Efficient Segmentation of a Breast in B-Mode Ultrasound Tomography Using Three-Dimensional GrabCut (GC3D)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Image Preprocessing
2.3. Benchmark Building
2.4. GC3D
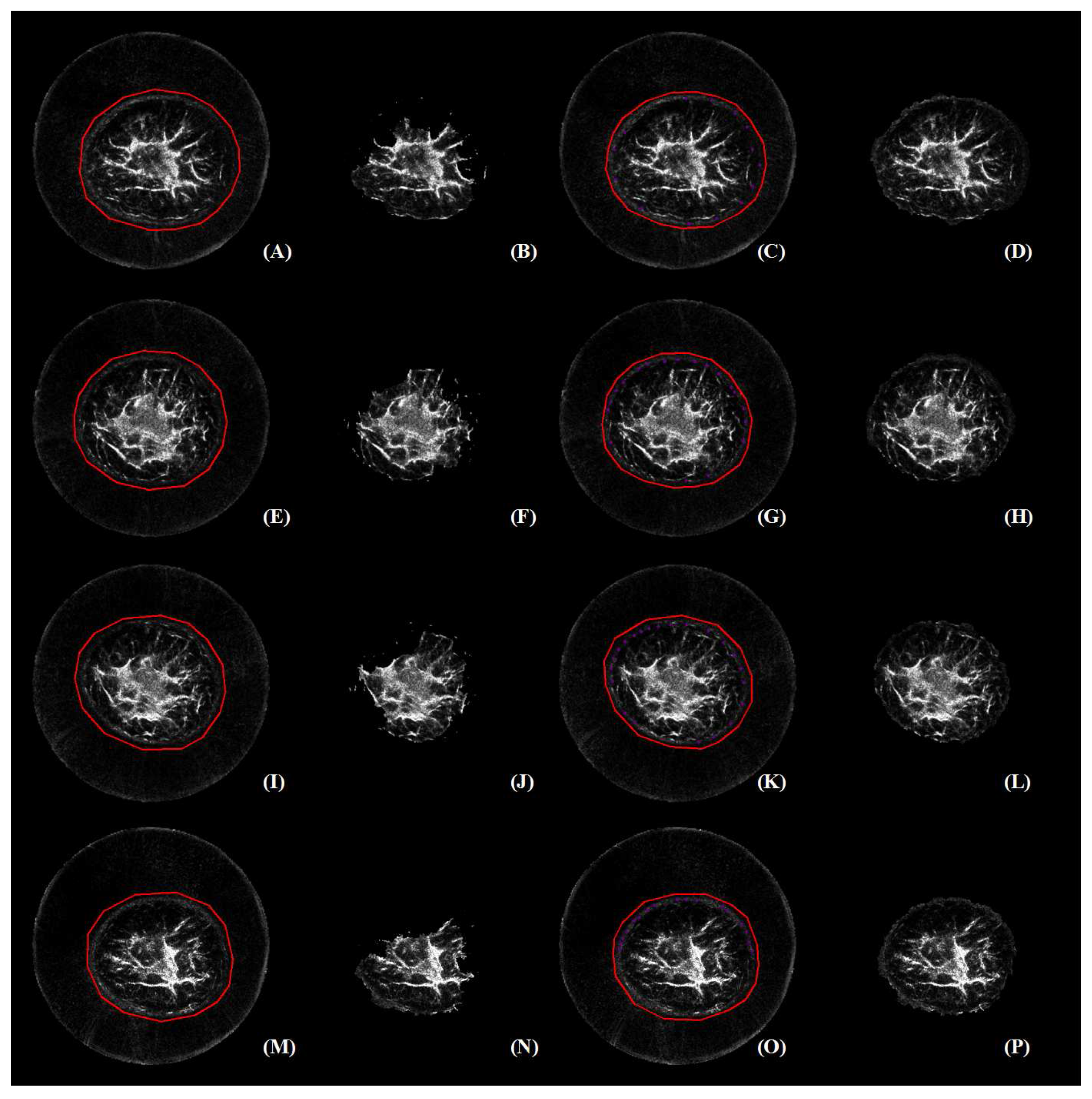
2.4.1. Image Segmentation
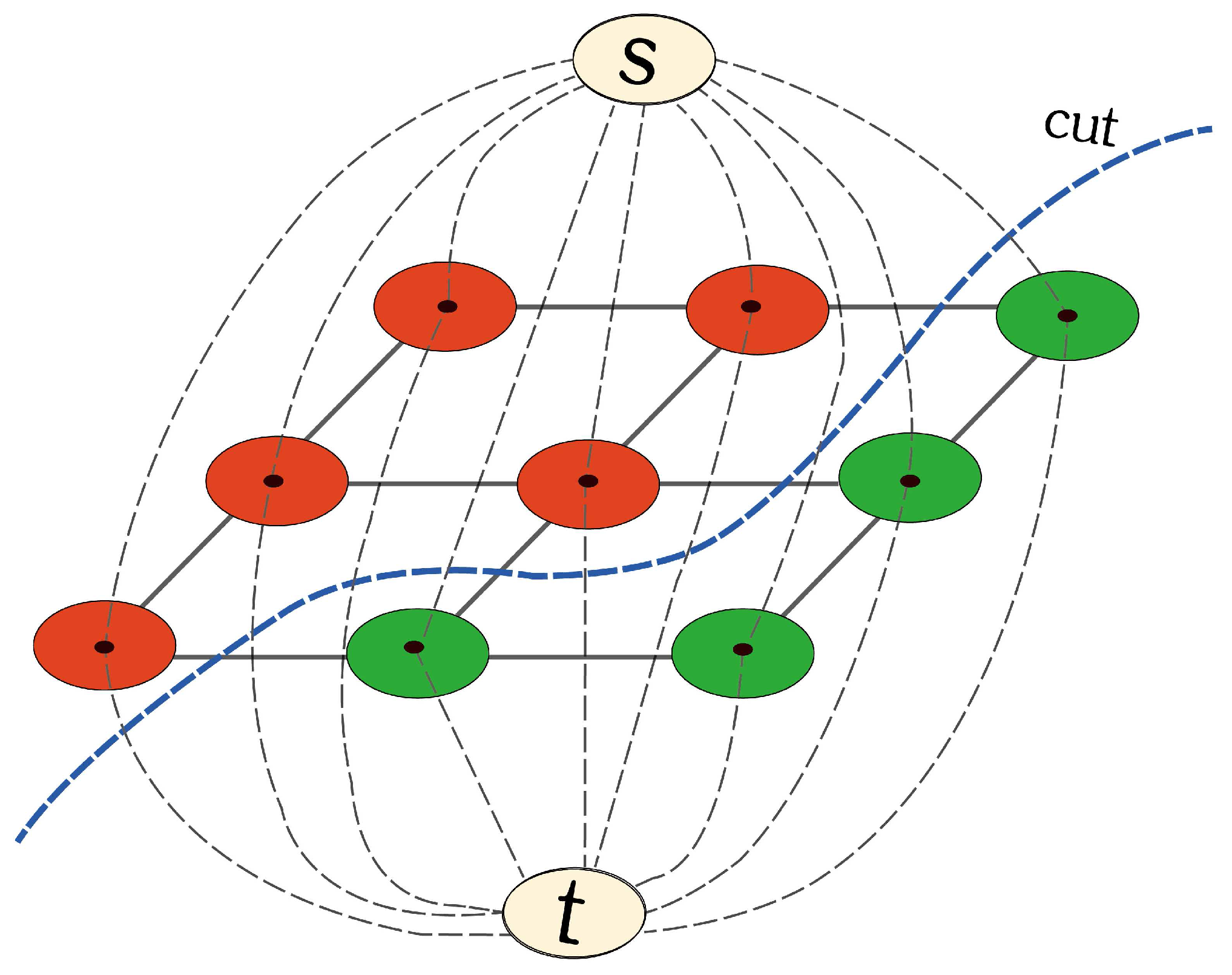
2.4.2. s-t Graph
2.4.3. Segmentation by Iterative Energy Minimization
2.4.4. Implementation of GC3D
2.5. Experiment Design
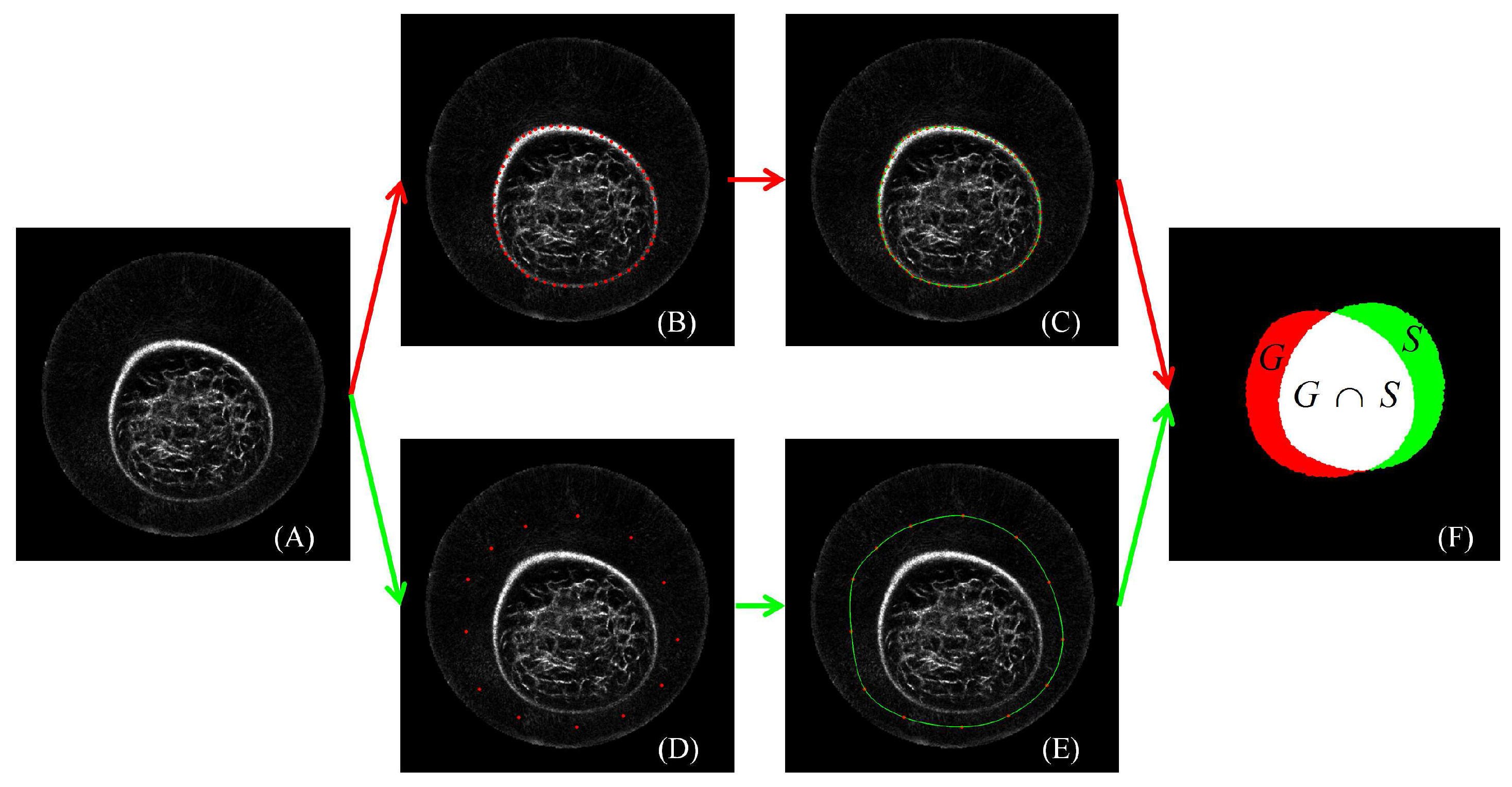
2.6. Performance Evaluation
2.7. Software Platform
3. Results
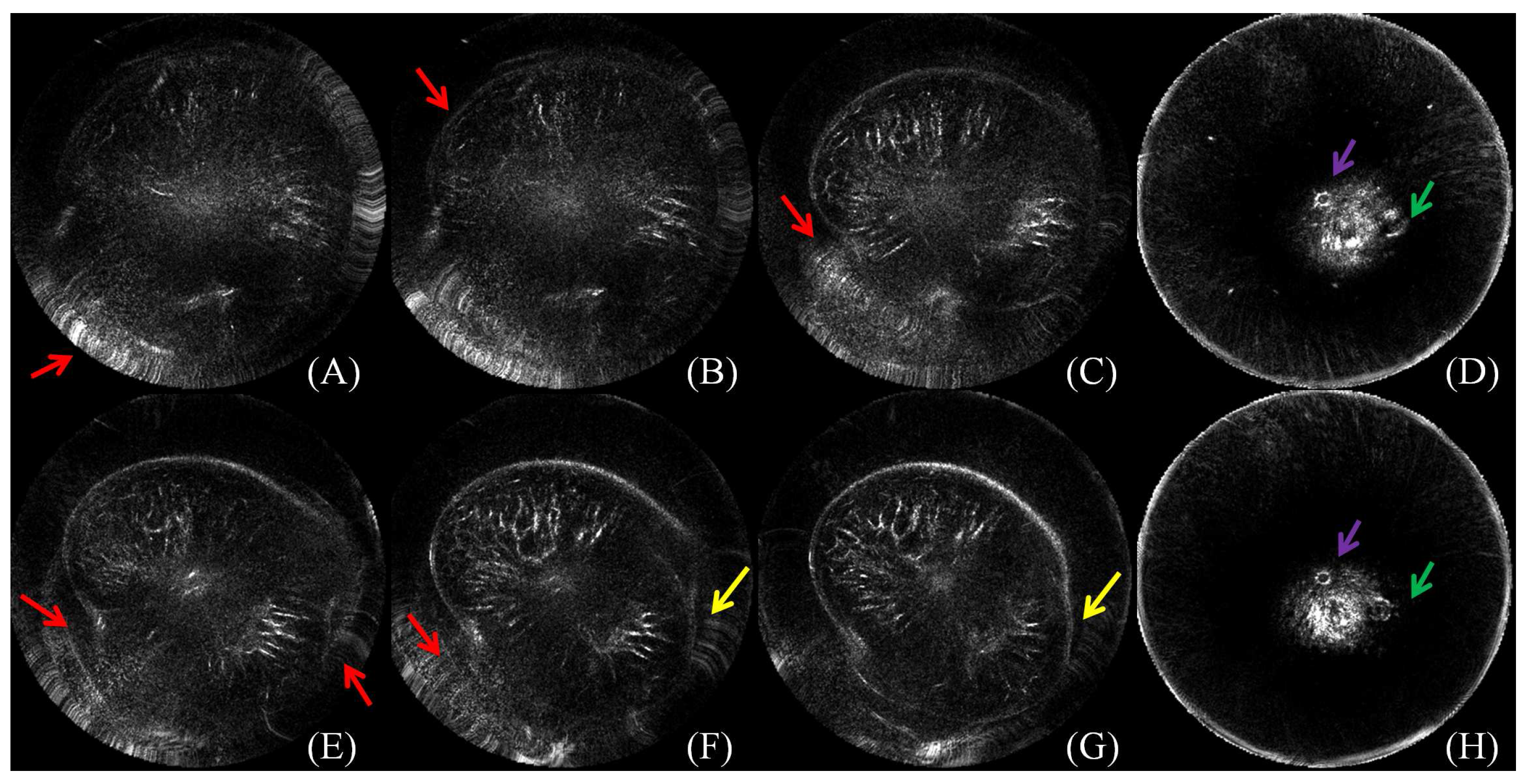
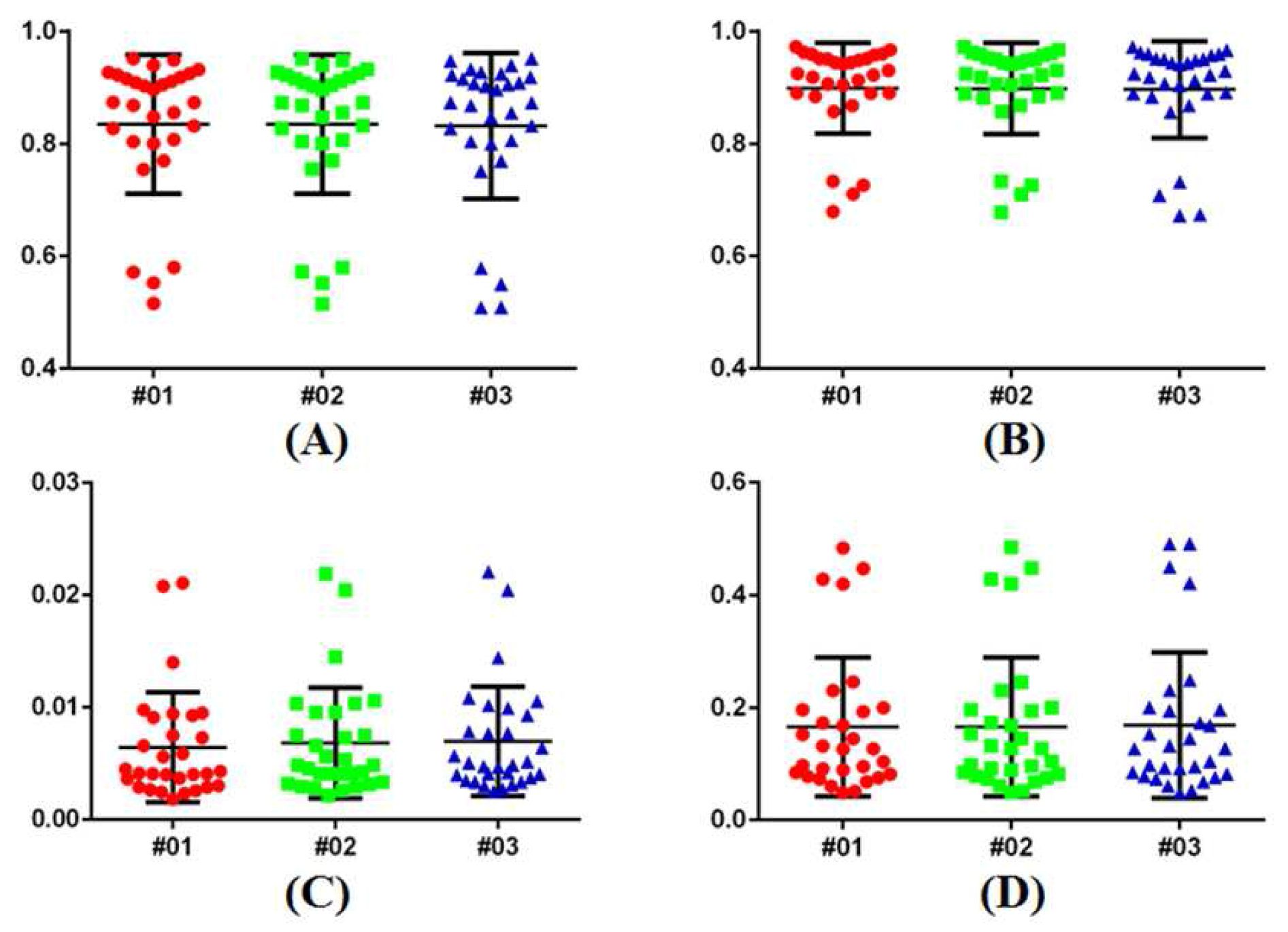
3.1. Reliability of the Ground Truth Data
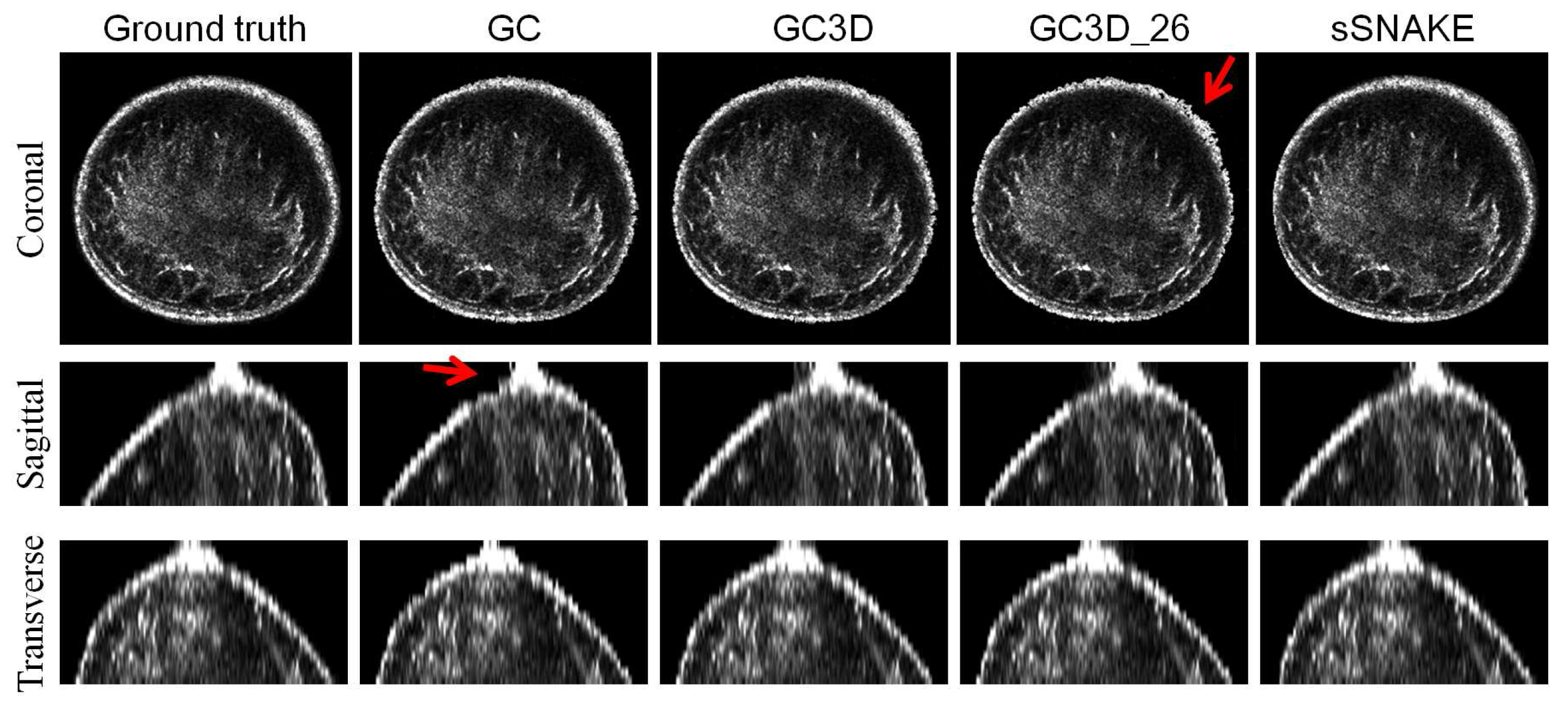
3.2. Perceived Evaluation
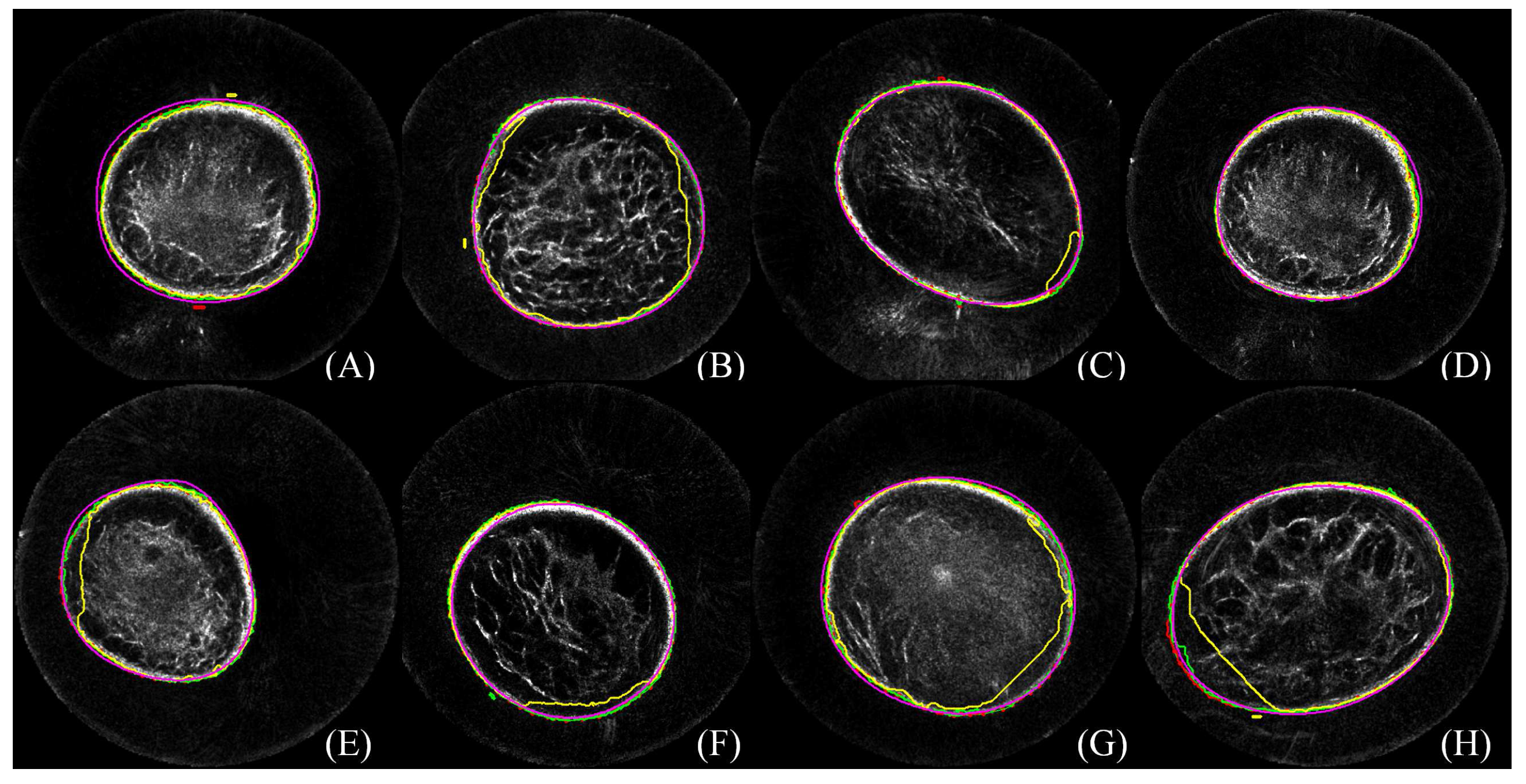
3.3. Quantitative Comparison
3.4. Ease-Of-Use
3.5. Robustness
3.6. Failure Case Analysis
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| UST | Ultrasound tomography |
| CT | Computerized tomography |
| MRI | Magnetic resonance imaging |
| GC3D | Three-dimensional GrabCut that takes six-connected neighboring voxels into computing |
| GC | GrabCut |
| GC3D_26 | Three-dimensional GrabCut that takes 26-connected neighboring voxels into computing |
| sSNAKE | A simplified SNAKE or active contour method |
| GMM | Gaussian mixture model |
| TO | Target overlap |
| MO | Mean overlap |
| FP | False positive |
| FN | False negative |
| TC | Time consumption |
References
- Siegel, R.; Miller, K.; Jemal, A. Cancer Statistics 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Strasser-Weippl, K.; Li, J.; St Louis, J.; Finkelstein, D.M.; Yu, K.; Chen, W.; Shao, Z.; Goss, P.E. Breast cancer in China. Lancet Oncol. 2014, 15, e279–e289. [Google Scholar] [CrossRef]
- Sak, M.A.; Littrup, P.J.; Duric, N.; Mullooly, M.; Sherman, M.E.; Gierach, G.L. Current and future methods for measuring breast density: A brief comparative review. Breast Cancer Manag. 2015, 4, 209–221. [Google Scholar] [CrossRef]
