Candidatus Scalindua, a Biological Solution to Treat Saline Recirculating Aquaculture System Wastewater
Abstract
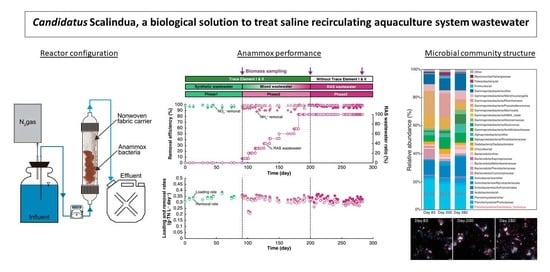
:Highlights
- The anammox process is a promising technique to treat nitrogen-rich marine RAS WW.
- The anammox strain Ca. Scalindua was successfully acclimated to high salinity marine RAS wastewater.
- Despite a slight decrease in population over the time, Ca. Scalindua remained the major species within the granules and was able to maintain a high nitrogen removal rate while exposed to RAS WW in the absence of TE supplementation.
Abstract
1. Introduction
2. Materials and Methods
2.1. RAS WW Collection and Characteristics
2.2. Reactors Operation
2.3. Analytical Methods
2.4. Microbial Community Analysis
2.5. Fluorescence In Situ Hybridization (FISH)
3. Results
3.1. Reactor Performance
3.2. Microbial Community Analysis and FISH
4. Discussion
4.1. Reactor Performance
4.2. Microbial Community
5. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects, Online Edition; Rev. 1; United Nations, Department of Economic and Social Affairs: New York, NY, USA, 2019. [Google Scholar]
- Béné, C.; Barange, M.; Subasinghe, R.; Pinstrup-Andersen, P.; Merino, G.; Hemre, G.-I.; Williams, M. Feeding 9 billion by 2050–Putting fish back on the menu. Food Secur. 2015, 7, 261–274. [Google Scholar] [CrossRef] [Green Version]
- FAO. The State of World Fisheries and Aquaculture (SOFIA) 2022; Food and Agriculture Organization of the United Nations: Rome, Italy, 2020. [Google Scholar]
- Pahri, S.D.R.; Mohamed, A.F.; Samat, A. LCA for open systems: A review of the influence of natural and anthropogenic factors on aquaculture systems. Int. J. Life Cycle Assess 2015, 20, 1324–1337. [Google Scholar] [CrossRef]
- Martins, C.; Eding, E.H.; Verdegem, M.C.; Heinsbroek, L.T.; Schneider, O.; Blancheton, J.-P.; d’Orbcastel, E.R.; Verreth, J. New developments in recirculating aquaculture systems in Europe: A perspective on environmental sustainability. Aquac. Eng. 2010, 43, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Kolarevic, J.; Baeverfjord, G.; Takle, H.; Ytteborg, E.; Reiten, B.K.M.; Nergård, S.; Terjesen, B.F. Performance and welfare of Atlantic salmon smolt reared in recirculating or flow through aquaculture systems. Aquaculture 2014, 432, 15–25. [Google Scholar] [CrossRef]
- Padervand, M.; Gholami, M.R. Removal of toxic heavy metal ions from waste water by functionalized magnetic core–zeolitic shell nanocomposites as adsorbents. Environ. Sci. Pollut. Res. 2013, 20, 3900–3909. [Google Scholar] [CrossRef]
- Abdelfatah, A.G.; Ali, M.A.; Abdelbary, K.M. Mechanical filtration pretreatment effect on ammonia biofiltration performance indicators in fish aquaculture wastewater. Misr J. Agric. Eng. 2022, 39, 555–570. [Google Scholar] [CrossRef]
- Øvrebø, T.K.; Balseiro, P.; Imsland, A.K.D.; Stefansson, S.O.; Tveterås, R.; Sveier, H.; Handeland, S. Investigation of growth performance of post-smolt Atlantic salmon (Salmo salar L.) in semi closed containment system: A big-scale benchmark study. Aquac. Res. 2022, 53, 4178–4189. [Google Scholar]
- Van Rijn, J. Waste treatment in recirculating aquaculture systems. Aquac. Eng. 2013, 53, 49–56. [Google Scholar] [CrossRef]
- Ahmed, N.; Turchini, G.M. Recirculating aquaculture systems (RAS): Environmental solution and climate change adaptation. J. Clean. Prod. 2021, 297, 126604. [Google Scholar] [CrossRef]
- Ahmad, A.; Abdullah, S.R.S.; Hasan, H.A.; Othman, A.R.; Ismail, N.I. Aquaculture industry: Supply and demand, best practices, effluent and its current issues and treatment technology. J. Environ. Manag. 2021, 287, 112271. [Google Scholar] [CrossRef]
