Identification of Multiple Replication Stages and Origins in the Nucleopolyhedrovirus of Anticarsia gemmatalis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell and Virus Stocks
2.2. Quantitative Real Time PCR Assays for AgMNPV and UFL-Ag-286 Cells
2.3. Replication Kinetics
2.4. AgMNPV Genomic Library
2.5. Transient Replication Assay
2.6. Bioinformatics Analysis
3. Results
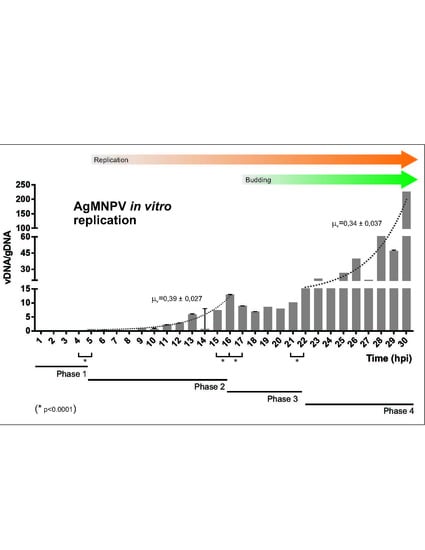
3.1. AgMNPV Kinetics of Replication
3.2. Discovering ORI Sequences in AgMNPV
3.3. Characterization of ORI Sequences from AgMNPV
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Genes Contained in the Different Clones (Table 1)
Early Promoters (Table 2)
Palindromes (Table 3)
References
- Miele, S.A.B.; Garavaglia, M.J.; Belaich, M.N.; Ghiringhelli, P.D. Baculovirus: Molecular Insights on Their Diversity and Conservation. Int. J. Evol. Biol. 2011, 2011, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Blissard, G.W.; Rohrmann, G.F. Baculovirus Diversity and Molecular Biology. Annu. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef]
- Braconi, C.T.; Ardisson-Araujo, D.M.P.; Leme, A.F.P.; Oliveira, J.V.; Pauletti, B.A.; Garcia-Maruniak, A.; Ribeiro, B.M.; Maruniak, J.E.; Zanotto, P.M. Proteomic analyses of baculovirus Anticarsia gemmatalis multiple nucleopolyhedrovirus budded and occluded virus. J. Gen. Virol. 2014, 95, 980–989. [Google Scholar] [CrossRef]
- Cheshenko, N.; Krougliak, N.; Eisensmith, R.C.; Krougliak, V.A. A novel system for the production of fully deleted adenovirus vectors that does not require helper adenovirus. Gene Ther. 2001, 8, 846–854. [Google Scholar] [CrossRef] [Green Version]
- Bieniossek, C.; Imasaki, T.; Takagi, Y.; Berger, I. MultiBac: Expanding the research toolbox for multiprotein complexes. Trends Biochem. Sci. 2012, 37, 49–57. [Google Scholar] [CrossRef]
- Gupta, K.; Tölzer, C.; Sari-Ak, D.; Fitzgerald, D.J.; Schaffitzel, C.; Berger, I. MultiBac: Baculovirus-Mediated Multigene DNA Cargo Delivery in Insect and Mammalian Cells. Viruses 2019, 11, 198. [Google Scholar] [CrossRef]
- Inceoglu, A.B.; Kamita, S.G.; Hinton, A.C.; Huang, Q.; Severson, T.F.; Kang, K.; Hammock, B.D. Recombinant baculoviruses for insect control. Pest Manag. Sci. 2001, 57, 981–987. [Google Scholar] [CrossRef]
- Inceoglu, A.B.; Kamita, S.G.; Hammock, B.D. Genetically Modified Baculoviruses: A Historical Overview and Future Outlook. Adv. Virus Res. 2006, 68, 323–360. [Google Scholar]
- Szewczyk, B.; Hoyos-Carvajal, L.; Paluszek, M.; Skrzecz, I.; Lobo de Souza, M. Baculoviruses — re-emerging biopesticides. Biotechnol. Adv. 2006, 24, 143–160. [Google Scholar] [CrossRef]
- Popham, H.J.R.; Nusawardani, T.; Bonning, B.C. Introduction to the Use of Baculoviruses as Biological Insecticides. Methods Mol. Biol. 2016, 1350, 383–392. [Google Scholar]
- Van der Merwe, M.; Jukes, M.D.; Rabalski, L.; Knox, C.; Opoku-Debrah, J.K.; Moore, S.D.; Krejmer-Rabalska, M.; Szewczyk, B.; Hill, M.P. Genome Analysis and Genetic Stability of the Cryptophlebia leucotreta Granulovirus (CrleGV-SA) after 15 Years of Commercial Use as a Biopesticide. Int. J. Mol. Sci. 2017, 18, 2327. [Google Scholar] [CrossRef]
- Kost, T.A.; Kemp, C.W. Fundamentals of Baculovirus Expression and Applications. Adv. Exp. Med. Biol. 2016, 896, 187–197. [Google Scholar]
- Irons, S.L.; Chambers, A.C.; Lissina, O.; King, L.A.; Possee, R.D. Protein Production Using the Baculovirus Expression System. In Current Protocols in Protein Science; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 5.5.1–5.5.22. [Google Scholar]
