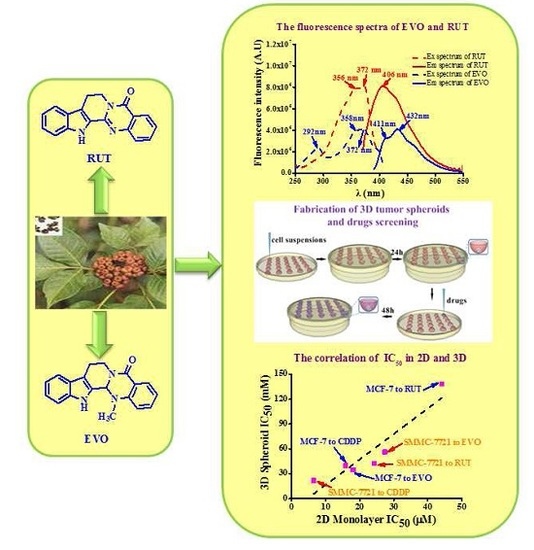
Antiproliferative Activity and Cellular Uptake of Evodiamine and Rutaecarpine Based on 3D Tumor Models
Abstract
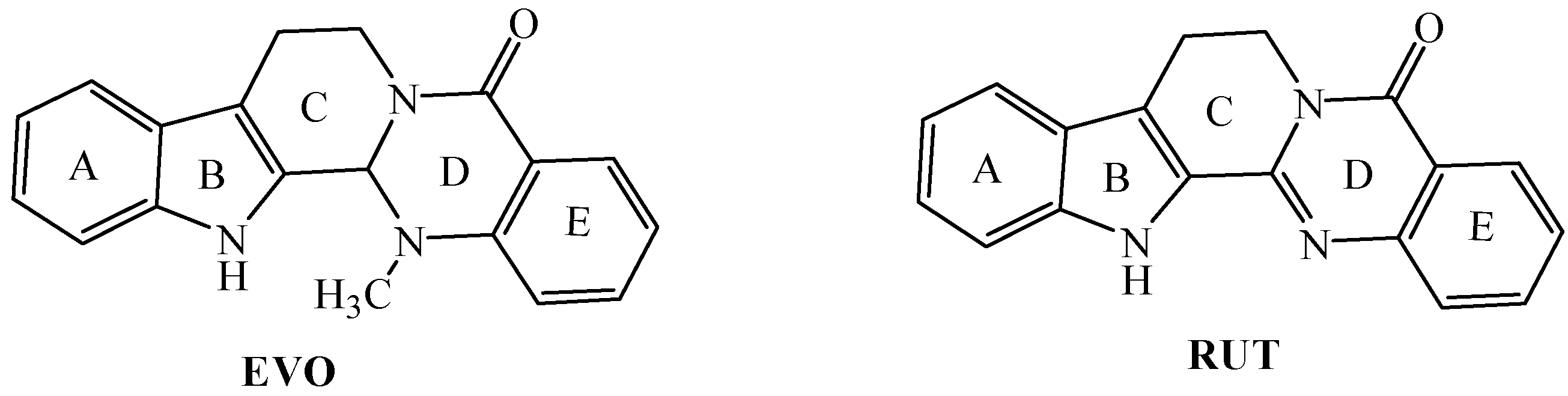
:1. Introduction
2. Results
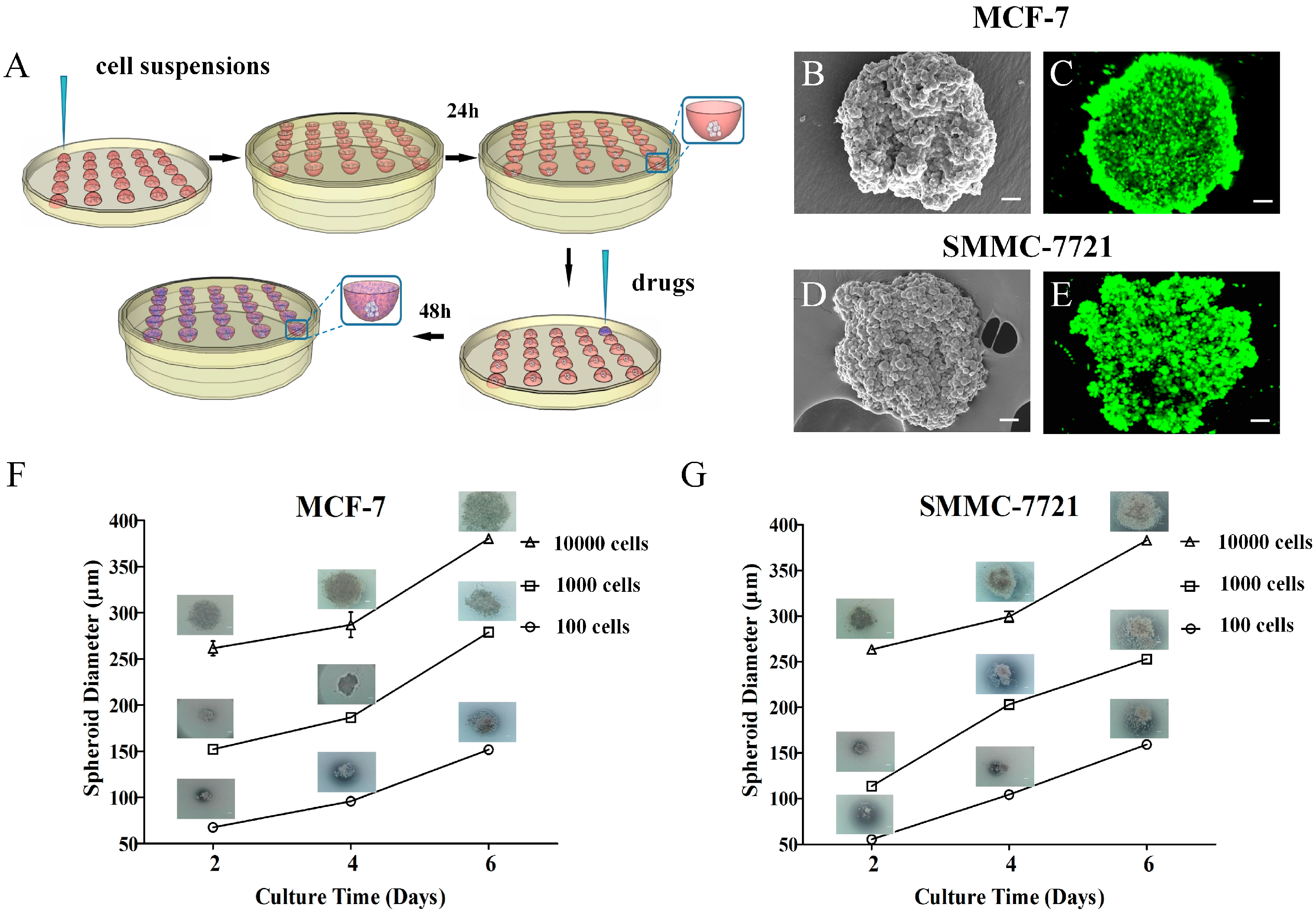
2.1. Fabrication and Characterization of 3D Tumor Spheroids
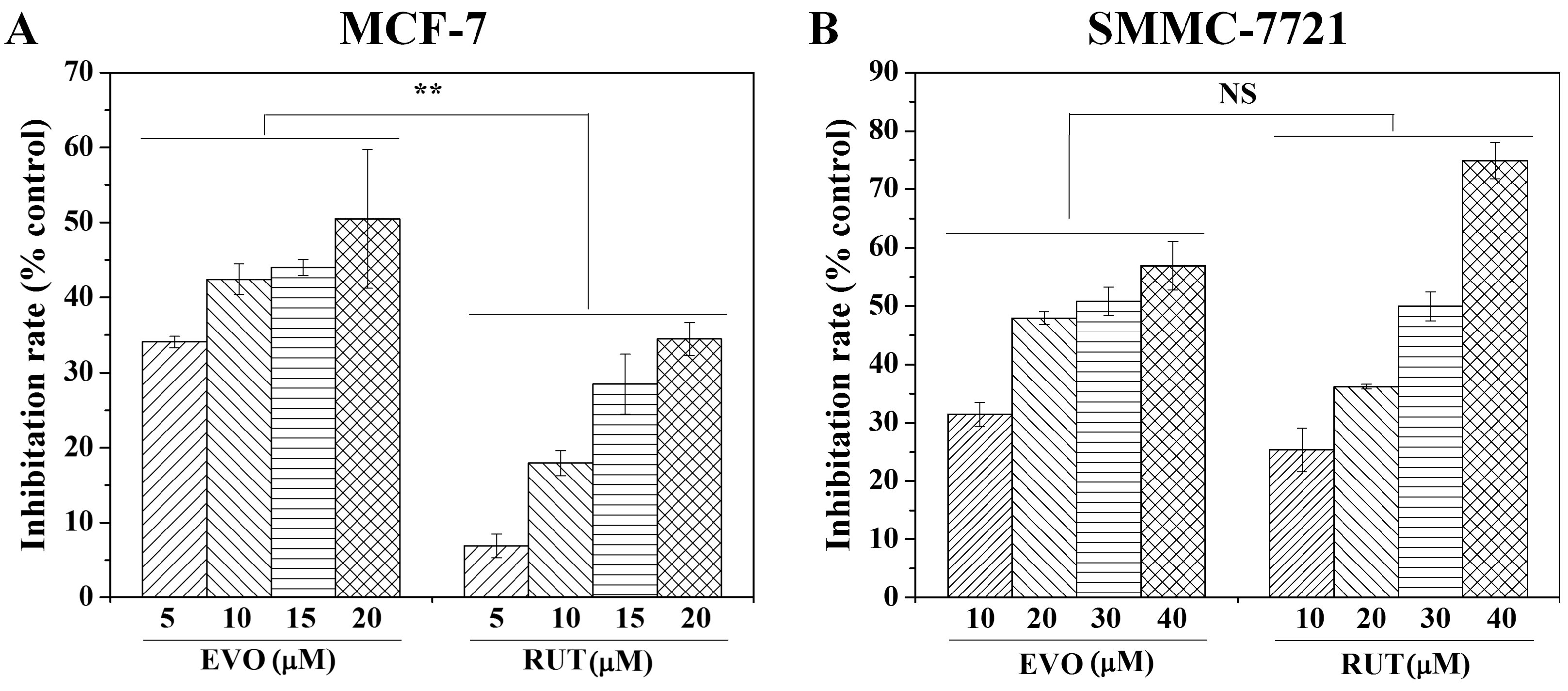
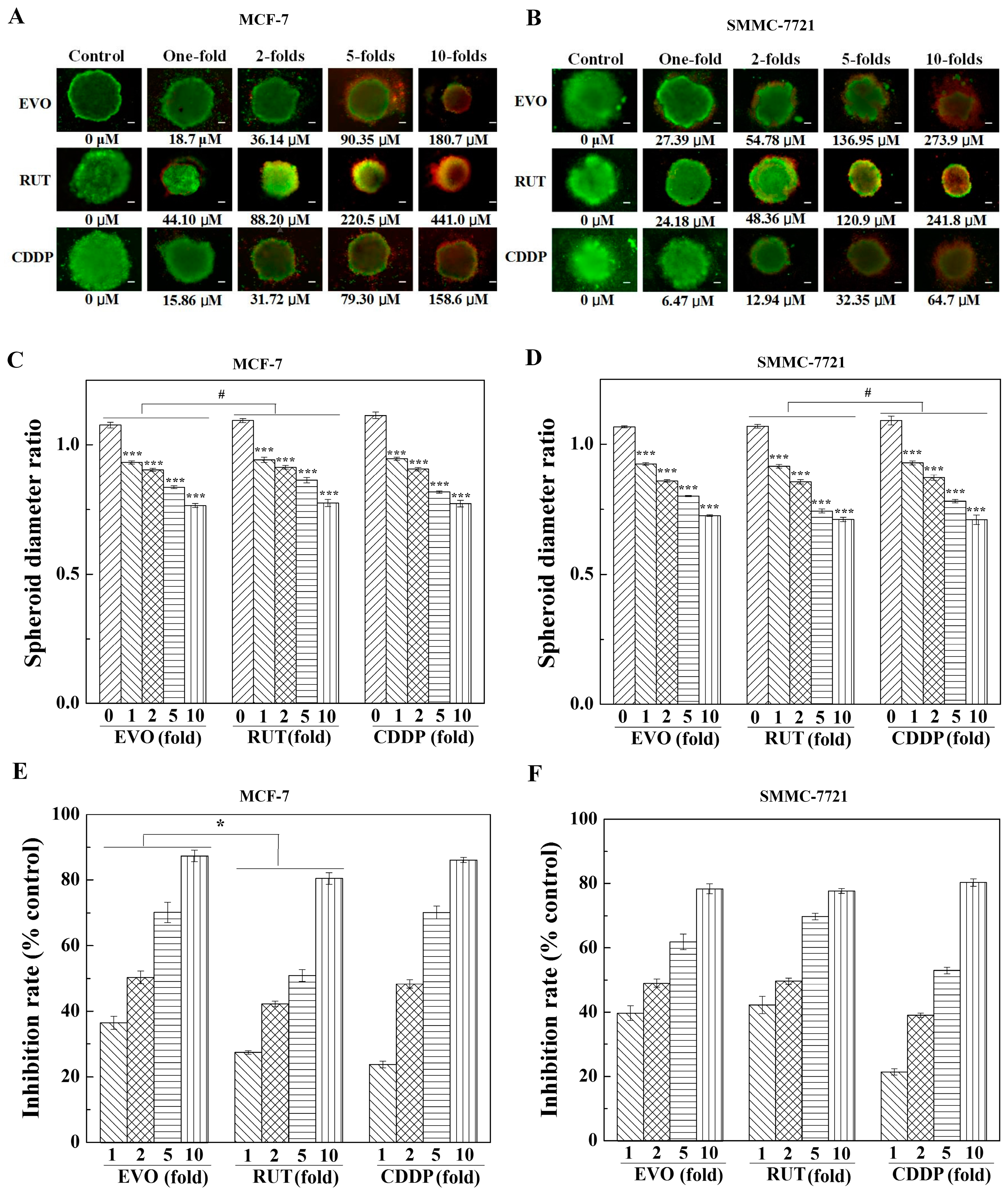
2.2. Anti-Tumor Activity Testing of Drugs in 2D and 3D Models
2.3. Cellular Uptake of EVO and RUT in in Vitro 2D and 3D Models
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. Fabrication of 3D Tumor Spheroids
4.4. MTT Assay
4.5. Cellular Uptake of EVO and RUT
4.6. Live/Dead Assay
4.7. Morphology, Physical and Optical Characterization
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yu, H.; Jin, H.; Gong, W.; Wang, Z.; Liang, H. Pharmacological actions of multi-target-directed evodiamine. Molecules 2013, 18, 1826–1843. [Google Scholar] [CrossRef] [PubMed]
- Mhaske, S.B.; Argade, N.P. The chemistry of recently isolated naturally occurring quinazolinone alkaloids. Tetrahedron 2006, 62, 9787–9826. [Google Scholar] [CrossRef]
- Liu, Y.N.; Pan, S.L.; Liao, C.H.; Huang, D.Y.; Guh, J.H.; Peng, C.Y.; Chang, Y.L.; Teng, C.M. Evodiamine represses hypoxia-induced inflammatory proteins expression and hypoxia-inducible factor 1alpha accumulation in RAW264.7. Shock 2009, 32, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, Y.; Kontani, Y.; Kobayashi, Y.; Sato, Y.; Mori, N.; Yamashita, H. Evodiamine improves diet-induced obesity in a uncoupling protein-1-independent manner: Involvement of antiadipogenic mechanism and extracellularly regulated kinase/mitogen-activated protein kinase signaling. Endocrinology 2008, 149, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hu, C. Evodiamine: A novel anti-cancer alkaloid from Evodia rutaecarpa. Molecules 2009, 14, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Wang, S.; Miao, Z.; Yao, J.; Zhang, Y.; Guo, Z.; Zhang, W.; Sheng, C. New tricks for an old natural product: Discovery of highly potent evodiamine derivatives as novel antitumor agents by systemic structure-activity relationship analysis and biological evaluations. J. Med. Chem. 2012, 55, 7593–7613. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.H.; Pan, S.L.; Guh, J.H.; Chang, Y.L.; Pai, H.C.; Lin, C.H.; Teng, C.M. Antitumor mechanism of evodiamine, a constituent from Chinese herb Evodiae fructus, in human multiple-drug resistant breast cancer NCI/ADR-RES cells in vitro and in vivo. Carcinogenesis 2005, 26, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Kan, S.F.; Yu, C.H.; Pu, H.F.; Hsu, J.M.; Chen, M.J.; Wang, P.S. Anti-proliferative effects of evodiamine on human prostate cancer cell lines DU145 and PC3. J. Cell. Biochem. 2007, 101, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.; Hsia, S.M.; Yeh, J.Y.; Cheng, S.C.; Wang, P.S.; Wang, S.W. Anti-Proliferative Effects of Evodiamine on Human Breast Cancer Cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Chao, D.C.; Lin, L.J.; Hsiang, C.Y.; Li, C.C.; Lo, H.Y.; Liang, J.A.; Kao, S.T.; Wu, S.L.; Ho, T.Y. Evodiamine inhibits 12-O-tetradecanoylphorbol-13-acetate-induced activator protein 1 transactivation and cell transformation in human hepatocytes. Phytother Res. 2011, 25, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.J.; Kim, E.J.; Kim, S.; Jung, E.M.; Park, J.W.; Jeong, S.H.; Park, S.E.; Yoo, Y.H.; Kwon, T.K. Caspase-dependent and caspase-independent apoptosis induced by evodiamine in human leukemic U937 cells. Mol. Cancer Ther. 2006, 5, 2398–2407. