Quorum Sensing Activity of Aeromonas Caviae Strain YL12, A Bacterium Isolated from Compost
Abstract
: Quorum sensing is a well-studied cell-to-cell communication method that involves a cell-density dependent regulation of genes expression mediated by signalling molecules. In this study, a bacterium isolated from a plant material compost pile was found to possess quorum sensing activity based on bioassay screening. Isolate YL12 was identified using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry and molecular typing using rpoD gene which identified the isolate as Aeromonas caviae. High resolution tandem mass spectrometry was subsequently employed to identify the N-acyl homoserine lactone profile of Aeromonas caviae YL12 and confirmed that this isolate produced two short chain N-acyl homoserine lactones, namely C4-HSL and C6, and the production was observed to be cell density-dependent. Using the thin layer chromatography (TLC) bioassay, both AHLs were found to activate C. violaceum CV026, whereas only C6-HSL was revealed to induce bioluminescence expression of E. coli [pSB401]. The data presented in this study will be the leading steps in understanding the role of quorum sensing in Aeromonas caviae strain YL12.1. Introduction
Composting is an aerobic process, which is mainly carried out by a rapid succession of mixed microbial populations, mainly constituted by bacteria and fungi. At different phases of composting, there are vast changes in the microbial diversity involved. The variations in microbial diversity are highly dependent on the types of raw material used in the composting environment as well as the temperature of each of the composting phases [1]. Through the synergistic action of the microorganisms, organic matters are transformed into a humus-like end product. A wide range of bacteria has been found in different compost environments, which includes species of Pseudomonas, Klebsiella, and Bacillus [2,3]. Composting is known to result in various environmental benefits, a number of which are made possible by the actions of the microorganisms within the compost itself, namely bioremediation of contaminated soil and suppression of plant diseases [4–7]. However, the roles of microflora with quorum sensing (QS) activity in composting environment have not been investigated in depth.
QS is an intercellular communication mechanism used by bacteria to control gene expression through the usage of small, diffusible signal molecules termed autoinducers [8]. QS allows behavioral regulation of bacteria according to population density as the transcription of the QS-regulated target genes are triggered only by a threshold level of exogenous autoinducers which indicates presence of a critical cell mass [9,10]. The autoinducers utilized by majority of gram-negative QS bacteria belongs to the N-acyl homoserine lactone (AHL) family [11]. Studies conducted have discovered that quorum sensing plays essential role in controlling expression of advantageous bacterial phenotype which complements to the environmental conditions. Several genes have been reported to be regulated by QS, namely conjugal transfers of Ti plasmid in Agrobacterium tumefaciens, production of extracellular toxins by Pseudomonas aeruginosa, generation of bioluminescence in Vibrio fischeri and Vibrio harveyi and production of plant tissue-degrading exoenzymes in Pectobacterium carotovora [11,12].
Aeromonas caviae is a mesophilic species of the genus Aeromonas and it is commonly known as an environmental strain that occur ubiquitously in aquatic environments. In recent years, A. caviae has gained a growing clinical recognition as one of the most prevalent causative agent of pediatric gastroenteritis [13,14]. Besides that, an isolated case of urinary tract infection caused by A. caviae have also been reported [15]. Besides its role as a clinical strain, A. caviae was also discovered to be a potential biocontrol agent against fungal pathogens with its chitinolytic activity [16]. In this study, we report the identification of a QS A. caviae strain isolated from compost.
2. Experimental Section
2.1. Compost Sampling and Processing
A compost sample was collected from a compost pile at Semenyih, Malaysia in 2014. The geographic coordinates of the sampling site is 2°58′18.8″N 101°50′33.5″E. A total of 10 subsamples were collected throughout the compost pile and subsequently inserted into sterile plastic tube. The compost sample was promptly processed upon arrival in the laboratory. Large particulates and coarse organic matters were removed using sterile spatula.
2.2. Isolation of Bacteria Strains
Briefly, 1 g of compost sample was mixed with sterile PBS buffer (100 mM, pH 6.5; 10 mL) via vigorous vortex to produce a suspension. The suspension was then serially diluted and subsequently spread onto Luria Bertani (LB) agar and MacConkey agar. Incubation of the agar plates was conducted at 28 °C for 24 h and colonies with observably different morphology was streaked onto new LB agar to obtain pure colonies.
2.3. Screening of Bacterial Isolates with Quorum Sensing Activity by Using C. Violaceum as Biosensor
Cross streaking of isolates with C. violaceum CV026 on LB agar was performed to screen for QS activity of bacteria isolates [17,18]. Inoculated plates were incubated for 24 h at 28 °C. After 24 h of incubation, development of purple pigmentation on the colonies of C. violaceum CV026 suggests that the tested isolate produces short chain exogenous AHLs. E. carotovora GS101 and E. carotovora PNP22 were used as positive and negative controls, respectively [19,20].
2.4. Bacterial Strains and Culture Conditions
The bacterial strains used in this study are as listed in Table 1.
All strains, exclusive of Escherichia coli [pSB401] are routinely cultured at 28 °C in Luria Bertani (LB) broth (1% peptone, 0.5% yeast extract, 0.5% NaCl, per 100 mL distilled water) with shaking (220 rpm) or on LB agar at 28 °C. Escherichia coli [pSB401] was cultured in LB broth supplemented with tetracycline (50 μL/mL) with shaking. For the purpose of AHL extraction, a modified LB medium buffered with 50 mM 3-[N-morpholino]propanesulfonic acid (MOPS) to pH 5.5 [22] was used to culture the bacteria strains.
2.5. Bacterial Strain Identification Using MALDI-TOF MS
The identification of bacterial isolate by MALDI-TOF MS [18,19,23] was performed using a Microflex MALDI-TOF (Bruker Daltonik GmbH, Leipzig, Germany) bench-top mass spectrometer equipped with built in software FlexControl software and Bruker MALI Biotyper Real Time Classification software (Version 3.1, Build 65). Freshly cultured bacterial isolates were processed using direct transfer procedure (DTP) in duplicates. Single colony of bacteria were smeared thinly onto a 96-well polished steel target plate followed by addition 1 μL of MALDI matrix (a concentrated solution composed of α-cyano-4-hydroxycinnamic acid in 50% v/v acetonitrile, 47.5% v/v water and 2.5% v/v trifluoroacetic acid) and air-dried. MALDI-TOF MS was performed using the manufacturer's suggested settings. In short, 337 nm nitrogen laser was used to generate ions in a mass range of 2 to 20 kDa. Analysis of the mass spectra was done by comparing to the MALDI Biotyper database. Based on the unique mass spectra generated, identification of microorganisms can be done. Identification results of samples were evaluated according to the spectra value obtained where spectra value between 2.3 and 3 indicates a species-level identification, whereas spectra value between 2 and 2.3 indicates a secure genus-level identification and spectra values between 1.7 and 2 indicates a probable genus-level identification. Spectra values obtained that were lower than 1.7 suggesting an unsuccessful identification. MALDI Biotyper MSP creation method (Bruker Daltonik) was used to assemble the dendrogram [24].