- Duric, N.; Littrup, P.; Schmidt, S.; Li, C.; Roy, O.; Bay-Knight, L.; Janer, R.; Kunz, D.; Chen, X.; Goll, J.; et al. Breast imaging with the softvue imaging system: First results. Med. Imaging Ultrason. Imaging Tomogr. Ther. 2013, 8675, 185–187. [Google Scholar]
- Duric, N.; Littrup, P.; Li, C.; Roy, O.; Schmidt, S.; Cheng, X.; Seamans, J.; Wallen, A.; Bay-Knight, L. Breast imaging with softvue: Initial clinical evaluation. SPIE Med. Imaging 2014, 2014, 252–260. [Google Scholar]
- Duric, N.; Littrup, P.; Poulo, L.; Babkin, A.; Pevzner, R.; Holsapple, E.; Rama, O.; Glide, C. Detection of breast cancer with ultrasound tomography: First results with the computed ultrasound risk evaluation (CURE) prototype. Med. Phys. 2007, 34, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Duric, N.; Littrup, P.; Huang, L. In vivo breast sound-speed imaging with ultrasound tomography. Ultrasound Med. Biol. 2009, 35, 1615–1628. [Google Scholar] [CrossRef] [PubMed]
- Ranger, B.; Littrup, P.; Duric, N.; Chandiwala-Mody, P.; Li, C.; Schmidt, S.; Lupinacci, J. Breast ultrasound tomography versus magnetic resonance imaging for clinical display of anatomy and tumor rendering: Preliminary results. Am. J. Roentgenol. 2012, 198, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Sak, M.; Duric, N.; Littrup, P.; Ali, H.; Vallieres, P.; Sherman, M.; Gierach, G. Using speed of sound imaging to characterize breast density. Ultrasound Med. Biol. 2017, 43, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Glide-Hurst, C.; Duric-Weippl, N. Novel approach to evaluating breast density utilizing ultrasound tomography. Med. Phys. 2007, 24, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Glide-Hurst, C.; Duric-Weippl, N.; Littrup, P. Volumetric breast density evaluation from ultrasound tomography images. Med. Phys. 2008, 35, 3988–3997. [Google Scholar] [CrossRef] [PubMed]
- Duric, N.; Norman, B.; Littrup, P.; Sak, M.; Myc, C.; Li, C.; West, E.; Minkin, S.; Martin, L.; Yaffe, M.; et al. Breast density measurements with ultrasound tomography: A comparison with film and digital mammography. Med. Phys. 2013, 40, 013501. [Google Scholar] [CrossRef] [PubMed]