- Ono, C.; Okamoto, T.; Abe, T.; Matsuura, Y. Baculovirus as a Tool for Gene Delivery and Gene Therapy. Viruses 2018, 10, 510. [Google Scholar] [CrossRef]
- Felberbaum, R.S. The baculovirus expression vector system: A commercial manufacturing platform for viral vaccines and gene therapy vectors. Biotechnol. J. 2015, 10, 702–714. [Google Scholar] [CrossRef]
- Mansouri, M.; Berger, P. Baculovirus for gene delivery to mammalian cells: Past, present and future. Plasmid 2018, 98, 1–7. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Keilwagen, J.; Jehle, J.A. Baculovirus Kimura two-parameter species demarcation criterion is confirmed by the distances of 38 core gene nucleotide sequences. J. Gen. Virol. 2018, 99, 1307–1320. [Google Scholar] [CrossRef]
- Thézé, J.; Lopez-Vaamonde, C.; Cory, J.S.; Herniou, E.A. Biodiversity, Evolution and Ecological Specialization of Baculoviruses: A Treasure Trove for Future Applied Research. Viruses 2018, 10, 366. [Google Scholar] [CrossRef]
- Gao, L.; Qi, J. Whole genome molecular phylogeny of large dsDNA viruses using composition vector method. BMC Evol. Biol. 2007, 7, 41. [Google Scholar] [CrossRef]
- Javed, M.A.; Biswas, S.; Willis, L.G.; Harris, S.; Pritchard, C.; van Oers, M.M.; Donly, B.C.; Erlandson, M.A.; Hegedus, D.D.; Theilmann, D.A. Autographa californica Multiple Nucleopolyhedrovirus AC83 is a Per Os Infectivity Factor (PIF) Protein Required for Occlusion-Derived Virus (ODV) and Budded Virus Nucleocapsid Assembly as well as Assembly of the PIF Complex in ODV Envelopes. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Garavaglia, M.J.; Miele, S.A.B.; Iserte, J.A.; Belaich, M.N.; Ghiringhelli, P.D. The ac53, ac78, ac101, and ac103 Genes Are Newly Discovered Core Genes in the Family Baculoviridae. J. Virol. 2012, 86, 12069–12079. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus Molecular Biology, 3rd ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013.
- Carstens, E.B. AcMNPV as a model for baculovirus DNA replication. Virol. Sin. 2009, 24, 243–267. [Google Scholar] [CrossRef]
- Mikhailov, V.S.; Rohrmann, G.F. Characterization of short-lived intermediates produced during replication of baculovirus DNA. Virus Res. 2009, 145, 106–111. [Google Scholar] [CrossRef] [Green Version]
- Anderson, S.; DePamphilis, M.L. Metabolism of Okazaki fragments during simian virus 40 DNA replication. J. Biol. Chem. 1979, 254, 11495–11504. [Google Scholar]
- Leisy, D.J.; Rohrmann, G.F. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology 1993, 196, 722–730. [Google Scholar] [CrossRef]
- Oppenheimer, D.I.; Volkman, L.E. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch. Virol. 1997, 142, 2107–2113. [Google Scholar] [CrossRef]
- Okano, K.; Vanarsdall, A.L.; Rohrmann, G.F. A baculovirus alkaline nuclease knockout construct produces fragmented DNA and aberrant capsids. Virology 2007, 359, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Liu, G.; Carstens, E.B. Replication, integration, and packaging of plasmid DNA following cotransfection with baculovirus viral DNA. J. Virol. 1999, 73, 5473–5480. [Google Scholar]
- Pearson, M.; Bjornson, R.; Pearson, G.; Rohrmann, G. The Autographa californica baculovirus genome: Evidence for multiple replication origins. Science 1992, 209, 1392–1396. [Google Scholar] [CrossRef]