2.6. Molecular Typing of Bacterial Strain via RpoD Gene Sequence
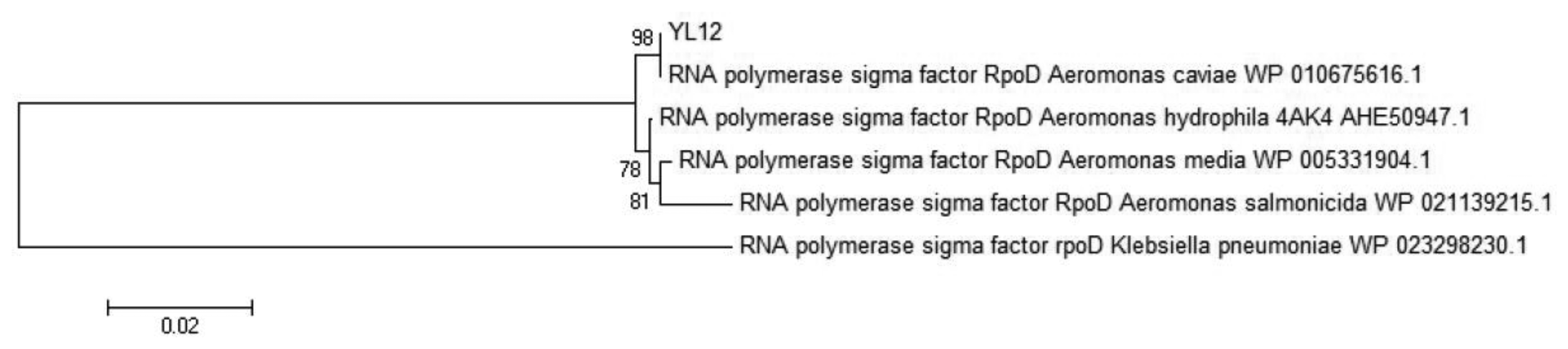
Whole genome sequencing of selected isolate was performed using the PacBio RS (Pacific Biosciences; Menlo Park, CA, USA) sequencing platform. Extracted genomic DNA was used to construct a 10-kb SMRTbell library per manufacturer′s instruction and was subsequently sequenced in 4 SMRT cells via Magbead loading. De novo assembly of the output sequencing data was performed via the hierarchical genome assembly process (HGap) approach. Furthermore, gene prediction of the assembled genomes was performed using Prodigal v2.60 [25] and the predicted translation product was further compared with NCBI non-redundant protein database to identify RpoD protein sequence. The identified RpoD protein sequence were then used as a query sequence to perform the Basic Local Alignment Search Tool (BLAST) search on the NCBI non-redundant protein sequences (nr) database using the blastp algorithm in order to acquire its homologous sequences. Homologous sequences which have maximum identity score between 97% and 100% were selected as reference sequences. Molecular Evolutionary Genetic Analysis (MEGA) version 5.2 [26] were used to align acquired sequences and also to construct phylogenetic tree from the alignment. The alignment of the query sequence with the acquired homologous sequences were done via the by CLUSTAL_W method whereas construction of the phylogeny trees were performed using the Neighbour-Joining method [27]. The RpoD protein reference sequences of the following strains were obtained from NCBI (accession numbers in parentheses): Aeromonas caviae (WP 010675616.1), Aeromonas hydrophila 4AK4 (AHE50947.1), Aeromonas media (WP 005331904.1), Aeromonas salmonicida (WP 021139215.1), Klebsiella pneumoniae (WP 023298230.1).
2.7. Extraction of AHL from Spent Supernatant
Bacteria cells diluted to turbidity of 0.1 at 600 nm were cultured in LB broth which was buffered to pH 6.5 with 50 mM of MOPS and was incubated in 28 °C with agitation at 220 rpm. The samples were prepared such that cultures in triplicates can be harvested at three time points (8 h, 16 h, 24 h) and be used for extraction. Spent supernatants from each culture of A. caviae strain YL 12 were mixed adequately with acidified (0.1% (v/v) glacial acetic acid) ethyl acetate at ratio of 1:1 as described previously [18,28]. The spent supernatants were extracted twice using this method. Following extraction, residual organic solvents were dried in the fume hood. The dried extracts were then reconstituted in 1 mL of acidified ethyl acetate and dried again. Lastly, 200 μL of HPLC grade acetonitrile were added into the dried extracts and vortexed for 3 min before leaving overnight at room temperature. The mixture was then centrifuged at 12,000 rpm for 10 min to remove any insoluble residues.
2.8. Bioluminescent Reporter Assay Using E. coli [pSB401] as Biosensor
Infinite M200 luminometer-spectrophotometer (Tecan, Männedorf, Switzerland) was used to perform cell density dependent bioluminescence quantification. Overnight culture of biosensor namely E. coli [pSB401] cells was diluted to an OD600nm of 0.1. Then, 250 μL of the diluted E. coli [pSB401] cells culture were mixed with 1 μL of extracted AHL and dispensed into a 96-well optical bottom microtitre plates following a previously described method with slight modification [18,29]. Synthetic 3-oxo-C6-HSL (250 pg/μL) was used as the positive control whereas sterile LB broth was used as the negative control in this assay. The bioluminescence assay was conducted for 24 h where bioluminescence and optical density (OD495nm) were recorded at every 60 min interval. Results were expressed as Relative Light Units (RLU)/OD495 against incubation time.
2.9. Characterization of AHL Profile of the Aeromonas Caviae via High Resolution Tandem Liquid Chromatography Quadrupole Mass Spectrometry (LC-MS/MS)
The LC delivery system used was the Agilent 1290 Infinity LC system (Agilent Technologies Inc., Santa Clara, CA, USA) with binary pumps, solvent degasser, thermostated column compartment and diode-array detector, injection volume (2 μL) and the settings used was as described previously [18,19,23]. The column used was an Agilent Zorbax Rapid Resolution High Definition SB-C18 Threaded Column (2.1 mm × 50 mm, 1.8 μm particle size) (Agilent Technologies Inc.). A gradient elution was performed with the mobile phase A being HPLC grade MilliQ water with 0.1% (v/v) formic acid and the mobile phase B being reagent grade acetonitrile added with 0.1 (v/v) formic acid. The gradient profiles used were 80% A and 20% B as the starting mobile phase used for 7 min to proportion of 50% A and 50% B, which continued to 20% A and 20% B at 12 min and progressed to a final proportion of 80% A and 20% B and ran 14 min. The flow rate was set at 0.5 mL/min. Subsequently, Agilent 6490 Triple Quadrupole LC/MS system (Agilent Technologies Inc.) was used to perform high-resolution tandem MS to study the AHL extract [18,20,23]. Electrospray ionization (ESI) was used as the ion source. Precursor ion scanning mode in positive ion mode was used for analysis, where Q1 was set to scan a mass range of m/z 80 to 400 kDa and Q3 was set to monitor for m/z 102, the product ion that indicates presence of lactone ring. Analysis of chromatograms and mass spectra were conducted using the Agilent MassHunter software where the extracted ion chromatogram were compared with reference peaks generated from synthetic AHLs standards.