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.; Yu, C.H.; Wang, S.W.; Pu, H.F.; Kan, S.F.; Lin, L.C.; Chi, C.W.; Ho, L.L.; Lee, C.H.; Wang, P.S. Anti-proliferative effects of evodiamine on human thyroid cancer cell line ARO. J. Cell. Biochem. 2010, 110, 1495–1503. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; DePinho, R.A.; Ernst, M.; Vousden, K. Cancer research: Past, present and future. Nat. Rev. Cancer 2011, 11, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Farach-Carson, M.C.; Jia, X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014, 32, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Nataliya, L.B.; Rimma, N.M.; Valeria, V.S.; Svetlana, F.K.; Vincent, C.L.; Rizvanov, A.A. Application of Cell and Tissue Culture Systems for Anticancer Drug Screening. World Appl. Sci. J. 2013, 23, 315–325. [Google Scholar]
- Yamada, K.M.; Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Wang, J.; Chen, D.Y.; Yang, J.; Yang, C.P.; Zhang, Y.X.; Zhang, H.Y.; Dou, J. Evaluation of characteristics of CD44+CD117+ ovarian cancer stem cells in three dimensional basement membrane extract scaffold versus two dimensional monocultures. BMC Cell Biol. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.C.; Hsiao, A.Y.; Allen, S.G.; Torisawa, Y.S.; Ho, M.; Takayama, S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011, 136, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Todorov, L.; VadeBoncouer, T. Etiology and use of the “hanging drop” technique: A review. Pain Res. Treat 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Choi, Y.S.; Seo, Y.J.; Lee, M.Y.; Jeon, S.Y.; Ku, B.; Kim, S.; Yi, S.H.; Nam, D.H. High-throughput screening (HTS) of anticancer drug