- Camargo, J.A.; Alonso, A.; Salamanca, A. Nitrate toxicity to aquatic animals: A review with new data for freshwater invertebrates. Chemosphere 2005, 58, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Roques, J.A.C. Apects of Fish Welfare in Aquaculture Practices; Radboud University Nijmegen: The Netherlands, 2013; p. 200. [Google Scholar]
- Preena, P.G.; Rejish Kumar, V.J.; Singh, I.S.B. Nitrification and denitrification in recirculating aquaculture systems: The processes and players. Rev. Aquac. 2021, 13, 2053–2075. [Google Scholar] [CrossRef]
- Chen, S. Recirculating Systems Effluents and Treatments; Aquaculture and the Environment in the United States, World Aquaculture Society: Baton Rouge, LA, USA, 2002; pp. 119–140. [Google Scholar]
- Stavrakidis-Zachou, O.; Ernst, A.; Steinbach, C.; Wagner, K.; Waller, U. Development of denitrification in semi-automated moving bed biofilm reactors operated in a marine recirculating aquaculture system. Aquac. Int. 2019, 27, 1485–1501. [Google Scholar] [CrossRef]
- Hu, Z.; Lee, J.W.; Chandran, K.; Kim, S.; Khanal, S.K. Nitrous oxide (N2O) emission from aquaculture: A review. Environ. Sci. Technol. 2012, 46, 6470–6480. [Google Scholar] [CrossRef]
- Kartal, B.; van Niftrik, L.; Keltjens, J.T.; den Camp, H.J.O.; Jetten, M.S. Anammox—Growth physiology, cell biology, and metabolism. Adv. Microb. Physiol. 2012, 60, 211–262. [Google Scholar]
- Dapena-Mora, A.; Campos, J.; Mosquera-Corral, A.; Jetten, M.; Méndez, R. Stability of the ANAMMOX process in a gas-lift reactor and a SBR. J. Biotechnol. 2004, 110, 159–170. [Google Scholar] [CrossRef]
- Strous, M.; Kuenen, J.G.; Jetten, M.S. Key physiology of anaerobic ammonium oxidation. Appl. Environ. Microbiol. 1999, 65, 3248–3250. [Google Scholar] [CrossRef] [Green Version]
- Strous, M.; Heijnen, J.; Kuenen, J.G.; Jetten, M. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl. Microbiol. Biotechnol. 1998, 50, 589–596. [Google Scholar] [CrossRef]
- Kartal, B.; Van Niftrik, L.; Rattray, J.; Van De Vossenberg, J.L.; Schmid, M.C.; Sinninghe Damsté, J.; Jetten, M.S.; Strous, M. Candidatu s ‘Brocadia fulgida’: An autofluorescent anaerobic ammonium oxidizing bacterium. FEMS Microbiol. Ecol. 2008, 63, 46–55. [Google Scholar] [CrossRef]
- Okabe, S.; Oshiki, M.; Takahashi, Y.; Satoh, H. N2O emission from a partial nitrification-anammox process and identification of a key biological process of N2O emission from anammox granules. Water Res. 2011, 45, 6461–6470. [Google Scholar] [CrossRef]
- Kuenen, J.G. Anammox bacteria: From discovery to application. Nat. Rev. Microbiol. 2008, 6, 320–326. [Google Scholar] [CrossRef]
- Van Dongen, U.; Jetten, M.S.; van Loosdrecht, M. The SHARON®-Anammox® process for treatment of ammonium rich wastewater. Water Sci. Technol. 2001, 44, 153–160. [Google Scholar] [CrossRef]
- Borin, S.; Mapelli, F.; Rolli, E.; Song, B.; Tobias, C.; Schmid, M.C.; De Lange, G.J.; Reichart, G.J.; Schouten, S.; Jetten, M. Anammox bacterial populations in deep marine hypersaline gradient systems. Extremophiles 2013, 17, 289–299. [Google Scholar] [CrossRef]
- Yokota, N.; Watanabe, Y.; Tokutomi, T.; Kiyokawa, T.; Hori, T.; Ikeda, D.; Song, K.; Hosomi, M.; Terada, A. High-rate nitrogen removal from waste brine by marine anammox bacteria in a pilot-scale UASB reactor. Appl. Microbiol. Biotechnol. 2018, 102, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Kawagoshi, Y.; Huang, X.; Hong, N.; Van Duc, L.; Yamashita, Y.; Hama, T. Nitrogen removal properties in a continuous marine anammox bacteria reactor under rapid and extensive salinity changes. Chemosphere 2016, 148, 444–451. [Google Scholar] [CrossRef]