2.10. Purification and Elucidation of Biosensor Induction Activity of C4-HSL and C6-HSL Using TLC- Bioassay
Purification of the extracted AHL molecules was conducted using thin-layer chromatography (TLC) as described previously [30]. Dried AHLs were dissolved in 100 μL reagent grade acetonitrile and applied to a reverse phase C18 TLC plate (TLC aluminium sheets 20 cm × 20 cm, RP-18 F254s, Merck, Darmstadt, Germany) in a volume of 20 μL per lane. Synthetic AHLs namely N-butanoyl-L-homoserine lactone (C4-HSL, 1.0 μg per lane) and N- hexanoyl-L-homoserine lactone (C6-HSL, 0.1 μg per lane) (Sigma–Aldrich, St Louis, MO, USA) were used as reference standards whereas the reporter strains used were C. violaceum and Escherichia coli [pSB401]. Subsequently, chromatography of the TLC plate were conducted using a methanol-water (60:40 [v/v]) solvent system followed by overlaying of LB agar seeded with overnight culture of selected reporter strain upon completion of the chromatography. The overlaid plates were then incubated overnight in a sealed container. Activation of biosensor by individual AHLs is indicated by manifestation of purple spots on C. violaceum-seeded LB agar and detection of bioluminescent spots on E. coli [pSB401]-seeded LB agar. ChemiDoc™ MP Imaging System (Bio-Rad, Hercules, CA, USA) was used to visualise the bioluminescence expression from TLC/E. coli [pSB401] bioassay. The Rf values of the AHL spots obtained were compared with those of the synthetic AHL standards to allow a conjectural identification of the types of AHLs produced.
3. Results and Discussion
3.1. Sampling and Screening for AHL-Producing Bacteria
Isolation and detection of bacterial isolates with QS was conducted by using compost as the sampling source. The mean temperature of compost was 28 °C during the time of sampling. The compost sample was collected from a compost pile where the raw material constituted mainly of plant materials.