- Kool, M.; van den Berg, P.M.M.M.; Tramper, J.; Goldbach, R.W.; Vlak, J.M. Location of Two Putative Origins of DNA Replication of Autographa californica Nuclear Polyhedrosis Virus. Virology 1993, 192, 94–101. [Google Scholar] [CrossRef]
- Lee, H.Y.; Krell, P.J. Generation and analysis of defective genomes of Autographa californica nuclear polyhedrosis virus. J. Virol. 1992, 66, 4339–4347. [Google Scholar] [Green Version]
- Pearson, M.N.; Bjornson, R.M.; Ahrens, C.; Rohrmann, G.F. Identification and characterization of a putative origin of DNA replication in the genome of a baculovirus pathogenic for Orgyia pseudotsugata. Virology 1993, 197, 715–725. [Google Scholar] [CrossRef]
- Wu, Y.; Carstens, E.B. Initiation of baculovirus DNA replication: Early promoter regions can function as infection-dependent replicating sequences in a plasmid-based replication assay. J. Virol. 1996, 70, 6967–6972. [Google Scholar]
- Wu, Y.-L.; Wu, C.-P.; Huang, Y.-H.; Huang, S.-P.; Lo, H.-R.; Chang, H.-S.; Lin, P.-H.; Wu, M.-C.; Chang, C.-J.; Chao, Y.-C. Identification of a high-efficiency baculovirus DNA replication origin that functions in insect and mammalian cells. J. Virol. 2014, 88, 13073–13085. [Google Scholar] [CrossRef]
- Guarino, L.A.; Summers, M.D. Interspersed Homologous DNA of Autographa californica Nuclear Polyhedrosis Virus Enhances Delayed-Early Gene Expression. J. Virol. 1986, 60, 215–223. [Google Scholar]
- Cochran, M.A.; Faulkner, P. Location of Homologous DNA Sequences Interspersed at Five Regions in the Baculovirus AcMNPV Genome. J. Virol. 1983, 45, 961–970. [Google Scholar] [Green Version]
- Kool, M.; Voeten, J.T.; Goldbach, R.W.; Tramper, J.; Vlak, J.M. Identification of seven putative origins of Autographa californica multiple nucleocapsid nuclear polyhedrosis virus DNA replication. J. Gen. Virol. 1993, 74 (Pt 12), 2661–2668. [Google Scholar] [CrossRef]
- Carstens, E.B.; Wu, Y. No single homologous repeat region is essential for DNA replication of the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Gen. Virol. 2007, 88, 114–122. [Google Scholar] [CrossRef]
- Friesen, P.D.; Miller, L.K. Divergent transcription of early 35- and 94-kilodalton protein genes encoded by the HindIII K genome fragment of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 1987, 61, 2264–2272. [Google Scholar] [Green Version]
- Oliveira, J.V.; Wolff, J.L.C.; Garcia-Maruniak, A.; Ribeiro, B.M.; de Castro, M.E.B.; de Souza, M.L.; Moscardi, F.; Maruniak, J.E.; Zanotto, P.M. Genome of the most widely used viral biopesticide: Anticarsia gemmatalis multiple nucleopolyhedrovirus. J. Gen. Virol. 2006, 87, 3233–3250. [Google Scholar] [CrossRef]
- Kool, M.; Goldbach, R.W.; Vlak, J.M. A putative non-hr origin of DNA replication in the HindIII-K fragment of Autographa californica multiple nucleocapsid nuclear polyhedrosis virus. J. Gen. Virol. 1994, 75 (Pt 12), 3345–3352. [Google Scholar] [CrossRef]
- Sieburth, P.J.; Maruniak, J.E. Growth characteristics of a continuous cell line from the velvetbean caterpillar, Anticarsia gemmatalis Hübner (Lepidoptera: Noctuidae). Vitr. Cell. Dev. Biol. 1988, 24, 195–198. [Google Scholar] [CrossRef]
- Zanotto, P.M.; Sampaio, M.J.; Johnson, D.W.; Rocha, T.L.; Maruniak, J.E. The Anticarsia gemmatalis nuclear polyhedrosis virus polyhedrin gene region: Sequence analysis, gene product and structural comparisons. J. Gen. Virol. 1992, 73 (Pt 5), 1049–1056. [Google Scholar] [CrossRef]