- Khodr, Z.; Sak, M.; Pfeiffer, R.; Duric, N.; Littrup, P.; Bey-Knight, L.; Ali, H.; Vallieres, P.; Sherman, M.; Gierach, G. Determinants of the reliability of ultrasound tomography sound speed estimates as a surrogate for volumetric breast density. Med. Phys. 2015, 42, 5671–5678. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, E.; Fromageau, J.; Ledger, J.; Messa, M.; D’Aquino, A.; Schoemaker, A. Breast density measurements with ultrasound tomography: A comparison with non-contrast MRI. Breast Cancer Res. 2015, 17 (Suppl. S1), O3. [Google Scholar]
- Ruiter, N.V.; Hopp, T.; Zapf, M.; Kretzek, E.; Gemmeke, A. Analysis of patient movement during 3D USCT data acquisition. SPIE Med. Imaging 2016, 2016, 979009. [Google Scholar]
- Sak, M.; Duric, N.; Littrup, P.; Li, C.; Bey-Knight, L.; Sherman, M.; Boyd, N.; Gierach, G. Breast density measurements using ultrasound tomography for patients undergoing tamoxifen treatment. SPIE Med. Imaging 2013, 2013, 86751. [Google Scholar]
- Ou, Y.; Weinstein, S.P.; Conant, E.F.; Englander, S.; Da, X.; Gaonkar, B.; Hsieh, M.K.; Rosen, M.; DeMichele, A.; Davatzikos, C.; et al. Deformable registration for quantifying longitudinal tumor changes during neoadjuvant chemotherapy. Magn. Reson. Med. 2015, 73, 2343–2356. [Google Scholar] [CrossRef] [PubMed]
- Hopp, T.; Zapf, M.; Kretzek, E.; Henrich, J.; Tukalo, A.; Gemmeke, H.; Kaiser, C.; Knaudt, J.; Ruiter, N.V. 3D Ultrasound Computer Tomography: Update from a clinical study. SPIE Med. Imaging 2016, 2016, 97900A. [Google Scholar]
- Sak, M.; Duric, N.; Littrup, P.; Sherman, M.; Gierach, G. Ultrasound tomography imaging with waveform sound speed: Parenchymal changes in women undergoing tamoxifen therapy. SPIE Med. Imaging 2017, 2017, 101390W. [Google Scholar]
- Hopp, T.; Dapp, R.; Zapf, M.; Kretzek, E.; Gemmeke, H.; Ruiter, N.V. Registration of 3D ultrasound computer tomography and MRI for evaluation of tissue correspondences. SPIE Med. Imaging 2015, 2015, 94190Q. [Google Scholar]