- Dapena-Mora, A.; Fernandez, I.; Campos, J.; Mosquera-Corral, A.; Mendez, R.; Jetten, M. Evaluation of activity and inhibition effects on Anammox process by batch tests based on the nitrogen gas production. Enzym. Microb. Technol. 2007, 40, 859–865. [Google Scholar] [CrossRef]
- Liu, C.; Yamamoto, T.; Nishiyama, T.; Fujii, T.; Furukawa, K. Effect of salt concentration in anammox treatment using non woven biomass carrier. J. Biosci. Bioeng. 2009, 107, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Malovanyy, A.; Plaza, E.; Trela, J.; Malovanyy, M. Ammonium removal by partial nitritation and Anammox processes from wastewater with increased salinity. Environ. Technol. 2015, 36, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Roques, J.A.C.; Micolucci, F.; Hosokawa, S.; Sundell, K.; Kindaichi, T. Effects of recirculating aquaculture system wastewater on anammox performance and community structure. Processes 2021, 9, 1183. [Google Scholar] [CrossRef]
- Van de Graaf, A.A.; de Bruijn, P.; Robertson, L.A.; Jetten, M.S.; Kuenen, J.G. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 1996, 142, 2187–2196. [Google Scholar] [CrossRef] [Green Version]
- Sikora, M.; Nowosad, J.; Kucharczyk, D. Nitrogen compound oxidation rate in recirculation systems using three biological filter medias in rearing common carp (Cyprinus carpio L.) juveniles. Aquaculture 2022, 547, 737532. [Google Scholar] [CrossRef]
- Brazil, B.L.; Summerfelt, S.T.; Libey, G.S. Application of Ozone to Recirculating Aquaculture Systems. In Successes and Failures in Commercial Recirculating Aquaculture (Conference Proceedings); Libey, G.S., Timmons, M.B., Eds.; Northeast Regional Agricultural Engineering Service: Ithaca, NY, USA, 1996; pp. 373–389. [Google Scholar]
- Krumins, V.; Ebeling, J.; Wheaton, F. Part-day ozonation for nitrogen and organic carbon control in recirculating aquaculture systems. Aquac. Eng. 2001, 24, 231–241. [Google Scholar] [CrossRef]
- Kindaichi, T.; Awata, T.; Tanabe, K.; Ozaki, N.; Ohashi, A. Enrichment of marine anammox bacteria in Hiroshima Bay sediments. Water Sci. Technol. 2011, 63, 964–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kindaichi, T.; Awata, T.; Suzuki, Y.; Tanabe, K.; Hatamoto, M.; Ozaki, N.; Ohashi, A. Enrichment using an up-flow column reactor and community structure of marine anammox bacteria from coastal sediment. Microbes Environ. 2011, 26, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mojiri, A.; Nishimoto, K.; Awata, T.; Aoi, Y.; Ozaki, N.; Ohashi, A.; Kindaichi, T. Effects of salts on the activity and growth of “Candidatus Scalindua sp. ”, a marine Anammox bacterium. Microbes Environ. 2018, 33, 336–339. [Google Scholar] [CrossRef]
- American Public Health Association. Standard Methods for the Examination of Water and Wastewater: Selected Analytical Methods Approved and Cited by the United States Environmental Protection Agency; APHA-AWWA-WEF: Washington, DC, USA, 2012. [Google Scholar]
- Mojiri, A.; Ohashi, A.; Ozaki, N.; Aoi, Y.; Kindaichi, T. Integrated anammox-biochar in synthetic wastewater treatment: Performance and optimization by artificial neural network. J. Clean. Prod. 2020, 243, 118638. [Google Scholar] [CrossRef]
- Shoiful, A.; Kambara, H.; Cao, L.T.T.; Matsushita, S.; Kindaichi, T.; Aoi, Y.; Ozaki, N.; Ohashi, A. Mn (II) oxidation and manganese-oxide reduction on the decolorization of an azo dye. Int. Biodeterior. Biodegrad. 2020, 146, 104820. [Google Scholar] [CrossRef]