- Mengual Gómez, D.L.; Belaich, M.N.; Rodríguez, V.A.; Ghiringhelli, P.D. Effects of fetal bovine serum deprivation in cell cultures on the production of Anticarsia gemmatalis multinucleopolyhedrovirus. BMC Biotechnol. 2010, 10, 68. [Google Scholar] [CrossRef]
- Kost, T.A.; Condreay, J.P. Recombinant baculoviruses as expression vectors for insect and mammalian cells. Curr. Opin. Biotechnol. 1999, 10, 428–433. [Google Scholar] [CrossRef]
- Green Michel R, S.J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012; ISBN 978-1-936113-42-2. [Google Scholar]
- Chen, Y.-R.; Zhong, S.; Fei, Z.; Hashimoto, Y.; Xiang, J.Z.; Zhang, S.; Blissard, G.W. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J. Virol. 2013, 87, 6391–6405. [Google Scholar] [CrossRef]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef] [Green Version]
- Carver, T.; Bleasby, A. The design of Jemboss: A graphical user interface to EMBOSS. Bioinformatics 2003, 19, 1837–1843. [Google Scholar] [CrossRef]
- Mathews, D.H. Using the RNAstructure Software Package to Predict Conserved RNA Structures. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; Volume 46, pp. 12.4.1–12.4.22. [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Tjia, S.T.; Carstens, E.B.; Doerfler, W. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus II. The viral DNA and the kinetics of its replication. Virology 1979, 99, 399–409. [Google Scholar] [CrossRef]
- Liu, X.; Yin, F.; Zhu, Z.; Hou, D.; Wang, J.; Zhang, L.; Wang, M.; Wang, H.; Hu, Z.; Deng, F. Genomic sequencing and analysis of Sucra jujuba nucleopolyhedrovirus. PLoS ONE 2014, 9, e110023. [Google Scholar] [CrossRef]
- Rasmussen, C.; Leisy, D.J.; Ho, P.S.; Rohrmann, G.F. Structure-function analysis of the Autographa californica multinucleocapsid nuclear polyhedrosis virus homologous region palindromes. Virology 1996, 224, 235–245. [Google Scholar] [CrossRef]
- Pearson, M.N.; Rohrmann, G.F. Lymantria dispar nuclear polyhedrosis virus homologous regions: Characterization of their ability to function as replication origins. J. Virol. 1995, 69, 213–221. [Google Scholar]
- Broer, R.; Heldens, J.G.; van Strien, E.A.; Zuidema, D.; Vlak, J.M. Specificity of multiple homologous genomic regions in Spodoptera exigua nucleopolyhedrovirus DNA replication. J. Gen. Virol. 1998, 79 (Pt 6), 1563–1572. [Google Scholar] [CrossRef]
- Heldens, J.G.; Broer, R.; Zuidema, D.; Goldbach, R.W.; Vlak, J.M. Identification and functional analysis of a non-hr origin of DNA replication in the genome of Spodoptera exigua multicapsid nucleopolyhedrovirus. J. Gen. Virol. 1997, 78 (Pt 6), 1497–1506. [Google Scholar] [CrossRef]
- Ahrens, C.H.; Pearson, M.N.; Rohrmann, G.F. Identification and Characterization of a Second Putative Origin of DNA Replication in a Baculovirus of Orgyia pseudotsugata. Virology 1995, 207, 572–576. [Google Scholar] [CrossRef] [Green Version]
- Jehle, J.A. The expansion of a hypervariable, non-hr ori-like region in the genome of Cryptophlebia leucotreta granulovirus provides in vivo evidence for the utilization of baculovirus non-hr oris during replication. J. Gen. Virol. 2002, 83, 2025–2034. [Google Scholar] [CrossRef]
- Ciardo, D.; Goldar, A.; Marheineke, K. On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway. Genes 2019, 10, 94. [Google Scholar] [CrossRef]
