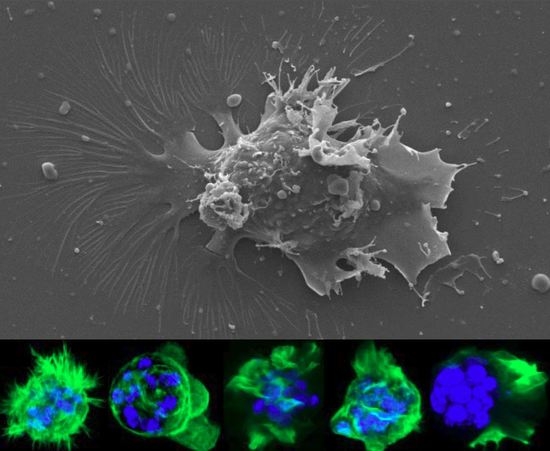
Myxozoan Adhesion and Virulence: Ceratonova shasta on the Move
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of C. shasta Genotypes IIR and I for Motility Studies
2.1.1. Light Microscopy and Time Lapse Series
2.1.2. Confocal Laser Scanning Microscopy
2.1.3. Electron Microscopy (SEM & TEM)
2.2. Surface Adhesion Experiment of Genotype IIR: 2D Environment
2.3. C. shasta Genotype 0 and IIR Infections for Transcriptomic Analysis
2.4. C. shasta Quantification
2.5. Motility Gene Mining from Reference Transcriptome
2.6. Motility Genes Expression Assays
3. Results
3.1. Motility Modes in C. shasta—Blebbing, Adhesion and Crawling
3.2. Myxozoan Blebbing Promotes Fast Amoeboid Motility and Spore Release
3.3. Filopodia and Lamellipodia Promote Myxozoan Adhesion While Adhesive Surfaces Promote the Formation of These Cell Protrusions
3.4. Crawling Stages: Fast and Directional Motility
3.5. Motility Mode Switching Optimizes C. shasta Migration in Complex Environments
3.6. Physical and Morphological Differences in Motility-Related Structures Exist Between C. shasta Genotypes
3.7. Low Proliferation and Delayed Spore Production Characterize Low Virulent Genotype Infections; Fast and Massive Proliferation Characterizes Virulent Genotypes
3.8. β-actin, Integrin-β, Talin and RhoA Are Upregulated in C. shasta Virulent Genotypes
4. Discussion
4.1. Fast Proliferation and Rapid Bleb-Based Migration Characterize Virulent C. shasta Strain Invasion
4.2. Virulent Genotypes Interact with and Disrupt ECM at Late Stage Infection
4.3. Early Direction-Driven Invasion Followed by Low Proliferation and Slow Mesenchymal Migration to Target Site Characterizes Low Virulent C. shasta
4.4. Moderate Exploitation of Target Tissues by Less Virulent Genotype
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedl, P.; Wolf, K. Plasticity of cell migration: A multiscale tuning model. J. Exp. Med. 2010, 207, 11–19. [Google Scholar] [CrossRef]
- Petrie, R.J.; Yamada, K.M. At the leading edge of three-dimensional cell migration. J. Cell Sci. 2012, 125, 5917–5926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrie, R.J.; Yamada, K.M. Multiple Mechanisms of 3D Migration: The Origins of Plasticity. Curr. Opin. Cell Boil. 2016, 42, 7–12. [Google Scholar] [CrossRef]
- Barragan, A.; Sibley, L.D. Transepithelial Migration of Toxoplasma gondii is Linked to Parasite Motility and Virulence. J. Exp. Med. 2002, 195, 1625–1633. [Google Scholar] [CrossRef] [PubMed]
- Marie, C.; Petri, W.A., Jr. Regulation of Virulence of Entamoeba histolytica. Annu. Rev. Microbiol. 2014, 68, 493–520. [Google Scholar] [CrossRef]
- Heaslip, A.T.; Nishi, M.; Stein, B.; Hu, K. The Motility of a Human Parasite, Toxoplasma gondii, is Regulated by a Novel Lysine Methyltransferase. PLoS Pathog. 2011, 7, 1002201. [Google Scholar] [CrossRef] [PubMed]
- McCammick, E.M.; McVeigh, P.; McCusker, P.; Timson, D.J.; Morphew, R.M.; Brophy, P.M.; Marks, N.J.; Mousley, A.; Maule, A.G. Calmodulin disruption impacts growth and motility in juvenile liver fluke. Parasites Vectors 2016, 9, 46. [Google Scholar] [CrossRef]
- Mejia, P.; Diez-Silva, M.; Kamena, F.; Lu, F.; Fernandes, S.M.; Seeberger, P.H.; Davis, A.E., III; Mitchell, J.R. Human C1-Inhibitor Suppresses Malaria Parasite Invasion and Cytoadhesion via Binding to Parasite Glycosylphosphatidylinositol and Host Cell Receptors. J. Infect. Dis. 2016, 213, 80–89. [Google Scholar] [CrossRef]