- Awata, T.; Goto, Y.; Kuratsuka, H.; Aoi, Y.; Ozaki, N.; Ohashi, A.; Kindaichi, T. Reactor performance and microbial community structure of single-stage partial nitritation anammox membrane bioreactors inoculated with Brocadia and Scalindua enrichment cultures. Biochem. Eng. J. 2021, 170, 107991. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Awata, T.; Oshiki, M.; Kindaichi, T.; Ozaki, N.; Ohashi, A.; Okabe, S. Physiological characterization of an anaerobic ammonium-oxidizing bacterium belonging to the “Candidatus Scalindua” group. Appl. Environ. Microbiol. 2013, 79, 4145–4148. [Google Scholar] [CrossRef] [Green Version]
- Daims, H.; Brühl, A.; Amann, R.; Schleifer, K.-H.; Wagner, M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 1999, 22, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.C.; Maas, B.; Dapena, A.; van de Pas-Schoonen, K.; van de Vossenberg, J.; Kartal, B.; van Niftrik, L.; Schmidt, I.; Cirpus, I.; Kuenen, J.G. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 2005, 71, 1677–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aklujkar, M.; Coppi, M.V.; Leang, C.; Kim, B.C.; Chavan, M.; Perpetua, L.; Giloteaux, L.; Liu, A.; Holmes, D. Proteins involved in electron transfer to Fe (III) and Mn (IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 2013, 159, 515–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Yao, H.; Zhang, D.; Zuo, L.; Ren, J.; Ma, J.; Pei, J.; Xu, Y.; Yang, C. Short-and long-term effects of manganese, zinc and copper ions on nitrogen removal in nitritation-anammox process. Chemosphere 2018, 193, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Gao, D.; Peng, S.; Tao, Y. Effects of ferrous and manganese ions on anammox process in sequencing batch biofilm reactors. J. Environ. Sci. 2014, 26, 1034–1039. [Google Scholar] [CrossRef]
- Lotti, T.; Cordola, M.; Kleerebezem, R.; Caffaz, S.; Lubello, C.; Van Loosdrecht, M. Inhibition effect of swine wastewater heavy metals and antibiotics on anammox activity. Water Sci. Technol. 2012, 66, 1519–1526. [Google Scholar] [CrossRef]
- Yang, G.-F.; Ni, W.-M.; Wu, K.; Wang, H.; Yang, B.-E.; Jia, X.-Y.; Jin, R.-C. The effect of Cu (II) stress on the activity, performance and recovery on the anaerobic ammonium-oxidizing (Anammox) process. Chem. Eng. J. 2013, 226, 39–45. [Google Scholar] [CrossRef]
- Kimura, Y.; Isaka, K. Evaluation of inhibitory effects of heavy metals on anaerobic ammonium oxidation (anammox) by continuous feeding tests. Appl. Microbiol. Biotechnol. 2014, 98, 6965–6972. [Google Scholar] [CrossRef]
- von Ahnen, M.; Aalto, S.L.; Suurnäkki, S.; Tiirola, M.; Pedersen, P.B. Salinity affects nitrate removal and microbial composition of denitrifying woodchip bioreactors treating recirculating aquaculture system effluents. Aquaculture 2019, 504, 182–189. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Liu, L.Z.; Wang, X.J.; Liu, Z.P. Heterotrophic bacterial community structure of multistage biofilters in a commercial pufferfish Takifugu rubripes RAS. In Advanced Materials Research; Trans Tech Publications: Zurich, Switzerland, 2013. [Google Scholar]
- Interdonato, F. Recirculating Aquaculture System (RAS) Biofilters: Focusing on Bacterial Communities Complexity and Activity; Università Degli Studi di Messina: Messina, Italy, 2012; p. 128. [Google Scholar]
- Sanchez, F.A.; Vivian-Rogers, V.R.; Urakawa, H. Tilapia recirculating aquaculture systems as a source of plant growth promoting bacteria. Aquac. Res. 2019, 50, 2054–2065. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Ng, H.J.; Webb, H.K. The Family Pseudoalteromonadaceae. In The Prokaryotes: Gammaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 575–582. [Google Scholar]