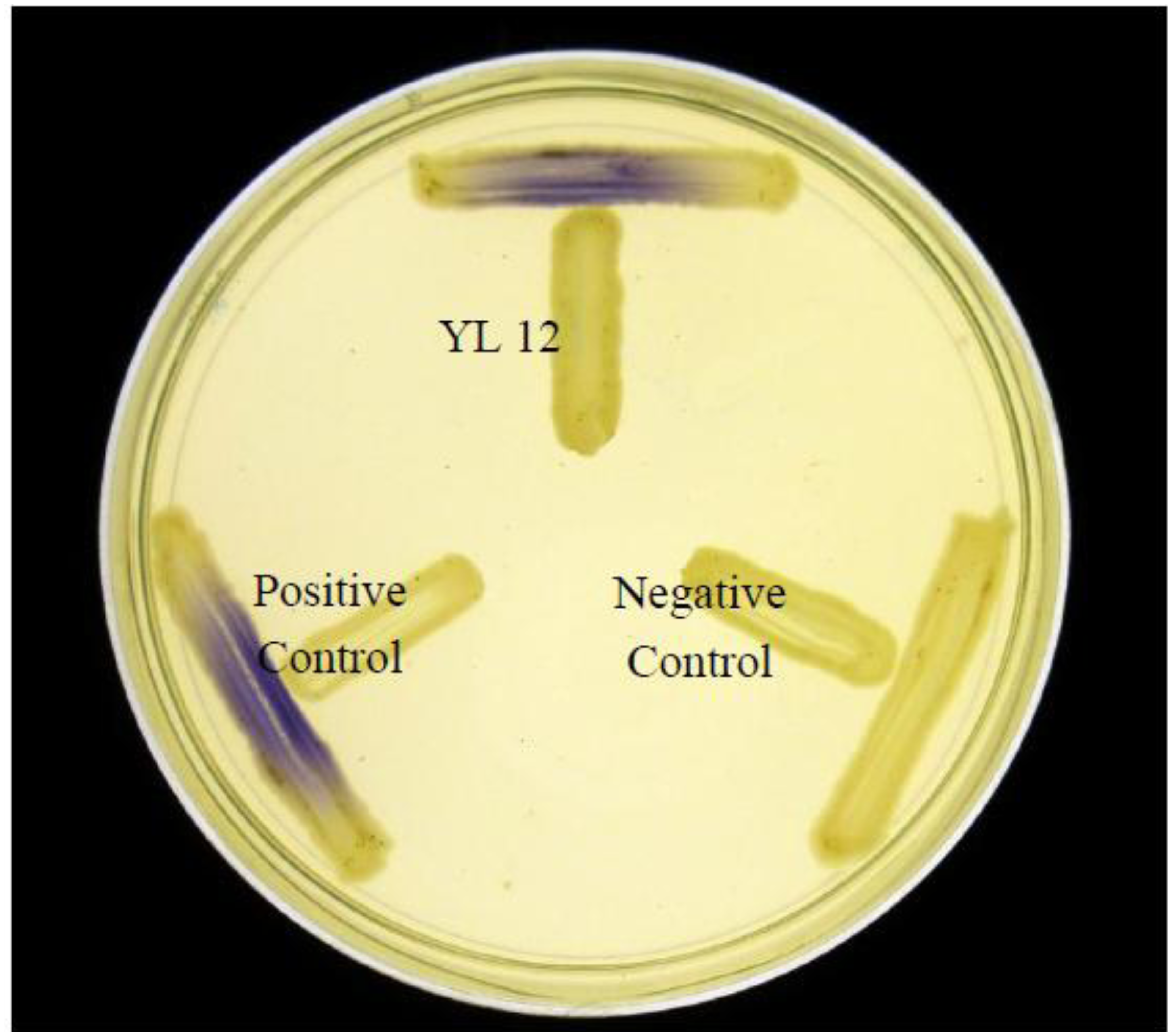
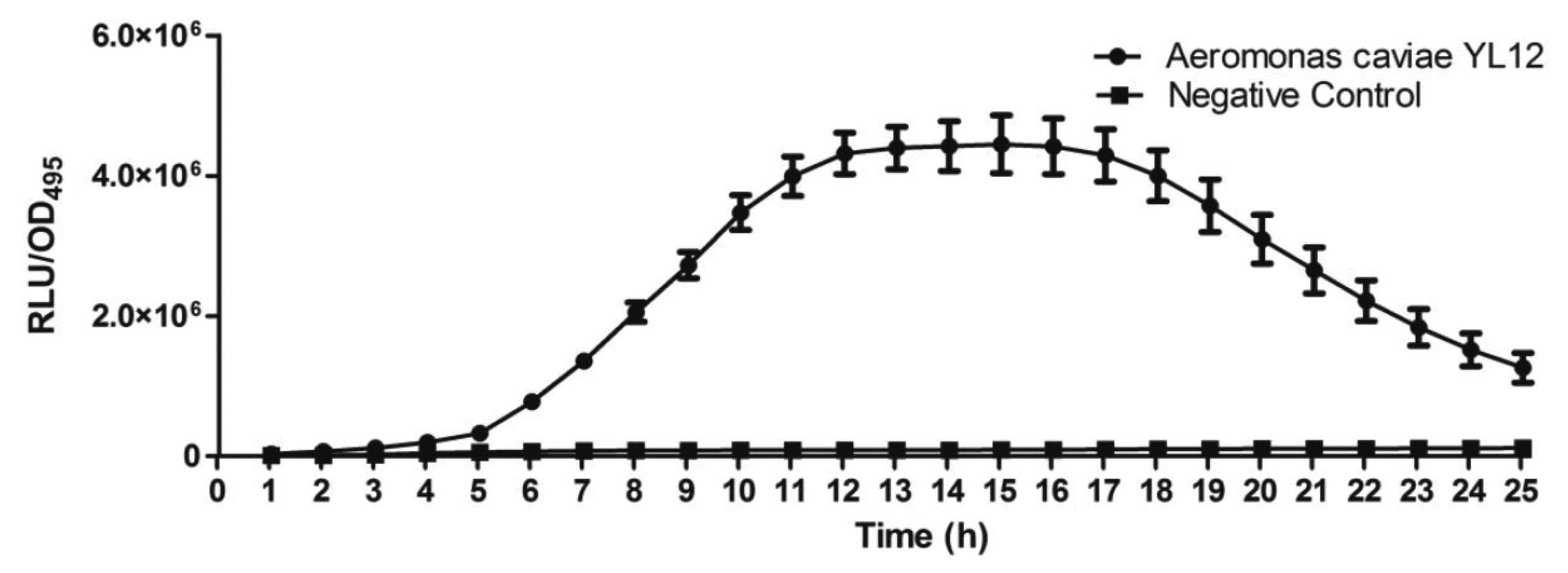
Approximately 80 strains of bacteria were isolated from the compost suspension and all of the strains were preliminarily screened for AHLs production by cross-streaking with C. violaceum CV026 as biosensor. C. violaceum CV026 is an AHL biosensor that was subjected to mini-Tn5 transposon mutagenesis which result in a mutation that is defective in production of AHL but retain the capability of producing violacein, a water-insoluble purple pigment, when exogenous short chain AHLs are detected [17]. In the cross-streaking experiment conducted, strain YL12 triggered CV026 violacein production (Figure 1), which suggests that YL12 produces diffusible short chain AHLs. In order to confirm QS activity of YL12, further QS analysis in the form of bioluminescent reporter assay was conducted and YL12 demonstrated a cell density-dependent induction of bioluminescence (Figure 2).
3.2. Strain Identification of Strain YL12 using Matrix-Assisted Desorption Ionization Time-of-Flight (MALDI-TOF)
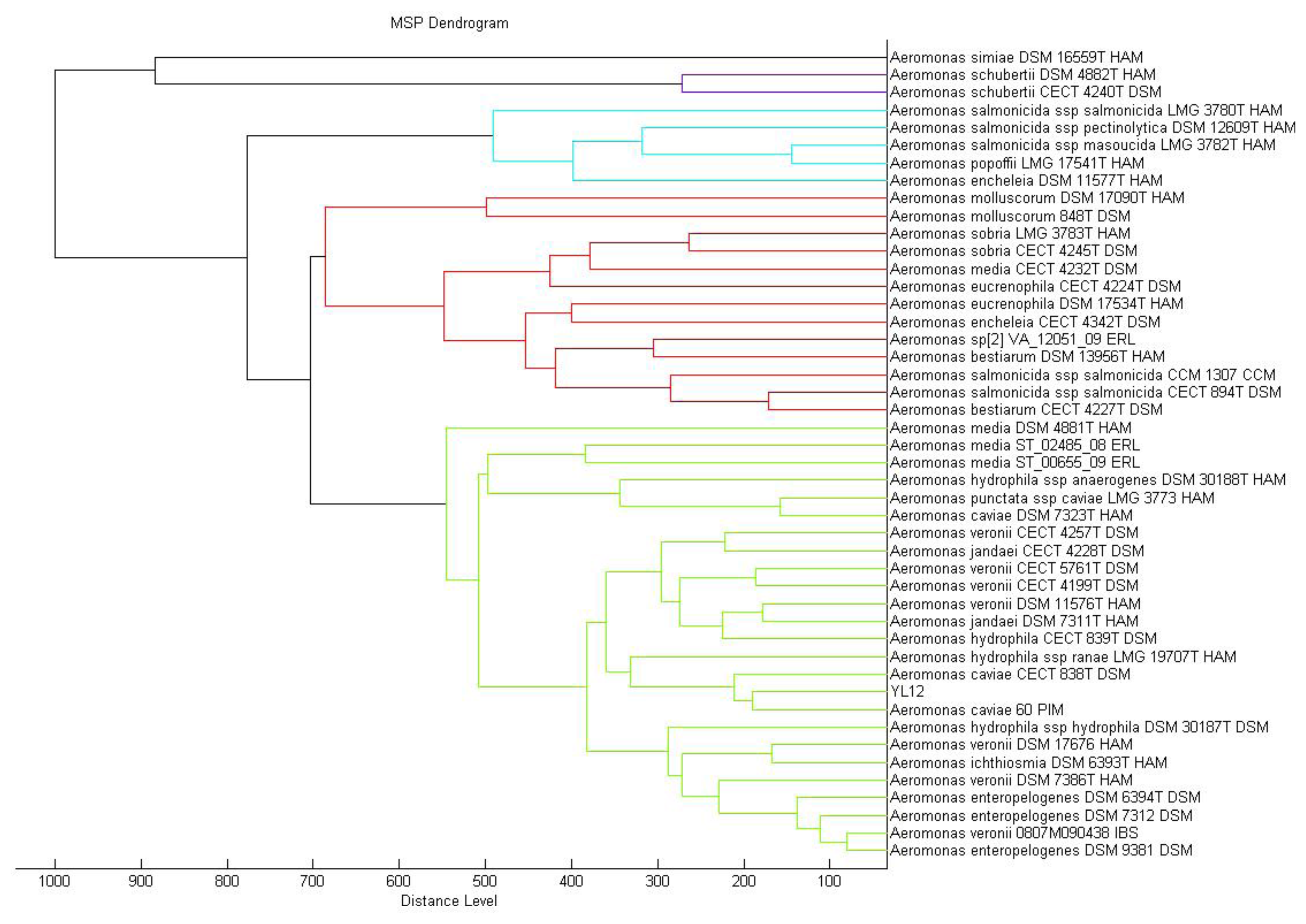
MALDI-TOF mass spectrometry is now routinely used for bacterial identification [23]. The approach to taxonomic characterization of bacteria using MALDI-TOF-MS is by comparing the ions observed in the resultant spectrum to the MALDI Biotyper database. Dendrogram generated from MALDI-TOF MS based on spectral analysis of strain YL12 along with other Aeromonas spp. within the Bruker database was used to visualise the taxonomic classification of strain YL12. Strain YL12 was observed to match closely to A. caviae 60 PIM. The different colours of the branches represent distinct clusters among the microorganisms in the database.