- Hopp, T.; Duric, N.; Ruiter, N.V. Automatic multimodal 2D/3D breast image registration using biomechanical FEM models and intensity-based optimization. Med. Image Anal. 2013, 17, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Hopp, T.; Duric, N.; Ruiter, N.V. Image fusion of ultrasound computer tomography volumes with X-ray mammograms using a biomechanical model based 2D/3D registration. Comput. Med. Imaging Gr. 2015, 40, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Majdouline, Y.; Ohayon, J.; Keshavarz-Motamed, Z.; Cardinal, M.H.C.; Garcia, D.; Allard, L.; Lerouge, S.; Arsenault, F.; Soulez, G. Endovascular shear strain elastography for the detection and characterization of the severity of atherosclerotic plaques: In vitro validation and in vivo evaluation. Ultrasound Med. Biol. 2014, 40, 890–903. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Guo, X.; Huang, Q. Systematic evaluation on speckle suppression methods in examination of ultrasound breast images. Appl. Sci. 2016, 7, 37. [Google Scholar] [CrossRef]
- Balic, I.; Goyal, P.; Roy, O.; Duric, N. Breast boundary detection with active contours. SPIE Med. Imaging 2014, 2014, 90400D. [Google Scholar]
- Delgado-Gonzalo, D.; Chenouard, N.; Unser, M. Spline-based deforming ellipsoids for interactive 3D bioimage segmentation. IEEE Trans. Image Process. 2013, 22, 3926–3940. [Google Scholar] [CrossRef] [PubMed]
- Hopp, T.; Zapf, M.; Ruiter, N.V. Segmentation of 3D ultrasound computer tomography reflection images using edge detection and surface fitting. SPIE Med. Imaging 2014, 2014, 90401R. [Google Scholar]
- Jain, A.K. Data clustering: 50 years beyond K-means. Pattern Recog. Lett. 2010, 31, 651–666. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. Automatica 1975, 11, 23–27. [Google Scholar] [CrossRef]
- Sak, M.; Duric, N.; Littrup, P.; Westerberg, K. A comparison of automated versus manual segmentation of breast UST transmission images to measure breast volume and sound speed. SPIE Med. Imaging 2017, 2017, 101391H. [Google Scholar]