- Oliveira, J.V.; de Brito, A.F.; Braconi, C.T.; de Melo Freire, C.C.; Iamarino, A.; de Andrade Zanotto, P.M. Modularity and evolutionary constraints in a baculovirus gene regulatory network. BMC Syst. Biol. 2013, 7, 87. [Google Scholar] [CrossRef]
- Habib, S.; Hasnain, S.E. Differential activity of two non-hr origins during replication of the baculovirus Autographa californica nuclear polyhedrosis virus genome. J. Virol. 2000, 74, 5182–5189. [Google Scholar] [CrossRef]
- Lange, M.; Jehle, J.A. The genome of the Cryptophlebia leucotreta granulovirus. Virology 2003, 317, 220–236. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Yu, J.; Wang, L.; Hu, X.; Bao, W.; Li, G.; Chen, C.; Han, H.; Hu, S.; Yang, H. Sequence Analysis of the Spodoptera litura Multicapsid Nucleopolyhedrovirus Genome. Virology 2001, 287, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Van Oers, M.M. Genome sequence of Chrysodeixis chalcites nucleopolyhedrovirus, a baculovirus with two DNA photolyase genes. J. Gen. Virol. 2005, 86, 2069–2080. [Google Scholar] [CrossRef] [Green Version]
- Willis, L.G.; Siepp, R.; Stewart, T.M.; Erlandson, M.A.; Theilmann, D.A. Sequence analysis of the complete genome of Trichoplusia ni single nucleopolyhedrovirus and the identification of a baculoviral photolyase gene. Virology 2005, 338, 209–226. [Google Scholar] [CrossRef] [Green Version]
- Habib, S.; Pandey, S.; Chatterji, U.; Burma, S.; Ahmad, R.; Jain, A.; Hasnain, S.E. Bifunctionality of the AcMNPV homologous region sequence (hr1): Enhancer and ori functions have different sequence requirements. DNA Cell Biol. 1996, 15, 737–747. [Google Scholar] [CrossRef]
- Xie, W.D.; Arif, B.; Dobos, P.; Krell, P.J. Identification and analysis of a putative origin of DNA replication in the Choristoneura fumiferana multinucleocapsid nuclear polyhedrosis virus genome. Virology 1995, 209, 409–419. [Google Scholar] [CrossRef]
- Levin, D.B.; Huang, J. Identification and functional analysis of a putative non-hr origin of DNA replication from the Spodoptera littoralis type B multinucleocapsid nucleopolyhedrovirus. J. Gen. Virol. 1999, 80, 2263–2274. [Google Scholar]
- Pijlman, G.P.; Dortmans, J.C.F.M.; Vermeesch, A.M.G.; Yang, K.; Martens, D.E.; Goldbach, R.W.; Vlak, J.M. Pivotal role of the non-hr origin of DNA replication in the genesis of defective interfering baculoviruses. J. Virol. 2002, 76, 5605–5611. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Vermeesch, A.M.G.; Vlak, J.M. Cell line-specific accumulation of the baculovirus non-hr origin of DNA replication in infected insect cells. J. Invertebr. Pathol. 2003, 84, 214–219. [Google Scholar] [CrossRef]
- Guarino, L.A.; Summers, M.D. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J. Virol. 1986, 57, 563–571. [Google Scholar] [Green Version]
- Guarino, L.A.; Gonzalez, M.A.; Summers, M.D. Complete Sequence and Enhancer Function of the Homologous DNA Regions of Autographa californica Nuclear Polyhedrosis Virus. J. Virol. 1986, 60, 224–229. [Google Scholar]
- Rodems, S.M.; Friesen, P.D. Transcriptional enhancer activity of hr5 requires dual-palindrome half sites that mediate binding of a dimeric form of the baculovirus transregulator IE1. J. Virol. 1995, 69, 5368–5375. [Google Scholar]
- Leisy, D.J.; Rasmussen, C.; Kim, H.T.; Rohrmann, G.F. The Autographa californica nuclear polyhedrosis virus homologous region 1a: Identical sequences are essential for DNA replication activity and transcriptional enhancer function. Virology 1995, 208, 742–752. [Google Scholar] [CrossRef]