In this study, the result generated from the MALDI-TOF-Bioyper Real Time Classification software showing the closest match to the M12 isolate is Aeromonas caviae with a best match score value of 2.430, while the second best match at a score value of 2.378 was Aeromonas hydrophila. A dendrogram created using the MALDI-Biotyper MSP software also showed that M12 isolates clustered closely to A. caviae strain 60 PIM (Figure 3). MALDI-TOF MS used with a reliable database was demonstrated to be a reliable method for the identification of aeromonads [31].
3.3. Molecular Identification of A. caviae YL12 via RpoD Gene Sequence
Identification of Aeromonas species had been reported to be a challenging task where several studies have highlighted the limitation of both phenotypic identification method as well as commercial identification systems in identification of aeromonads till the species level, particularly in the environmental isolates [32–35]. In view of this problem, molecular approaches had become the preferred tool to perform species identification of the aeromonads. RpoD gene, the single-copy housekeeping gene which encodes the sigma70 factor, was demonstrated in several literatures to be a better molecular marker to be used in the study of phylogenetic and taxonomic relationships [35–37]. The deduced RpoD protein sequence (619 amino acids) was used as query sequence to build a phylogenetic tree. Based on the phylogenetic analysis, the closest relative to YL12 was A. caviae with very high bootstrap value (Figure 4). RpoD protein sequence of Klebsiella pneumoniae was used as an outgroup. The optimal tree with the sum of branch length = 0.20177266 was constructed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (out of 1,000 replicates) was shown next to the branches [38]. The evolutionary distances were computed using the Poisson correction method [39] and are in the units of the number of amino acid substitutions per site. The analysis involved 6 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 611 positions in the final dataset. Evolutionary analyses were conducted in MEGA5.2 [40]. The evolutionary history was inferred using the Neighbor-Joining method [27].
3.4. AHLs Identification by LC-MS/MS
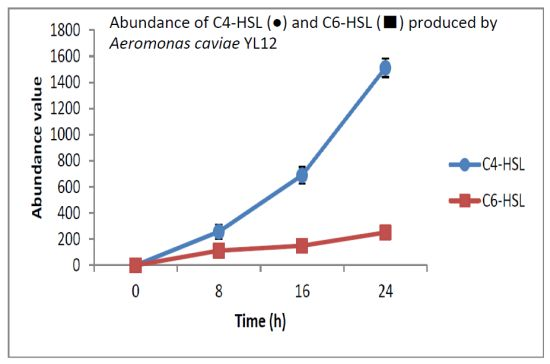
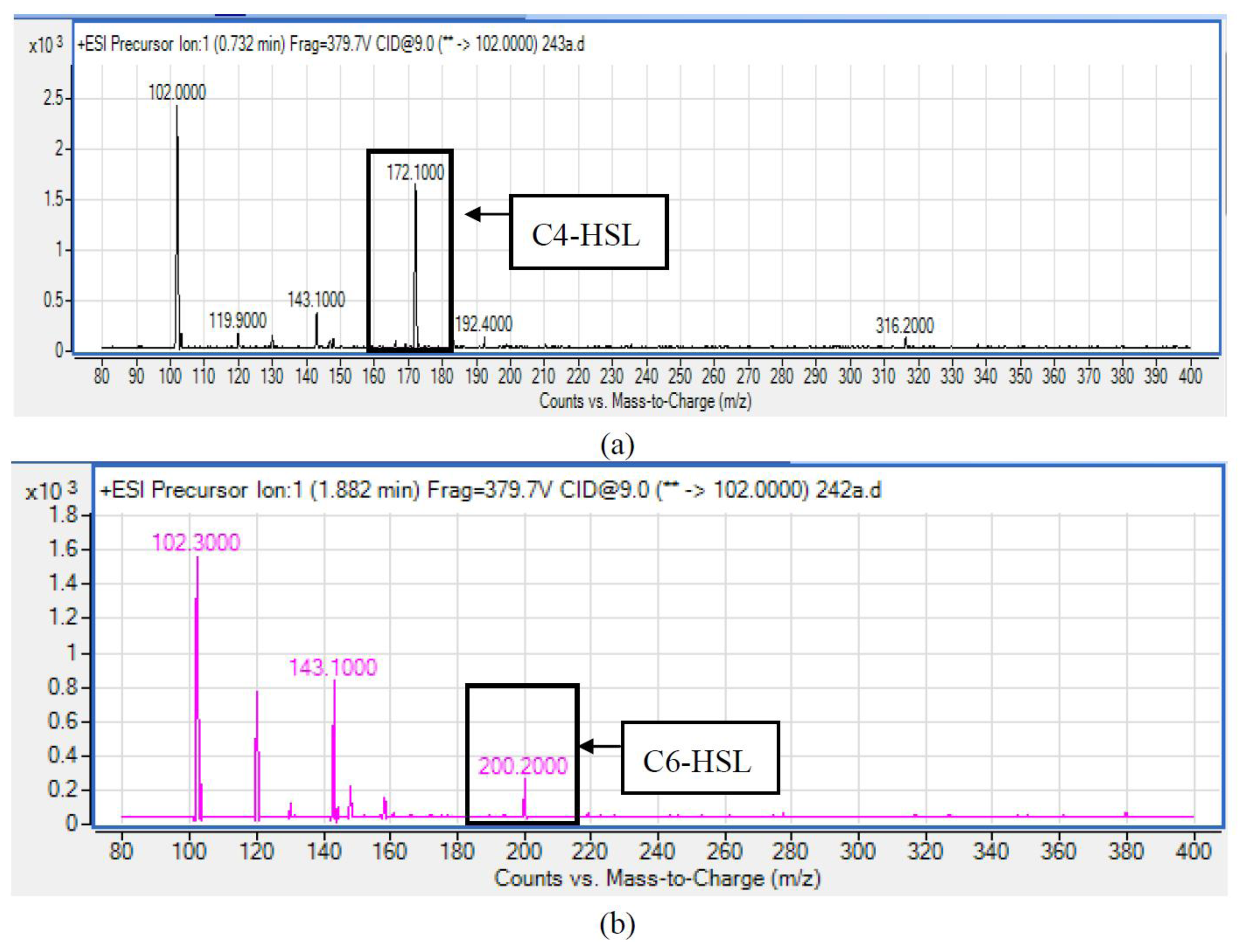
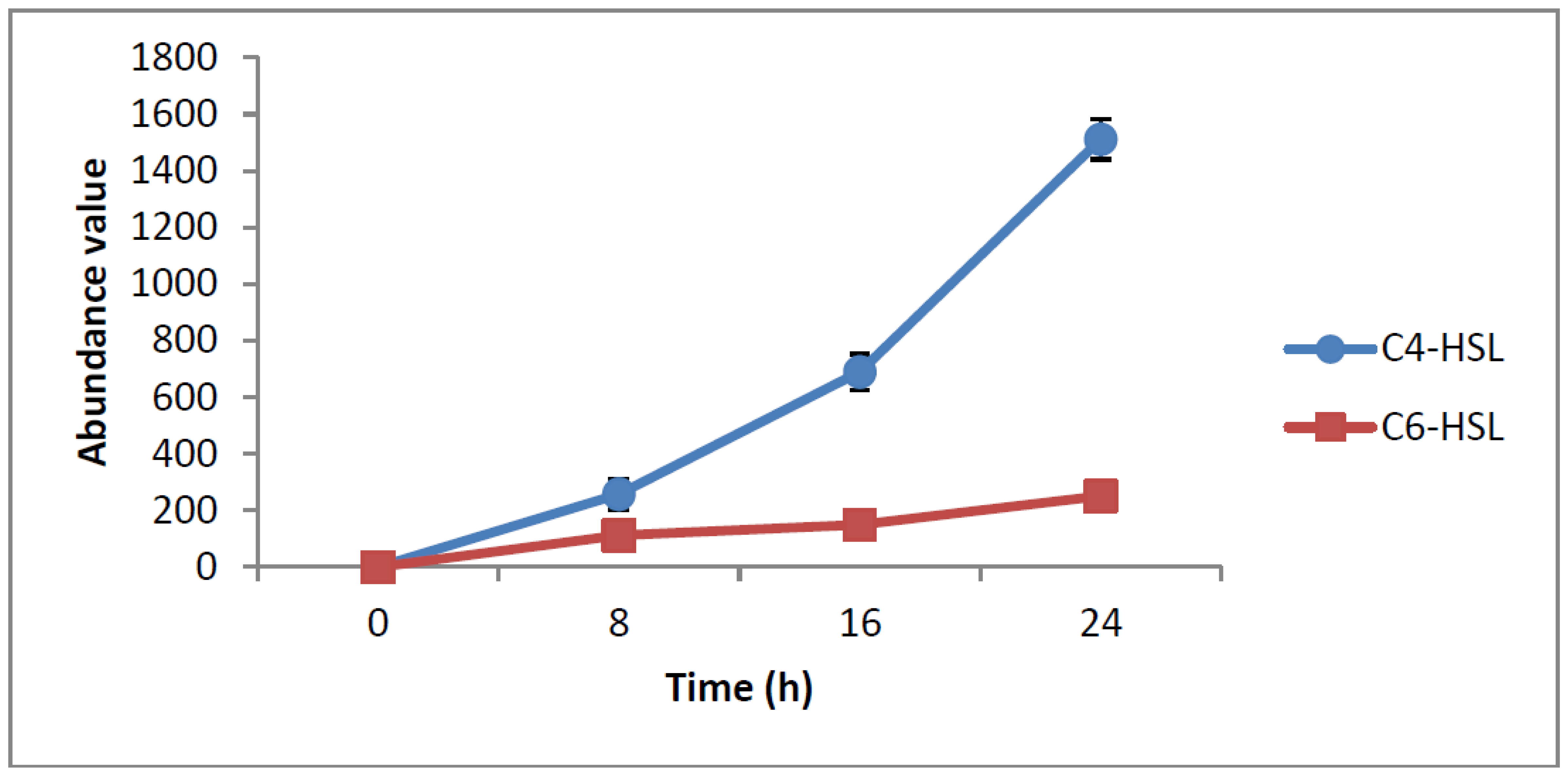
HPLC fractionation followed by MS analysis of the spent culture supernatants extracts of strain YL12 collected were performed to identify the types of AHLs secreted as well as to determine the effect of increased cell density on the accumulated amount of each AHLs produced. All samples were detected to contain two fractions with retention times corresponding to the synthetic C4-HSL (N-butyryl-L-homoserine lactone) and C6-HSL (N-hexanoyl-L-homoserine lactone) (Figure 5). In addition, analysis of the collisionally induced spectra of all the identified fractions demonstrated the presence of the major product ion (m/z value 102) of most AHLs, this detection further affirmed that the fractions detected are AHL molecules (Figure 5). Further, analysis of the abundance value of C4-HSL and C6-HSL detected in each sample showed that the abundance of both AHLs are relatively dependent to the length of incubation period of the respective culture where a longer incubation period coincides with