efficacy on a micropillar/microwell chip platform. Anal. Chem. 2014, 86, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Matsusaki, M.; Sakaue, K.; Kadowaki, K.; Akashi, M. Three-dimensional human tissue chips fabricated by rapid and automatic inkjet cell printing. Adv. Healthc. Mater. 2013, 2, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yao, R.; Ouyang, L.; Ding, H.; Zhang, T.; Zhang, K.; Cheng, S.; Sun, W. Three-dimensional printing of Hela cells for cervical tumor model in vitro. Biofabrication 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Moon, S.J.; Emre, A.E.; Turali, E.S.; Song, Y.S.; Hacking, S.A.; Nagatomi, J.; Demirci, U. A droplet-based building block approach for bladder smooth muscle cell (SMC) proliferation. Biofabrication 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Archacka, K.; Pozzobon, M.; Repele, A.; Rossi, C.A.; Campanella, M.; De Coppi, P. Culturing muscle fibres in hanging drop: A novel approach to solve an old problem. Biol. Cell 2014, 106, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Foty, R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011, 51, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Qi, Y.; Lv, L.; Xu, Y.; Zheng, L.; Yin, L.; Liu, K.; Han, X.; Zhao, Y.; Peng, J. In vitro anti-proliferative effects of Zuojinwan on eight kinds of human cancer cell lines. Cytotechnology 2014, 66, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Croce, A.C.; Bottiroli, G.; Supino, R.; Favini, E.; Zuco, V.; Zunino, F. Subcellular localization of the camptothecin analogues, topotecan and gimatecan. Biochem. Pharmacol. 2004, 67, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Natesan, S.; Sugumaran, A.; Ponnusamy, C.; Jeevanesan, V.; Girija, G.; Palanichamy, R. Development and evaluation of magnetic microemulsion: Tool for targeted delivery of camptothecin to BALB/c mice-bearing breast cancer. J. Drug Target. 