- Wormleaton, S.; Kuzio, J.; Winstanley, D. The complete sequence of the Adoxophyes orana granulovirus genome. Virology 2003, 311, 350–365. [Google Scholar] [CrossRef] [Green Version]
- Crook, N.; Luque, T.; O’Reilly, D.R.; Winstanley, D.; Finch, R. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 2001, 82, 2531–2547. [Google Scholar]
- Wang, Y.; Choi, J.Y.; Roh, J.Y.; Liu, Q.; Tao, X.Y.; Park, J.B.; Kim, J.S.; Je, Y.H. Genomic Sequence Analysis of Granulovirus Isolated from the Tobacco Cutworm, Spodoptera litura. PLoS ONE 2011, 6, e28163. [Google Scholar] [CrossRef]
- Ki, J.-S.; Hop, H.; Kim, S.-J.; Kim, I.-C.; Park, H.G.; Lee, J.-S. Complete mitochondrial genome sequence of the Arctic gammarid, Onisimus nanseni (Crustacea; Amphipoda): Novel gene structures and unusual control region features. Comp. Biochem. Physiol. Part D Genom. Proteom. 2010, 5, 105–115. [Google Scholar] [CrossRef]
- Cheung, A.K. Porcine circovirus: Transcription and DNA replication. Virus Res. 2012, 164, 46–53. [Google Scholar] [CrossRef]
- Macao, B.; Olsson, M.; Elias, P. Functional Properties of the Herpes Simplex Virus Type I Origin-binding Protein Are Controlled by Precise Interactions with the Activated Form of the Origin of DNA Replication. J. Biol. Chem. 2004, 279, 29211–29217. [Google Scholar] [CrossRef] [Green Version]
- Bikard, D.; Loot, C.; Baharoglu, Z.; Mazel, D. Folded DNA in action: Hairpin formation and biological functions in prokaryotes. Microbiol. Mol. Biol. Rev. 2010, 74, 570–588. [Google Scholar] [CrossRef]
- Sun, W.; Godson, G.N. Structure of the Escherichia coli primase/single-strand DNA-binding protein/phage G4oric complex required for primer RNA synthesis. J. Mol. Biol. 1998, 276, 689–703. [Google Scholar] [CrossRef]
- Masai, H.; Nomura, N.; Arai, K. The ABC-primosome. A novel priming system employing dnaA, dnaB, dnaC, and primase on a hairpin containing a dnaA box sequence. J. Biol. Chem. 1990, 265, 15134–15144. [Google Scholar]
- Miao, D.M.; Honda, Y.; Tanaka, K.; Higashi, A.; Nakamura, T.; Taguchi, Y.; Sakai, H.; Komano, T.; Bagdasarian, M. A base-paired hairpin structure essential for the functional priming signal for DNA replication of the broad host range plasmid RSF1010. Nucleic Acids Res. 1993, 21, 4900–4903. [Google Scholar] [CrossRef] [Green Version]
- Noirot, P.; Bargonetti, J.; Novick, R.P. Initiation of rolling-circle replication in pT181 plasmid: Initiator protein enhances cruciform extrusion at the origin. Proc. Natl. Acad. Sci. USA 1990, 87, 8560–8564. [Google Scholar] [CrossRef]
- Lin Guangyun, B.G.W. Analysis of an Autographa californica Nucleopolyhedrovirus lef-11 Knockout: LEF-11 Is Essential for Viral DNA Replication. J. Virol. 2002, 76, 2770–2779. [Google Scholar]




| ORI-hr | Clone Number | Position (bp) | Identity Sequence | Repetitions | ||||||
| 39 | 107,445–106,982 | hr9 | 8× GCTTTWCRARYACMRTYRWTHKTGWAAARCD | |||||||
| 113 | 64,519–63,441 | hr7 | 1× GTTTTACAAAAACAATCGTATGTGAAAAAC | |||||||
| ORI-Igr | Number | Position | Surrounding ORFs | Shared by | ||||||
| 27 | 45,424–44,663 | ORF60 (he65-like), ORF58 | ORF57 (pnk-pnl) | Most GI, some GII and 3 Beta | 5 baculovirus genomes | |||||
| 119/141 | 69,305–69,564 | ORF85 (ac78-like) | ORF86 (vlf1) | Baculoviridae | Baculoviridae | |||||
| 133 | 102,699–101,360 | ORF126 | ORF125 | ORF124 (lef-6) | ORF123 (iap-1) | GI | GI, most GII and Beta | Alpha and Beta | GI | |