- Hopp, T.; You, W.; Zapf, M.; Tan, W.Y.; Gemmeke, H.; Ruiter, N.V. Automated breast segmentation in Ultrasound Computer Tomography SAFT images. SPIE Med. Imaging 2017, 2017, 101390G. [Google Scholar]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vision 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Xu, C.; Prince, J.L. Snakes, shapes, and gradient vector flow. IEEE Trans. Image Process. 1998, 7, 359–369. [Google Scholar] [PubMed]
- Cabezas, M.; Oliver, A.; Llado, X.; Freixenet, J.; Cuadra, M.B. A review of atlas-based segmentation for magnetic resonance brain images. Comput. Methods Progr. Biomed. 2011, 104, e158–e177. [Google Scholar] [CrossRef] [PubMed]
- Heimann, T.; Meinzer, H.P. Statistical shape models for 3D medical image segmentation: A review. Med. Image Anal. 2009, 13, 543–563. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Zhang, L.; Zhang, D. A survey of graph theoretical approaches to image segmentation. Pattern Recogn. 2013, 46, 1020–1038. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, S.; Zhou, Q. A novel gradient vector flow Snake model based on convex function for infrared image segmentation. Sensors 2016, 16, 1756. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tang, Z.; Cui, Y.; Wu, G. Local competition-based superpixel segmentation algorithm in remote sensing. Sensors 2017, 17, 1364. [Google Scholar] [CrossRef] [PubMed]
- Grau, V.; Mewes, A.U.J.; Alcaniz, M.; Kikinis, R.; Warfield, S.K. Improved watershed transform for medical image segmentation using prior information. IEEE Trans. Med. Imaging 2004, 23, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, F.; Li, G.; Gao, Y.; Lin, W.; Gilmore, J.H.; Shen, D. Segmentation of neonatal brain MR images using patch-driven level sets. NeuroImage 2014, 84, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Boykov, Y.; Funka-Lea, G. Graph cuts and efficient N-D image segmentation. Int. J. Comput.Vision 2006, 70, 109–131. [Google Scholar]
- Rother, C.; Kolmogorov, V.; Blake, A. Grabcut: Interactive foreground extraction using iterated graph cuts. ACM Trans. Gr. 2004, 23, 309–314. [Google Scholar] [CrossRef]
- Blake, A.; Rother, C.; Brown, M.; Perez, P.; Torr, P. Interactive image segmentation using an adaptive GMMRF model. ECCV 2004, 2004, 428–441. [Google Scholar]