2014, 22, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Feng, S.S. The drug encapsulation efficiency, in vitro drug release, cellular uptake and cytotoxicity of paclitaxel-loaded poly(lactide)-tocopheryl polyethylene glycol succinate nanoparticles. Biomaterials 2006, 27, 4025–4033. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Biswas, R.; Moon, J.H.; Chung, P.S. Cellular uptake of 9-hydroxypheophorbide-alpha and its photoactivation to induce ER stress-related apoptosis in human cervical cancer cells. Lasers Med. Sci. 2014, 29, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Lukka, P.B.; Joseph, W.R.; Finlay, G.J.; Paxton, J.W.; McKeage, M.J.; Baguley, B.C. Selective cellular uptake and retention of SN 28049, a new DNA-binding topoisomerase II-directed antitumor agent. Cancer Chemother. Pharmacol. 2014, 74, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Dvoranová, D.; Brezová, V.; Valko, M.; Staško, A. Photoinduced transformation of camptothecin in the presence of iron(III) ions. J. Photochem. Photobio. A Chem. 2007, 185, 172–180. [Google Scholar]
- Gonzalez-Ruiz, V.; Mussardo, P.; Corda, E.; Girotti, S.; Olives, A.I.; Martin, M.A. Liquid chromatographic analysis of the anticancer alkaloid luotonin A and some new derivatives in human serum samples. J. Sep. Sci. 2010, 33, 2086–2093. [Google Scholar] [CrossRef] [PubMed]
- Laskar, P.; Samanta, S.; Ghosh, S.K.; Dey, J. In vitro evaluation of pH-sensitive cholesterol-containing stable polymeric micelles for delivery of camptothecin. J. Colloid Interface Sci. 2014, 430, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Huang, F.; Ding, S.; Feng, G. Near-infrared fluorescent probe for detection of thiophenols in water samples and living cells. Anal. Chem. 2014, 86, 8835–8841. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Choi, S.J.; Kim, Y.C.; Kuh, H.J. Anti-tumor activity of noble indirubin derivatives in human solid tumor models in vitro. Arch. Pharm. Res. 2009, 32, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Kunjithapatham, R.; Karthikeyan, S.; Geschwind, J.F.; Kieserman, E.; Lin, M.; Fu, D.X.; Ganapathy-Kanniappan, S. Reversal of anchorage-independent multicellular spheroid into a monolayer mimics a metastatic model. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Q.H.; Wu, L.J.; Tashiro, S.; Onodera, S.; Ikejima, T. Atypical apoptosis in L929 cells induced by evodiamine isolated from Evodia rutaecarpa. J. Asian. Nat. Prod. Res. 2004, 6, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Zi, C.T.; Xu, F.Q.; Li, G.T.; Li, Y.; Ding, Z.T.; Zhou, J.; Jiang, Z.H.; Hu, J.M. Synthesis and anticancer activity of glucosylated podophyllotoxin derivatives linked via 4beta-triazole rings. Molecules 2013, 18, 13992–14012. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ding, Y.; Sun, X.S.; Nguyen, T.A. Peptide hydrogelation and cell encapsulation for 3D culture of MCF-7 breast cancer cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Man, S.; Gao, W.; Wei, C.; Liu, C. Anticancer drugs from traditional toxic chinese medicines. Phytother. Res. 2012, 26, 1449–1465. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A.; Freyer, J.P.; Hofstaedter, F.; Ebner, R. The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. J. Biomol. Screen 2004, 9, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Mellor, H.R.; Ferguson, D.J.; Callaghan, R. A model of quiescent tumour microregions for evaluating multicellular resistance to chemotherapeutic drugs. Br. J. Cancer 2005, 93, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.