| 150 | 95,156–94,754 | ORF115 | ORF114 (odv-e66) | GI and SeMNPV | GI, most GII, Beta | |||||
| 33/162 | 52,036–52,450 | ORF67 (lef-5) | ORF68 (38k) | Baculoviridae | Baculoviridae | |||||
| ORI-gr | Number | Position | ORF | Shared by | ||||||
| 26 | 118,470–118,641 | egt | GI, GII, and most Beta | |||||||
| 104 | 69,883–70,458 | vlf-1 | Baculoviridae | |||||||
| 131 | 60,314–60,131 | vlf-1 | Baculoviridae | |||||||
| 136 | 90,683–91,393 | lef-8 | Baculoviridae | |||||||
| 144 | 74,559–74,844 | lef-3 | Alpha and Beta | |||||||
| 153 | 50,359–50,236 | p40 | Baculoviridae | |||||||
| 168 | 66,087–65,983 | vp91 | Baculoviridae | |||||||
| ORI | Clone Number | Promoter Pattern | Palindrome Pattern | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | Early | Late | INR | TATAAA | Perfect | Imperfect | |||||||||||||||
| + | - | + | - | + | - | + | - | 5 nt | 6 nt | 7 nt | 8 nt | 11 nt | 5 nt | 6 nt | 7 nt | 8 nt | 9 nt | >10 nt | |||
| ORI-hr | 39 | 833 | 7 | 10 | 1 | - | - | - | - | - | 11 | 4 | 2 | - | - | 90 | 23 | 10 | 4 | 5 | 2 |
| 113 | 1080 | 9 (3*) | 11 (3*) | - | - | 1* | 1 | - | - | 30 | 13 | 2 | - | 1 | 147 | 43 | 25 | 10 | 3 | 2 | |
| ORI-igr | 27 | 763 | 4* | 3* | - | - | - | - | - | 1* | 17 | 4 | 1 | - | - | 88 | 32 | 10 | 4 | - | - |
| 119/141 | 260 | 3 | 1 | 1* | - | - | - | - | - | 5 | 1 | - | - | - | 21 | 10 | 3 | 2 | - | - | |
| 133 | 1337 | 11 (1*) | 11 (2*) | 3 (1*) | - | - | - | - | - | 28 | 9 | 3 | 1 | - | 150 | 68 | 13 | 4 | 2 | 2 | |
| 150 | 403 | 5 (2*) | - | 1* | - | 1* | - | 1* | 1 | 14 | 1 | 2 | 1 | - | 46 | 19 | 16 | 1 | 3 | 2 | |
| 33/162 | 415 | 2 (1*) | 3* | - | - | 1* | - | - | - | 10 | - | 1 | - | - | 45 | 18 | 4 | 3 | 1 | - | |
| ORI-gr | 26 | 172 | 2 | 2 | - | - | - | - | - | - | 2 | - | - | - | - | 13 | 5 | 2 | 1 | 1 | - |
| 104 | 576 | 1 | 4 | - | - | 1 | - | - | - | 6 | - | 1 | 1 | - | 57 | 31 | 8 | - | 1 | 1 | |
| 131 | 318 | 1 | 1 | 1 | - | - | - | - | - | 9 | - | - | - | - | 39 | 10 | 8 | 1 | - | - | |
| 136 | 711 | 7 | 10 | 1 | 2 | - | 1 | - | - | 13 | - | 5 | 1 | - | 69 | 30 | 6 | 3 | - | - | |
| 144 | 286 | 2 | 5 | - | - | - | - | - | - | 7 | - | 4 | 3 | - | 45 | 14 | 8 | 2 | 1 | - | |
| 153 | 124 | - | 2 | - | - | - | - | - | - | 2 | - | - | - | - | 6 | 2 | - | 1 | - | - | |
| 168 | 105 | - | 2 | - | - | - | - | - | 2 | - | - | - | - | 5 | 1 | 1 | - | - | - | ||
| Clone Number | Number of Structures | Start (bp) | Length | AT% | End (bp) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 (bp) | Linker 1 (nt) | S2 (bp) | Linker 2 (nt) | S3 (bp) | S1 | Linker 1 | S2 | Linker 2 | S3 | |||||
| ORI-hr | 39 | 5 | 107,396 | 3 | 2 | 6 | 2 | 4 | 77.7 | 100 | 65.2 | 0 | 53.8 | 107,348 |
| 107,360 | 4 | 2 | 10 | 4 | 2 | 53.8 | 50 | 56.2 | 50 | 50 | 107,294 | |||
| 107,309 | 2 | 2 | 8 | 3 | 8 | 50 | 100 | 60.5 | 33.3 | 56.5 | 107,227 | |||
| 107,204 | 5 | 0 | 6 | 2 | 4 | 61.5 | 0 | 65.2 | 0 | 53.8 | 107,151 | |||
| 107,121 | 14 | 6 | 15 | 6 | 3 | 50 | 66.7 | 62.5 | 66.7 | 45.4 | 107,010 | |||
| 113 | 8 | 64,384 | 6 | 5 | 9 | 3 | 10 | 40.9 | 80 | 36.7 | 66.7 | 40 | 64,295 | |
| 64,266 | 5 | 5 | 10 | 3 | 4 | 81.2 | 80 | 72.5 | 0 | 76.9 | 64,187 | |||
| 64,169 | 4 | 2 | 5 | 3 | 2 | 100 | 50 | 57.1 | 66.7 | 75 | 64,130 | |||
| 63,982 | 4 | 4 | 8 | 4 | 4 | 84.6 | 50 | 96.1 | 100 | 63.6 | 63,923 | |||
| 63,872 | 4 | 2 | 11 | 1 | 6 | 63.2 | 100 | 96.8 | 100 | 88.2 | 63,803 | |||
| 63,784 | 3 | 3 | 7 | 5 | 4 | 54.5 | 66.7 | 65.2 | 80 | 76.9 | 63,730 | |||