- Brailo, M.; Schreier, H.J.; McDonald, R.; Maršić-Lučić, J.; Gavrilović, A.; Pećarević, M.; Jug-Dujaković, J. Bacterial community analysis of marine recirculating aquaculture system bioreactors for complete nitrogen removal established from a commercial inoculum. Aquaculture 2019, 503, 198–206. [Google Scholar] [CrossRef]
- Brettar, I.; Christen, R.; Höfle, M.G. Rheinheimera baltica gen. nov., sp. nov., a blue-coloured bacterium isolated from the central Baltic Sea. Int. J. Syst. Evol. Microbiol. 2002, 52, 1851–1857. [Google Scholar]
- Zhong, Z.-P.; Liu, Y.; Liu, L.-Z.; Wang, F.; Zhou, Y.-G.; Liu, Z.-P. Rheinheimera tuosuensis sp. nov., isolated from a saline lake. Int. J. Syst. Evol. Microbiol. 2014, 64, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Jeon, C.O. Rheinheimera aestuari sp. nov., a marine bacterium isolated from coastal sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 2640–2645. [Google Scholar] [CrossRef]
- Chen, W.-M.; Chen, W.-T.; Young, C.-C.; Sheu, S.-Y. Rheinheimera riviphila sp. nov., isolated from a freshwater stream. Arch. Microbiol. 2019, 201, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, J.-T.; Xu, C.-J.; Liu, Y.-H.; Song, X.-F.; Li, H.; Liu, Z.-P. Rheinheimera longhuensis sp. nov., isolated from a slightly alkaline lake, and emended description of genus Rheinheimera Brettar et al. 2002. Int. J. Syst. Evol. Microbiol. 2012, 62, 2927–2933. [Google Scholar] [CrossRef] [Green Version]
- Sorokin, D.Y.; Mosier, D.; Zorz, J.K.; Dong, X.; Strous, M. Wenzhouxiangella strain AB-CW3, a proteolytic bacterium from hypersaline soda lakes that preys on cells of gram-positive bacteria. Front. Microbiol. 2020, 11, 597686. [Google Scholar] [CrossRef]
- Wang, G.; Tang, M.; Li, T.; Dai, S.; Wu, H.; Chen, C.; He, H.; Fan, J.; Xiang, W.; Li, X. Wenzhouxiangella marina gen. nov, sp. nov, a marine bacterium from the culture broth of Picochlorum sp. 122, and proposal of Wenzhouxiangellaceae fam. nov. in the order Chromatiales. Antonie Van Leeuwenhoek 2015, 107, 1625–1632. [Google Scholar] [CrossRef]






| TE | B | Co | Cu | Fe | Mn | Mo | Ni | Se | Zn |
|---|---|---|---|---|---|---|---|---|---|
| Concentration (µg L−1) | 2.4 | 28.3 | 63.6 | 1840 | 274.8 | 96.4 | 46.9 | 47.9 | 97.8 |
| Parameter | Salinity (‰) | NH4+ (mg-N L−1) | NO2− (mg-N L−1) | NO3− (mg-N L−1) | pH | TSS 1 (mg L−1) | COD 2 (mg-C L−1) | Total P 3 (mg-P L−1) |
|---|---|---|---|---|---|---|---|---|
| Value | 29 ± 1 | 6.22 ± 2.34 | 2.24 ± 0.69 | 1.81 ± 0.88 | 7.3 ± 0.3 | 58.8 ± 5.6 | 39.5 ± 5.3 | 2.6 ± 0.7 |
| Phase | Period (d) | AS/RAS 1 | TE 2 | HRT (h) | Nitrogen Loading Rate (g-TN L−1 day−1) 3 |
|---|---|---|---|---|---|
| 1 | 0–83 | AS | + | 4.6 | 0.38 ± 0.03 |
| 2 | 84–200 | RAS | + | 4.4 | 0.32 ± 0.06 |
| 3 | 201–280 | RAS | − | 4.8 | 0.35 ± 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Micolucci, F.; Roques, J.A.C.; Ziccardi, G.S.; Fujii, N.; Sundell, K.; Kindaichi, T. Candidatus Scalindua, a Biological Solution to Treat Saline Recirculating Aquaculture System Wastewater. Processes 2023, 11, 690. https://doi.org/10.3390/pr11030690
Micolucci F, Roques JAC, Ziccardi GS, Fujii N, Sundell K, Kindaichi T. Candidatus Scalindua, a Biological Solution to Treat Saline Recirculating Aquaculture System Wastewater. Processes. 2023; 11(3):690. https://doi.org/10.3390/pr11030690
Chicago/Turabian StyleMicolucci, Federico, Jonathan A. C. Roques, Geoffrey S. Ziccardi, Naoki Fujii, Kristina Sundell, and Tomonori Kindaichi. 2023. "Candidatus Scalindua, a Biological Solution to Treat Saline Recirculating Aquaculture System Wastewater" Processes 11, no. 3: 690. https://doi.org/10.3390/pr11030690