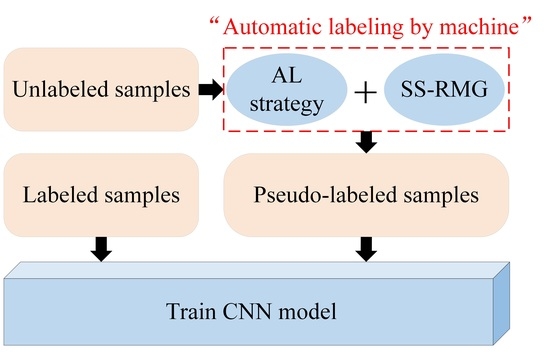
3.2.1. Active Deep Learning (ADL) Module
The paper proposes an improved ADL semi-supervised classification framework based on the traditional ADL classification model shown in
Figure 2, where the red rectangle indicates the deficiency of traditional ADL. The following is a detailed theoretical introduction of the active deep learning module in
Figure 1.
Data augmentation (DA) is widely used in the field of hyperspectral image data classification. It is often used as a means to expand the dataset and improve the accuracy. In this paper, the DA method was utilized to expand training samples by flipping horizontally or vertically in the literature [
30], and the specific rotation angle can be adjusted according to the characteristics of the actual training sample. DA will also refine the classification accuracy by adding an appropriate number of samples to assist the training model during each iteration.
The objective function of the CNN classification model is defined as shown in Equation (1):
where
FΘ is a nonlinear function of the CNN using parameters Θ,
is the unary term that predicts the samples by using CNN, and
can make adjacent samples to have the similar label. Here, the detailed explanation of label Y and the parameter Θ refers to the ADL classification method from [
33]. The data input to the convolutional neural network is provided in the form of a three-dimensional cubic block with a three-dimensional convolutional kernel for convolution and pooling. Subsequently, the output of the network is selected to be a Softmax function defined as:
where
is the output value of the
ith node,
C is the number of output nodes (e.g., the number of categories of the classification) The output value of the multiclassification is converted into a probability distribution in the range of [0, 1], with a sum of 1, by the Softmax function.
AL is a widely used strategy that selects the sample with the least confidence in an iterative manner. As we can retrieve category membership probability estimates from CNNs, this paper uses some AL metrics about the class probabilities as well (e.g., the Entropy (EP) metric and the Best-versus-Second-Best (BVSB) metric in probability-based heuristics). Both of these criteria can provide a basis for selecting the informative candidates for querying annotations. These two learning strategies are introduced in details below.
(1) EP measure: EP measures the uncertainty of class membership, and the sample has a higher EP value with the greater the uncertainty of class. Each unlabeled sample corresponds to a variable
, and we obtain
z distribution of
P with the calculated estimated probability of class affiliation, that is
, where
denotes the
element of the variable
. The specific calculation of EP value is shown in Equation (3):
(2) BVSB measure: by calculating the difference between the most similar categories, the information content of the sample is determined. If the BVSB value of the sample is small, the sample has a large amount of information. The formula is shown in Equation (4) below:
3.2.2. Spatial and Spectral Random Multi-Graph (SS-RMG) Module
This paper uses the SS-RMG module to improve the expert marking action in traditional ADL, shown in the red box in
Figure 2. The following is a detailed theoretical introduction of the Random multi-graph module in
Figure 1.
Spectral features of finite labelled samples and the spatial features of hyperspectral data, extracted by local binary patterns (LBP) [
35], are extracted separately, according to the Li et al. [
36] methodology. Then, the spatial and spectral joint features are utilized as the input into random multi-graphs (RMG) for classification. In this study, the graph was constructed by randomly selecting a subset of features by using the anchor graph algorithm. Then, labels of unlabeled samples were predicted based on the objective function.