| 63,662 | 3 | 3 | 6 | 2 | 2 | 83.3 | 100 | 52.4 | 0 | 55.6 | 63,616 | |||
| 63,567 | 3 | 2 | 7 | 5 | 3 | 66.7 | 100 | 73.5 | 80 | 40.0 | 63,505 | |||
| ORI-Igr | 27 | 3 | 45,405 | 3 | 2 | 10 | 1 | 5 | 33.3 | 100.0 | 63.0 | 100.0 | 64.3 | 45,353 |
| 44,997 | 3 | 4 | 6 | 2 | 2 | 50.0 | 75.0 | 70.6 | 100.0 | 53.8 | 44,952 | |||
| 44,764 | 3 | 3 | 8 | 3 | 4 | 53.8 | 66.7 | 65.5 | 33.3 | 66.7 | 44,709 | |||
| 119/141 | 1 | 69,393 | 4 | 1 | 9 | 3 | 6 | 92.3 | 100.0 | 69.7 | 66.7 | 47.4 | 69,464 | |
| 133 | 8 | 102,620 | 3 | 3 | 10 | 3 | 8 | 60.0 | 66.7 | 44.1 | 66.7 | 33.3 | 102,547 | |
| 102,443 | 6 | 2 | 8 | 2 | 4 | 64.7 | 100.0 | 43.3 | 0.0 | 45.5 | 102,382 | |||
| 102,333 | 5 | 1 | 7 | 2 | 3 | 86.7 | 0.0 | 63.2 | 50.0 | 60.0 | 102,287 | |||
| 102,084 | 3 | 3 | 11 | 5 | 4 | 90.9 | 100.0 | 81.3 | 100.0 | 82.4 | 102,017 | |||
| 101,984 | 3 | 4 | 5 | 3 | 3 | 40.0 | 50.0 | 45.8 | 33.3 | 61.1 | 101,921 | |||
| 101,854 | 6 | 5 | 6 | 0 | 5 | 56.3 | 80.0 | 40.9 | 0 | 36.8 | 101,793 | |||
| 101,727 | 12 | 4 | 11 | 4 | 10 | 51.4 | 75.0 | 42.5 | 75.0 | 36.7 | 101,619 | |||
| 101,468 | 7 | 5 | 11 | 0 | 4 | 77.8 | 60.0 | 44.4 | 0 | 27.3 | 101,394 | |||
| 150 | 1 | 95,126 | 4 | 2 | 5 | 3 | 6 | 50.0 | 0.0 | 70.0 | 100.0 | 75.0 | 95,074 | |
| 33/162 | 2 | 52,068 | 5 | 5 | 13 | 2 | 3 | 56 | 100 | 75 | 50.0 | 44 | 52,137 | |
| 52,246 | 8 | 3 | 16 | 6 | 10 | 66.7 | 33.3 | 71.1 | 83.3 | 41.7 | 52,355 | |||
| ORI-gr | 26 | 1 | 118,483 | 4 | 2 | 7 | 4 | 3 | 18.2 | 50.0 | 28.6 | 50.0 | 54.5 | 118,531 |
| 104 | 3 | 70,228 | 4 | 5 | 6 | 0 | 5 | 41.7 | 40 | 47.4 | 0 | 33.3 | 70,281 | |
| 70,264 | 5 | 3 | 8 | 2 | 7 | 33.3 | 100 | 64.5 | 50 | 45 | 70,337 | |||
| 70,379 | 6 | 4 | 13 | 5 | 2 | 65.2 | 75.0 | 30 | 60.0 | 30.0 | 70,457 | |||
| 131 | 2 | 69,698 | 3 | 3 | 5 | 3 | 5 | 0.8 | 66.7 | 39 | 66.7 | 66.7 | 69,750 | |
| 69,820 | 4 | 4 | 6 | 3 | 5 | 36.4 | 50 | 66.7 | 66.7 | 52.9 | 69,881 | |||
| 136 | 4 | 90,742 | 7 | 3 | 11 | 4 | 5 | 70 | 66.7 | 51.3 | 25 | 46.7 | 90,822 | |
| 90,808 | 5 | 2 | 6 | 4 | 2 | 47 | 50.0 | 40.9 | 75 | 22.2 | 90,859 | |||
| 90,972 | 2 | 5 | 9 | 3 | 2 | 40 | 60 | 67 | 66.7 | 44 | 91,025 | |||
| 91,079 | 5 | 4 | 6 | 5 | 3 | 20 | 50 | 37 | 60.0 | 70 | 91,131 | |||
| 144 | 1 | 74,567 | 4 | 5 | 14 | 3 | 10 | 83.3 | 60.0 | 75.0 | 66.7 | 16.1 | 74,657 | |
| 153 | 1 | 50,358 | 3 | 4 | 7 | 3 | 4 | 30.0 | 50.0 | 41.4 | 66.7 | 82.4 | 50,296 | |
| 168 | 1 | 66,071 | 4 | 2 | 6 | 4 | 3 | 37.5 | 50.0 | 36.8 | 100.0 | 72.7 | 66,020 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miele, S.A.B.; Cerrudo, C.S.; Parsza, C.N.; Nugnes, M.V.; Mengual Gómez, D.L.; Belaich, M.N.; Ghiringhelli, P.D. Identification of Multiple Replication Stages and Origins in the Nucleopolyhedrovirus of Anticarsia gemmatalis. Viruses 2019, 11, 648. https://doi.org/10.3390/v11070648
Miele SAB, Cerrudo CS, Parsza CN, Nugnes MV, Mengual Gómez DL, Belaich MN, Ghiringhelli PD. Identification of Multiple Replication Stages and Origins in the Nucleopolyhedrovirus of Anticarsia gemmatalis. Viruses. 2019; 11(7):648. https://doi.org/10.3390/v11070648
Chicago/Turabian StyleMiele, Solange A.B., Carolina S. Cerrudo, Cintia N. Parsza, María Victoria Nugnes, Diego L. Mengual Gómez, Mariano N. Belaich, and P. Daniel Ghiringhelli. 2019. "Identification of Multiple Replication Stages and Origins in the Nucleopolyhedrovirus of Anticarsia gemmatalis" Viruses 11, no. 7: 648. https://doi.org/10.3390/v11070648