increased abundance of both AHLs. Interestingly, abundance of C4-HSL was shown to increase exponentially from 8 h to 24 h whereas the abundance value of C6-HSL only increased slightly over the same period of time (Figure 6). This finding is in agreement with previously reported studies which demonstrated that C4-HSL is the principal AHL produced in aeromonads, where both LuxI homologue quorum sensing proteins of this species, AhyI and AsaI, were found to produce C4-HSL while C6-HSL is produced in a relatively lower level [30,41]. The pattern of elevation in amount of C4-HSL correlates with its reported role in regulation of cell-density dependent expression of pathogenesis-related phenotypes in this species such as biofilm production, serine protease and metalloprotease activities [42–44]. Regulation of the coordinated expression of these virulence factors via QS system is imperative to enhance the pathogenicity of this bacterium by allowing the bacterium to evade host's defense response while mounting effective attacks. Despite the extensive investigation of QS regulated expression of virulence factors in clinical isolates, the role of QS in aeromonads from environmental isolates, particularly from compost, are scarce. Understanding of AHL profile of A. caviae from compost isolates can lead to further inspection of the role of QS in the biology of this bacteria and its interaction with its niche.
3.5. Characterization of Biosensor Induction Activity of C4-HSL and C6-HSL Using TLC Bioassay
To determine whether AHLs extracted could induce activation of biosensors, bioassay of the AHL extracts were done using C. violaceum CV026 and E. coli [pSB401] used in conjunction with TLC (Figure 7).
The result obtained revealed that both C4-HSL and C6-HSL produced by YL12 were able to activate CV026, a biosensor which responds to presence of short chain AHLs rapidly (with four to eight side chains) with production of purple violacein pigments [17]. The assay also further confirmed the results obtained from LC-MS/MS where C6-HSL is produced in relatively lower abundance compared to C4-HSL as visualized by the vast difference in the size of the spots produced. On the contrary, the E. coli [pSB401]/TLC bioassay shows that only C6-HSL in the solvent extracts of YL12 were detected to be able to activate bioluminescence expression of this biosensor. This finding is in agreement with the previously reported characterization of E. coli [pSB401], a biosensor developed from luxCDABE operon of Photorhabdus luminescences, where the biosensor express good sensitivity towards C6-HSL, even at low concentrations, and have minimal efficacies towards C4-HSL as well as AHLs with long side chains (C10- and longer) [45].
4. Conclusions
We report here the identification of a compost isolate, A. caviae strain YL12 which produced two short chain AHLs, namely C4-HSL and C6-HSL, and the production of each AHL was discovered to be cell-density dependent. This leads to the hypothesis that the QS system in A. caviae YL12 is likely to mirror the QS systems of other Aeromonas sp., hence suggesting possible similarities in QS-regulated gene expression. Further research is underway to identify the gene responsible for the QS activity of A. caviae YL12 as well as study the physiological activity regulated by the signalling molecules on this bacterium.