(1) Data feature extraction: spectral feature extraction is all spectral bands in HSI data. After the principal component analysis, the spatial information of the spectral bands was extracted using LBP. This was an operator for describing the local texture features of an image. Here, we opted for the improvement of the original LBP, namely, equivalence pattern. The improved equivalent mode of LBP implied that, when the cyclic binary number, corresponding to certain LBP, changes from 0 to 1 or from 1 to 0, in at most two transitions, the binary corresponding to the LBP became an equivalent mode.
For the center pixel
, the center pixel was applied as the threshold, while the position of the pixel was marked as “1” or “0”. This depends on whether the neighboring pixel value is greater than the threshold or not. By assuming that
p adjacent pixels were generated by a circle with
as the center and radius
r, the LBP code of the central pixel can be expressed by Equation (5):
The LBP algorithm expresses the spatial information characteristics of the image with a local region of size
, where the local area
k is a custom parameter. Then, the histogram of each area is calculated, and the statistical histogram is connected to a feature vector, that is, the spatial feature vector. Schematic diagram of LBP is shown in
Figure 3. Such improvements greatly reduce the number of binary patterns from
to
. Subsequently, the information is preserved, while the dimensionality of the feature vector is reduced.
(2) Random multi-graph algorithm defined the following symbols: a weighted graph can be obtained from a dataset consisting of labeled data and unlabeled data . Here, the vertices of the graph represent N data points, and the edges of the graph are formalized by a matrix of weights , which expresses the similarity between the associated nodes, and if the weight is larger, the adjacent vertices and are considered to have the same label.
The traditional graph-based semi-supervised learning approach is formulated as the following quadratic optimization problem, whereas the objective function of
C-classification semi-supervised learning can be defined by Equation (6):
where
is the tracking function,
is the label prediction matrix,
,
is a diagonal matrix with the
diagonal element.
is the regularization matrix, representing the Laplacian graph, defined as
, where
W is the weight matrix of the graph.
The anchor graph algorithm was used according to the high dimensional characteristics of hyperspectral data in this study to construct and to learn the graph. The anchor graph algorithm makes the label prediction function a weighted average of the labels on a subset of anchor point samples. Thus, the label prediction function can be represented by a subset
. The
is an anchor point, and the label prediction function can be formalized by Equation (7):
where
is the data adaptive weight, defining the vector
and the vector
, so that Equation (7) is rewritten to Equation (8):
Equation (8) indicates that the solution space for the unknown labels is reduced from
to
, and these anchor points are used as
K-means clustering centers to realistically represent the data stream shape. The matrix
is the data-anchor mapping matrix to be learned, and the matrix
P is written in the paper, using the definition of kernel-based functions from the Nadaraya–Watson kernel regression, as shown in Equation (9):
where a Gaussian kernel function
with bandwidth σ is used as the kernel regression,
is the
r-nearest neighbor anchor point of
. It is usually considered that
for
, where it is considered that
is larger if
and
are close to each other and vice versa.
A previous study [
31] applied the Local Anchor Embedding algorithm to retrieve anchor points, and the data-anchor mapping problem can be formulated by Equation (10):
where
is the data matrix, each row in the matrix is a data sample,
is the anchor matrix, each row in the matrix represents an anchor,
is the learnable data-anchor mapping matrix.
According to the matrix
P, its adjacency matrix
W can be designed, as indicated by Equation (11):
The diagonal matrix
is defined by Equation (12):
According to the anchor’s label prediction model, the
is solved first, and then, the labels of other data points can be found using Equation (8), and the objective function of
is defined by Equation (13):
where
is the submatrix of
P,
is the label of the labeled data,
is the Frobenius norm of the matrix, and γ is the regularization factor. If we let
, according to Equation (11), the graph Laplacian matrix is
, then:
Letting the partial derivative w.r.t
equal to 0, we obtain:
The label of the unlabeled sample can be predicted by the following Equation (16):
where
represents the
row of the
P matrix and
represents the
column of the
matrix.
Figure 4 shows the SS-RMG module of classification model.