. Microbes Environ. 2018, 33, 230–233. [Google Scholar] [CrossRef] [Green Version]
- Walther, B.; Karl, J.P.; Booth, S.L.; Boyaval, P. Menaquinones, Bacteria, and the Food Supply: The Relevance of Dairy and Fermented Food Products to Vitamin K Requirements. Adv. Nutr. Int. Rev. J. 2013, 4, 463–473. [Google Scholar] [CrossRef]
- Richts, B.; Rosenberg, J.; Commichau, F.M. A Survey of Pyridoxal 5’-Phosphate-Dependent Proteins in the Gram-Positive Model Bacterium Bacillus subtilis. Front. Mol. Biosci. 2019, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Waller, J.C.; Alvarez, S.; Naponelli, V.; Lara-Nuñez, A.; Blaby, I.K.; Da Silva, V.; Ziemak, M.J.; Vickers, T.J.; Beverley, S.M.; Edison, A.S.; et al. A role for tetrahydrofolates in the metabolism of iron-sulfur clusters in all domains of life. Proc. Natl. Acad. Sci. USA 2010, 107, 10412–10417. [Google Scholar] [CrossRef] [PubMed]
- Electroactive microorganisSun, S.; Jones, R.B.; Fodor, A.A. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome 2020, 8, 1–9. [Google Scholar] [CrossRef]
- Toole, D.R.; Zhao, J.; Martens-Habbena, W.; Strauss, S.L. Bacterial functional prediction tools detect but underestimate metabolic diversity compared to shotgun metagenomics in southwest Florida soils. Appl. Soil Ecol. 2021, 168, 104129. [Google Scholar] [CrossRef]
- Ishii, S.; Suzuki, S.; Norden-Krichmar, T.M.; Phan, T.; Wanger, G.; Nealson, K.H.; Sekiguchi, Y.; Gorby, Y.A.; Bretschger, O. Microbial population and functional dynamics associated with surface potential and carbon metabolism. ISME J. 2013, 8, 963–978. [Google Scholar] [CrossRef]
- Ishii, S.; Suzuki, S.; Tenney, A.; Nealson, K.H.; Bretschger, O. Comparative metatranscriptomics reveals extracellular electron transfer pathways conferring microbial adaptivity to surface redox potential changes. ISME J. 2018, 12, 2844–2863. [Google Scholar] [CrossRef]






| Samples | Soil Type | Location | Voltage, mV | Groups * | References |
|---|---|---|---|---|---|
| D1 | nonhydric | 24°00′ N, 120°36′ E | none | L | [14] |
| D2 | nonhydric | 40.92 | H2 | ||
| D3 ** | nonhydric | 105.88 | H1 | ||
| D4 | nonhydric | 0.2908 | H2 | ||
| D5 | nonhydric | 328.89 | H1 | ||
| S1 | hydric | 24°00′ N, 120°36′ E | none | L | |
| S2 | hydric | 64.605 | H1 | ||
| S3 ** | hydric | 16.573 | H2 | ||
| S4 | hydric | 153.11 | H1 | ||
| S5 | hydric | 179.6 | H1 | ||
| D | drainage ditch | 24°00′ N, 120°33′ E | 49 | H2 | [15] |
| DA*** | drainage ditch | 37 | H2 | ||
| RF | rice paddy field | 24°00′ N, 120°34′ E | 76 | H1 | |
| RFA *** | rice paddy field | 22 | H2 | ||
| Nr1 | rice paddy field | 23°58′ N, 120°35′ E | None | L | Manuscript in preparation |
| Nr2 | rice paddy field | None | L | ||
| Nr3 | rice paddy field | None | L |
| Program | Reference Database | Metabolic Pathway Database | Functional Features * |
|---|---|---|---|
| BugBase | Greengenes | KEGG module | 67 (574) |
| FAPROTAX | Silva | 9 (92) | |
| PICRUSt2 | IMG | MetaCys | 37 (440) |
| Tax4Fun2 | Ref99NR (NCBI RefSeq) | KEGG pathway | 38 (374) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, J.; Liu, D.; Lin, C.-H. Functional Prediction of Microbial Communities in Sediment Microbial Fuel Cells. Bioengineering 2023, 10, 199. https://doi.org/10.3390/bioengineering10020199
Kuo J, Liu D, Lin C-H. Functional Prediction of Microbial Communities in Sediment Microbial Fuel Cells. Bioengineering. 2023; 10(2):199. https://doi.org/10.3390/bioengineering10020199
Chicago/Turabian StyleKuo, Jimmy, Daniel Liu, and Chorng-Horng Lin. 2023. "Functional Prediction of Microbial Communities in Sediment Microbial Fuel Cells" Bioengineering 10, no. 2: 199. https://doi.org/10.3390/bioengineering10020199