- Liu, J.; Pan, T.; You, X.; Xu, Y.; Liang, J.; Limpanont, Y.; Sun, X.; Okanurak, K.; Zheng, H.; Wu, Z.; et al. SjCa8, a calcium-binding protein from Schistosoma japonicum, inhibits cell migration and suppresses nitric oxide release of RAW264.7 macrophages. Parasites Vectors 2015, 8, 513. [Google Scholar] [CrossRef]
- Feist, S.W.; Morris, D.J.; Alama-Bermejo, G.; Holzer, A.S. Cellular Processes in Myxozoans. In Myxozoan Evolution, Ecology and Development; Okamura, B., Gruhl, A., Bartholomew, J., Eds.; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Gruhl, A.; Okamura, B. Development and myogenesis of the vermiform Buddenbrockia (Myxozoa) and implications for cnidarian body plan evolution. EvoDevo 2012, 3, 10. [Google Scholar] [CrossRef]
- Alama-Bermejo, G.; Bron, J.E.; Raga, J.A.; Holzer, A.S. 3D Morphology, Ultrastructure and Development of Ceratomyxa puntazzi Stages: First Insights into the Mechanisms of Motility and Budding in the Myxozoa. PLoS ONE 2012, 7, e32679. [Google Scholar] [CrossRef] [PubMed]
- Noble, E.R. Nuclear cycles in the life history of the protozoan genus Ceratomyxa. J. Morphol. 1941, 69, 455–479. [Google Scholar] [CrossRef]
- Meglitsch, P. Some coelozoic myxosporidia from New Zealand fishes I. General, and family Ceratomyxidae. Trans. Proc. R. Soc. N. Z. 1960, 88, 265–356. [Google Scholar]
- Sitjà-Bobadilla, A.; Palenzuela, O.; Alvarez-Pellitero, P. Ceratomyxa sparusaurati n. sp. (Myxosporea: Bivalvulida), a new parasite from cultured gilthead seabream (Sparus aurata L.) (Teleostei: Sparidae): Light and electron microscopic description. J. Eukaryot. Microbiol. 1995, 42, 529–539. [Google Scholar] [CrossRef]
- Cho, J.B.; Kwon, S.R.; Kim, S.K.; Nam, Y.K.; Kim, K.H. Ultrastructure and development of Ceratomyxa protopsettae Fujita, 1923 (Myxosporea) in the gallbladder of cultured olive flounder, Paralichthys olivaceus. Acta Protozool. 2004, 43, 241–250. [Google Scholar]
- Hartigan, A.; Estensoro, I.; Vancová, M.; Bílý, T.; Patra, S.; Eszterbauer, E.; Holzer, A.S. New cell motility model observed in parasitic cnidarian Sphaerospora molnari (Myxozoa:Myxosporea) blood stages in fish. Sci. Rep. 2016, 6, 39093. [Google Scholar] [CrossRef] [PubMed]
- Adriano, E.; Okamura, B. Motility, morphology and phylogeny of the plasmodial worm, Ceratomyxa vermiformis n. sp. (Cnidaria: Myxozoa: Myxosporea). Parasitology 2017, 144, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Bjork, S.J.; Bartholomew, J.L. Invasion of Ceratomyxa shasta (Myxozoa) and comparison of migration to the intestine between susceptible and resistant fish hosts. Int. J. Parasitol. 2010, 40, 1087–1095. [Google Scholar] [CrossRef]
- Bartholomew, J.L.; Whipple, M.J.; Stevens, D.G.; Fryer, J.L. The Life Cycle of Ceratomyxa shasta, a Myxosporean Parasite of Salmonids, Requires a Freshwater Polychaete as an Alternate Host. J. Parasitol. 1997, 83, 859. [Google Scholar] [CrossRef]
- Atkinson, S.D.; Bartholomew, J.L. Disparate infection patterns of Ceratomyxa shasta (Myxozoa) in rainbow trout (Oncorhynchus mykiss) and Chinook salmon (Oncorhynchus tshawytscha) correlate with internal transcribed spacer-1 sequence variation in the parasite. Int. J. Parasitol. 2010, 40, 599–604. [Google Scholar] [CrossRef]