- Lee, G.; Lee, S.; Kim, G.; Park, J.; Park, Y. A modified GrabCut using a clustering technique to reduce image noise. Symmetry 2016, 8, 64. [Google Scholar] [CrossRef]
- Chen, D.; Li, G.; Sun, Y.; Kong, J.; Jiang, G.; Tang, H.; Ju, Z.; Yu, H.; Liu, H. An interactive image segmentation method in hand gesture recognition. Sensors 2017, 17, 253. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Xie, Y. Interactive contour delineation and refinement in treatment planning of image-guided radiation therapy. J. Appl. Clin. Med. Phys. 2014, 15, 4499. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef]
- Ciccone, L.; Guay, M.; Sumner, R. Flow Curves: An Intuitive Interface for Coherent Scene Deformation. Comput. Gr. Forum 2016, 35, 247–256. [Google Scholar] [CrossRef]
- Boykov, Y.; Veksler, O.; Zabih, R. Fast approximate energy minimization via graph cuts. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 1222–1239. [Google Scholar] [CrossRef]
- Carreira, J.; Sminchisescu, C. CPMC: Automatic object segmentation using constrained parametric min-cuts. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 1312–1328. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.F.; Xu, X. Implementing Grabcut. Brigh. Young Univ. 2006, 3, 1–4. [Google Scholar]
- Lawler, E.L. Combinatorial Optimization: Networks and Matroids; Courier Corporation: Mineola, NY, USA, 2001. [Google Scholar]
- Boykov, Y.; Kolmogorov, V. An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.; Andersson, J.; Ardekani, B.A.; Ashburner, J.; Avants, B.; Chiang, M.C.; Christensen, G.E.; Collins, D.L.; Gee, J.; Hellier, P.; et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 2009, 46, 786–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shrout, P.E.; Fleiss, J.L. Intraclass correlations: Uses in assessing rater reliability. Psychol. Bulletin 1979, 86, 420–428. [Google Scholar] [CrossRef]
- McGraw, K.O.; Wong, S.P. Forming inferences about some intraclass correlation coefficients. Psychol. Methods 1996, 1, 30–46. [Google Scholar] [CrossRef]