Acknowledgments
This work was supported by the University of Malaya for High Impact Research Grant (UM-MOHE HIR Grant UM.C/625/1/HIR/MOHE/CHAN/01, No. A000001-50001) awarded to Kok-Gan Chan.
Author Contributions
YL Lim collected the sample, and YL Lim, R Ee and WF Yin performed the experiments, analyzed the data. All authors contributed the writing of the paper, KG Chan supervised the whole project, conceived the idea, applied the funding and managed the whole project and did the final approval of the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tuomela, M.; Vikman, M.; Hatakka, A.; Itävaara, M. Biodegradation of Lignin in A Compost Environment: A Review. Bioresour. Technol. 2000, 72, 169–183. [Google Scholar]
- Atkinson, C.F.; Jones, D.D.; Gauthier, J.J. Biodegradabilities and Microbial Activities during Composting of Oxidation Ditch Sludge. Compost. Sci. Util. 1996, 4, 84–96. [Google Scholar]
- Kutzner, H.J. Microbiology of Composting. In Biotechnology Set, 2nd ed.; Wiley-VCH Verlag GmbH: Berlin, Germany, 2008; pp. 35–100. [Google Scholar]
- Postma, J.; Montanari, M.; van den Boogert, P.H.J.F. Microbial Enrichment to Enhance the Disease Suppressive Activity of Compost. Eur. J. Soil. Biol. 2003, 39, 157–163. [Google Scholar]
- Namkoong, W.; Hwang, E.Y.; Park, J.S.; Choi, J.Y. Bioremediation of Diesel-Contaminated Soil with Composting. Environ. Pollut. 2002, 119, 23–31. [Google Scholar]
- Noble, R.; Coventry, E. Suppression of Soil-Borne Plant Diseases with Composts: A Review. Biocontrol. Sci. Technol. 2005, 15, 3–20. [Google Scholar]
- Semple, K.T.; Reid, B.J.; Fermor, T.R. Impact of Composting Strategies on the Treatment of Soils Contaminated with Organic Pollutants. Environ. Pollut. 2001, 112, 269–283. [Google Scholar]
- Salmond, G.P.; Bycroft, B.W.; Stewart, G.S.; Williams, P. The Bacterial ‘Enigma’: Cracking the Code of Cell-Cell Communication. Mol. Microbiol. 1995, 16, 615–624. [Google Scholar]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum Sensing in Bacteria: The LuxR-LuxI Family of Cell Density-Responsive Transcriptional Regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar]
- Bassler, B.L. How Bacteria Talk to Each Other: Regulation of Gene Expression by Quorum Sensing. Curr. Opin. Microbiol. 1999, 2, 582–587. [Google Scholar]
- Swift, S.; Throup, J.P.; Williams, P.; Salmond, G.P.C.; Stewart, G.S.A.B. Quorum Sensing: A Population-Density Component in the Determination of Bacterial Phenotype. Trends. Biochem. Sci. 1996, 21, 214–219. [Google Scholar]
- De Kievit, T.R.; Iglewski, B.H. Bacterial Quorum Sensing in Pathogenic Relationships. Infect. Immun. 2000, 68, 4839–4849. [Google Scholar]
- Namdari, H.; Bottone, E.J. Microbiologic and Clinical Evidence Supporting the Role of Aeromonas Caviae as a Pediatric Enteric Pathogen. J. Clin. Microbiol. 1990, 28, 837–840. [Google Scholar]
- Rabaan, A.A.; Gryllos, I.; Tomas, J.M.; Shaw, J.G. Motility and the Polar Flagellum are Required for Aeromonas caviae Adherence to HEp-2 cells. Infect. Immun. 2001, 69, 4257–4267. [Google Scholar]
- Al-Benwan, K.; Abbott, S.; Janda, J.M.; Huys, G.; Albert, M.J. Cystitis Caused by. Aeromonas caviae. J. Clin. Microbiol. 2007, 45, 2348–2350. [Google Scholar]
- Inbar, J.; Chet, I. Evidence that Chitinase Produced by Aeromonas caviae is Involved in the Biological Control of Soil-Borne Plant Pathogens by this Bacterium. Soil. Biol. Biochem. 1991, 23, 973–978. [Google Scholar]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum Sensing and Chromobacterium Violaceum: Exploitation of Violacein Production and Inhibition for the Detection of N-Acylhomoserine Lactones. Microbiol. 1997, 143 (Pt 12), 3703–3711. [Google Scholar]
- Han-Jen, R.; Wai-Fong, Y.; Kok-Gan, C. Pandoraea sp. RB-44, A Novel Quorum Sensing Soil Bacterium. Sensors 2013, 13, 14121–14132. [Google Scholar]
- Lau, Y.; Sulaiman, J.; Chen, J.; Yin, W.F.; Chan, K.G. Quorum Sensing Activity of Enterobacter asburiae Isolated from Lettuce Leaves. Sensors 2013, 13, 14189–14199. [Google Scholar]
- Ngeow, Y.; Cheng, H.; Chen, J.; Yin, W.F.; Chan, K.G. Short Chain N-Acylhomoserine Lactone Production by Clinical Multidrug Resistant Klebsiella pneumoniae Strain CSG20. Sensors 2013, 13, 15242–15251. [Google Scholar]
- Winson, M.K.; Swift, S.; Fish, L.; Throup, J.P.; Jorgensen, F.; Chhabra, S.R.; Bycroft, B.W.; Williams, P.; Stewart, G.S. Construction and Analysis of luxCDABE-Based Plasmid Sensors for Investigating N-Acyl Homoserine Lactone-Mediated Quorum Sensing. FEMS. Microbiol. Lett. 1998, 163, 185–192. [Google Scholar]
- Yates, E.A.; Philipp, B.; Buckley, C.; Atkinson, S.; Chhabra, S.R.; Sockett, R.E.; Goldner, M.; Dessaux, Y.; Camara, M.; Smith, H.; et al. N-Acylhomoserine Lactones Undergo Lactonolysis in a pH-, Temperature-, and Acyl Chain Length-Dependent Manner During Growth of Yersinia pseudotuberculosis and. Pseudomonas aeruginosa. Infect. Immun. 2002, 70, 5635–5646. [Google Scholar]
- Chen, J.; Koh, C.L.; Sam, C.K.; Yin, W.F.; Chan, K.G. Short Chain N-Acyl Homoserine Lactone Production By Soil Isolate Burkholderia sp. Strain A9. Sensors 2013, 13, 13217–13227. [Google Scholar]
- Saffert, R.T.; Cunningham, S.A.; Ihde, S.M.; Jobe, K.E.; Mandrekar, J.; Patel, R. Comparison of Bruker Biotyper Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometer to BD Phoenix Automated Microbiology System for Identification of Gram-Negative Bacilli. J. Clin. Microbiol. 2011, 49, 887–892. [Google Scholar]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC. Bioinform. 2010, 11, 119. [Google Scholar]