-P.; Xie, Y.-L.; Tian, Z.-Y.; Xu, R.; Qin, Y.; Li, L.; Zhu, J.-H.; Zhang, X.-H. Solubilities of Rutaecarpine in Twelve Organic Solvents from (283.2 to 323.2) K. J. Chem. Eng. Data 2013, 58, 1969–1973. [Google Scholar] [CrossRef]
- Antoon, J.W.; Liu, J.; Ponnapakkam, A.P.; Gestaut, M.M.; Foroozesh, M.; Beckman, B.S. Novel d-erythro N-octanoyl sphingosine analogs as chemo- and endocrine-resistant breast cancer therapeutics. Cancer Chemother. Pharmacol. 2010, 65, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.

| Cells | EVO | RUT | CDDP | |||
|---|---|---|---|---|---|---|
| 2D | 3D | 2D | 3D | 2D | 3D | |
| MCF-7 | 18.1 | 34.6 | 44.1 | 138.0 | 15.9 [40] | 39.9 |
| SMMC-7721 | 27.4 | 56.2 | 24.2 | 42.3 | 6.4 [40] | 21.8 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Liu, D.; Gao, B.; Zhang, X.; You, M.; Ren, H.; Zhang, H.; Santos, H.A.; Xu, F. Antiproliferative Activity and Cellular Uptake of Evodiamine and Rutaecarpine Based on 3D Tumor Models. Molecules 2016, 21, 954. https://doi.org/10.3390/molecules21070954
Guo H, Liu D, Gao B, Zhang X, You M, Ren H, Zhang H, Santos HA, Xu F. Antiproliferative Activity and Cellular Uptake of Evodiamine and Rutaecarpine Based on 3D Tumor Models. Molecules. 2016; 21(7):954. https://doi.org/10.3390/molecules21070954
Chicago/Turabian StyleGuo, Hui, Dongmei Liu, Bin Gao, Xiaohui Zhang, Minli You, Hui Ren, Hongbo Zhang, Hélder A. Santos, and Feng Xu. 2016. "Antiproliferative Activity and Cellular Uptake of Evodiamine and Rutaecarpine Based on 3D Tumor Models" Molecules 21, no. 7: 954. https://doi.org/10.3390/molecules21070954