- Lin, H.S.; Chen, Y.J.; Lu, H.L.; Lu, T.W.; Chen, C.C. Test–retest reliability of mandibular morphology measurements on cone-beam computed tomography-synthesized cephalograms with random head positioning errors. Biomed. Eng. Online 2017, 16, 62. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Temoche, P.; Carmona, R. A volume segmentation approach based on GrabCut. CLEI Electron. J. 2013, 16, 4–14. [Google Scholar]
- Chen, L.; Wu, S.; Zhang, Z.; Yu, S.; Xie, Y.; Zhang, H. Real-Time Patient Table Removal in CT Images; Springer HIS: Cham, Switzerland, 2016; pp. 1–8. [Google Scholar]
- Liang, X.; Zhang, Z.; Niu, T.; Yu, S.; Wu, S.; Li, Z.; Zhang, H.; Xie, Y. Iterative image-domain ring artifact removal in cone-beam CT. Phys. Med. Biol. 2017, 62, 5276–5292. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, G.; Li, C.; Roy, O.; Schmidt, S.; Duric, N. Frequency domain ultrasound waveform tomography: Breast imaging using a ring transducer. Phys. Med. Biol. 2015, 60, 5381. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Matthews, T.; Anis, F.; Li, C.; Duric, N.; Anastasio, M. Waveform inversion with source encoding for breast sound speed reconstruction in ultrasound computed tomography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2015, 62, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiao, L.; Shang, R.; Stolkin, R. Dynamic-context cooperative quantum-behaved particle swarm optimization based on multilevel thresholding applied to medical image segmentation. Inf. Sci. 2015, 294, 408–422. [Google Scholar] [CrossRef]
- Torbati, N.; Ayatollahi, A.; Kermani, A. An efficient neural network based method for medical image segmentation. Compute. Biol. Med. 2014, 44, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, R.; Deng, H.; Wang, L.; Lin, W.; Ji, S.; Shen, D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 2015, 108, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Shao, L.; Li, X.; Wang, L. Targeting accurate object extraction from an image: A comprehensive study of natural image matting. IEEE Trans. Neural Netw. Learn. Syst. 2015, 26, 185–207. [Google Scholar] [PubMed]