- Kumar, S.; Tamura, K.; Nei, M. Mega: Molecular Evolutionary Genetics Analysis Software for Microcomputers. Comput. Appl. Biosci. 1994, 10, 189–191. [Google Scholar]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Chong, T.M.; Koh, C.L.; Sam, C.K.; Choo, Y.M.; Yin, W.F.; Chan, K.G. Characterization of Quorum Sensing and Quorum Quenching Soil Bacteria Isolated from Malaysian Tropical Montane Forest. Sensors 2012, 12, 4846–4859. [Google Scholar]
- Tan, L.Y.; Yin, W.F.; Chan, K.G. Piper nigrum, Piper betle and Gnetum gnemon-Natural Food Sources with Anti-Quorum Sensing Properties. Sensors 2013, 13, 3975–3985. [Google Scholar]
- Chan, K.G.; Puthucheary, S.D.; Chan, X.Y.; Yin, W.F.; Wong, C.S.; Too, W.S.; Chua, K.H. Quorum Sensing in Aeromonas Species Isolated from Patients in Malaysia. Curr. Microbiol. 2011, 62, 167–172. [Google Scholar]
- Benagli, C.; Demarta, A.; Caminada, A.; Ziegler, D.; Petrini, O.; Tonolla, M. A Rapid MALDI-TOF MS Identification Database at Genospecies Level for Clinical and Environmental Aeromonas Strains. PLoS One 2012, 7, e48441. [Google Scholar]
- Abbott, S.L.; Cheung, W.K.; Janda, J.M. The Genus Aeromonas: Biochemical Characteristics, Atypical Reactions, and Phenotypic Identification Schemes. J. Clin. Microbiol. 2003, 41, 2348–2357. [Google Scholar]
- Beaz-Hidalgo, R.; Alperi, A.; Bujan, N.; Romalde, J.L.; Figueras, M.J. Comparison of Phenotypical and Genetic Identification of Aeromonas Strains Isolated from Diseased Fish. Syst. Appl. Microbiol. 2010, 33, 149–153. [Google Scholar]
- Lamy, B.; Laurent, F.; Verdier, I.; Decousser, J.W.; Lecaillon, E.; Marchandin, H.; Roger, F.; Tigaud, S.; de Montclos, H.; Kodjo, A. Accuracy of 6 Commercial Systems for Identifying Clinical Aeromonas Isolates. Diagn. Microbiol. Infect. Dis. 2010, 67, 9–14. [Google Scholar]
- Nagar, V.; Shashidhar, R.; Bandekar, J. Characterization of Aeromonas Strains Isolated from Indian Foods Using Rpod Gene Sequencing and Whole Cell Protein Analysis. World. J. Microbiol. Biotechnol. 2013, 29, 745–752. [Google Scholar]
- Soler, L.; Yanez, M.A.; Chacon, M.R.; Aguilera-Arreola, M.G.; Catalan, V.; Figueras, M.J.; Martinez-Murcia, A.J. Phylogenetic Analysis of the Genus Aeromonas Based on Two Housekeeping Genes. Int. J. Syst. Evol. Microbiol. 2004, 54, 1511–1519. [Google Scholar]
- Aravena-Roman, M.; Harnett, G.B.; Riley, T.V.; Inglis, T.J.; Chang, B.J. Aeromonas aquariorum Is Widely Distributed in Clinical and Environmental Specimens and Can be Misidentified as. Aeromonas hydrophila. J. Clin. Microbiol 2011, 49, 3006–3008. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins. Evol. Genes. Protein 1965, 97–166. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. Mega5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar]
- Swift, S.; Karlyshev, A.V.; Fish, L.; Durant, E.L.; Winson, M.K.; Chhabra, S.R.; Williams, P.; Macintyre, S.; Stewart, G.S. Quorum Sensing in Aeromonas hydrophila and Aeromonas salmonicida: Identification of the LuxRI Homologs AhyRI and AsaRI and Their Cognate N-Acylhomoserine Lactone Signal Molecules. J. Bacteriol. 1997, 179, 5271–5281. [Google Scholar]
- Khajanchi, B.K.; Sha, J.; Kozlova, E.V.; Erova, T.E.; Suarez, G.; Sierra, J.C.; Popov, V.L.; Horneman, A.J.; Chopra, A.K. N-Acylhomoserine Lactones Involved in Quorum Sensing Control the Type VI Secretion System, Biofilm Formation, Protease Production, and in vivo Virulence in a Clinical Isolate of. Aeromonas hydrophila. Microbiology 2009, 155, 3518–3531. [Google Scholar]
- Swift, S.; Lynch, M.J.; Fish, L.; Kirke, D.F.; Tomas, J.M.; Stewart, G.S.; Williams, P. Quorum Sensing-Dependent Regulation and Blockade of Exoprotease Production in. Aeromonas hydrophila. Infect. Immun. 1999, 67, 5192–5199. [Google Scholar]
- Lynch, M.J.; Swift, S.; Kirke, D.F.; Keevil, C.W.; Dodd, C.E.; Williams, P. The Regulation of Biofilm Development by Quorum Sensing in. Aeromonas hydrophila. Environ. Microbiol. 2002, 4, 18–28. [Google Scholar]
- Steindler, L.; Venturi, V. Detection of Quorum-Sensing N-Acyl Homoserine Lactone Signal Molecules by Bacterial Biosensors. FEMS Microbiol. Lett. 2007, 266, 1–9. [Google Scholar]

| Strain | Description | Source/Reference |
|---|---|---|
| Chromobacterium violaceum CV026 | Double mini-Tn5 mutant derived from C. violaceum ATCC31532 serves as a biosensor which formation of purple violacein on its colonies indicate presence of short chain exogenous AHL molecules. | [17] |
| Erwinia carotovora GS101 | Served as positive control in AHL production screening test due to its capability of producing AHL molecules that is detectable by C. violaceum CV026 | Gift from Prof. Paul Williams |
| Erwinia carotovora PNP22 | Served as negative control in AHL production screening test as it lacks AHL molecules production. | Gift from Prof. Paul Williams |
| Escherichia coli [pSB401] | A pACYC184-derived mutant developed from Photobacterium fischeri [ATCC 7744] which functions as a AHL biosensor as it produce bioluminescence in the presence of short chain AHL. | [21] |
| Aeromonas caviae strain YL 12 | Compost isolates | This study |
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lim, Y.-L.; Ee, R.; Yin, W.-F.; Chan, K.-G. Quorum Sensing Activity of Aeromonas Caviae Strain YL12, A Bacterium Isolated from Compost. Sensors 2014, 14, 7026-7040. https://doi.org/10.3390/s140407026
Lim Y-L, Ee R, Yin W-F, Chan K-G. Quorum Sensing Activity of Aeromonas Caviae Strain YL12, A Bacterium Isolated from Compost. Sensors. 2014; 14(4):7026-7040. https://doi.org/10.3390/s140407026
Chicago/Turabian StyleLim, Yan-Lue, Robson Ee, Wai-Fong Yin, and Kok-Gan Chan. 2014. "Quorum Sensing Activity of Aeromonas Caviae Strain YL12, A Bacterium Isolated from Compost" Sensors 14, no. 4: 7026-7040. https://doi.org/10.3390/s140407026