- Matthews, T.P.; Wang, K.; Li, C.; Duric, N.; Anastasio, M.A. Regularized dual averaging image reconstruction for full-wave ultrasound computed tomography. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2017, 64, 811–825. [Google Scholar] [CrossRef] [PubMed]
- Birk, M.; Kretzek, E.; Figuli, P.; Weber, M.; Bacjer, J.; Ruiter, N.V. High-speed medical imaging in 3D ultrasound computer tomography. IEEE Trans. Parallel Distrib. Syst. 2016, 27, 455–467. [Google Scholar] [CrossRef]


| SS’ | SJ | S’J | |
|---|---|---|---|
| 0.9762 ± 0.0417 | 0.9641 ± 0.0432 | 0.9657 ± 0.0556 | |
| p value | 0.4359 | 0.3934 | 0.7649 |
| ICC | 0.9517 | 0.9588 | 0.9757 |
| TO | MO | FN | FP | TC (Min) | No. of Points Localized | |
|---|---|---|---|---|---|---|
| Benchmark | 11.8 ± 4.82 | 423 ± 87 | ||||
| GC | 0.82 | 0.90 | 0.005 | 0.18 | 2.37 ± 0.84 | 98 ± 21 |
| GC3D | 0.84 | 0.91 | 0.006 | 0.16 | 1.23 ± 0.62 | 12 ± 3 |
| GC3D_26 | 0.65 | 0.75 | 0.023 | 0.35 | 1.48 ± 0.65 | 12 ± 3 |
| sSNAKE | 0.89 | 0.93 | 0.029 | 0.11 | 36.8 ± 5.16 | 226 ± 32 |
| TO | MO | FN | FP | TC (Min) | |
|---|---|---|---|---|---|
| Rectangle | 0.83 | 0.88 | 0.021 | 0.17 | 1.85 ± 1.12 |
| Polygon | 0.84 | 0.91 | 0.006 | 0.16 | 1.23 ± 0.62 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Wu, S.; Zhuang, L.; Wei, X.; Sak, M.; Neb, D.; Hu, J.; Xie, Y. Efficient Segmentation of a Breast in B-Mode Ultrasound Tomography Using Three-Dimensional GrabCut (GC3D). Sensors 2017, 17, 1827. https://doi.org/10.3390/s17081827
Yu S, Wu S, Zhuang L, Wei X, Sak M, Neb D, Hu J, Xie Y. Efficient Segmentation of a Breast in B-Mode Ultrasound Tomography Using Three-Dimensional GrabCut (GC3D). Sensors. 2017; 17(8):1827. https://doi.org/10.3390/s17081827
Chicago/Turabian StyleYu, Shaode, Shibin Wu, Ling Zhuang, Xinhua Wei, Mark Sak, Duric Neb, Jiani Hu, and Yaoqin Xie. 2017. "Efficient Segmentation of a Breast in B-Mode Ultrasound Tomography Using Three-Dimensional GrabCut (GC3D)" Sensors 17, no. 8: 1827. https://doi.org/10.3390/s17081827