- Atkinson, S.D.; Bartholomew, J.L. Spatial, temporal and host factors structure the Ceratomyxa shasta (Myxozoa) population in the Klamath River basin. Infect. Genet. Evol. 2010, 10, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.N.; Bartholomew, J.L. Ceratomyxa shasta genotypes cause differential mortality in their salmonid hosts. J. Fish Dis. 2012, 35, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Stinson, M.E.T.; Atkinson, S.D.; Bartholomew, J.L. Widespread Distribution of Ceratonova shasta (Cnidaria: Myxosporea) Genotypes Indicates Evolutionary Adaptation to its Salmonid Fish Hosts. J. Parasitol. 2018, 104, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Gall, G.; Hedrick, R. Susceptibility of two strains of rainbow trout Oncorhynchus mykiss to experimentally induced infections with the myxosporean Ceratomyxa Shasta. Dis. Aquat. Org. 1991, 10, 191–194. [Google Scholar] [CrossRef]
- Stocking, R.W.; Holt, R.A.; Foott, J.S.; Bartholomew, J.L. Spatial and Temporal Occurrence of the Salmonid Parasite Ceratomyxa shasta in the Oregon–California Klamath River Basin. J. Aquat. Anim. Health 2006, 18, 194–202. [Google Scholar] [CrossRef]
- Hallett, S.; Bartholomew, J. Application of a real-time PCR assay to detect and quantify the myxozoan parasite Ceratomyxa shasta in river water samples. Dis. Aquat. Org. 2006, 71, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Hallett, S.L.; Ray, R.A.; Hurst, C.N.; Holt, R.A.; Buckles, G.R.; Atkinson, S.D.; Bartholomew, J.L. Density of the Waterborne Parasite Ceratomyxa shasta and Its Biological Effects on Salmon. Appl. Environ. Microbiol. 2012, 78, 3724–3731. [Google Scholar] [CrossRef]
- Atkinson, S.D.; Hallett, S.L.; Bartholomew, J.L. Genotyping of individual Ceratonova shasta (Cnidaria: Myxosporea) myxospores reveals intra-spore ITS-1 variation and invalidates the distinction of genotypes II and III. Parasitology 2018, 145, 1588–1593. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Paluch, E.K.; Raz, E. The role and regulation of blebs in cell migration. Curr. Opin. Cell Boil. 2013, 25, 582–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charras, G.; Paluch, E. Blebs lead the way: How to migrate without lamellipodia. Nat. Rev. Mol. Cell Boil. 2008, 9, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Maugis, B.; Brugues, J.; Nassoy, P.; Guilĺen, N.; Sens, P.; Amblard, F. Dynamic instability of the intracellular pressure drives bleb-based motility. J. Cell Sci. 2010, 123, 3884–3892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charras, G.; Charras, G. A short history of blebbing. J. Microsc. 2008, 231, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.E. Circus movements and blebbing locomotion in dissociated embryonic cells of an amphibian, Xenopus laevis. J. Cell Sci. 1976, 22, 575–583. [Google Scholar] [PubMed]
- Olson, E.C.E. Onset of Electrical Excitability during a Period of Circus Plasma Membrane Movements in Differentiating Xenopus neurons. J. Neurosci. 1996, 16, 5117–5129. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Boil. 2008, 9, 446–454. [Google Scholar] [CrossRef]
- Paňková, K.; Rösel, D.; Novotný, M.; Brábek, J. The molecular mechanisms of transition between mesenchymal and amoeboid invasiveness in tumor cells. Cell. Mol. Life Sci. 2010, 67, 63–71. [Google Scholar] [CrossRef]
- Galbraith, C.G.; Yamada, K.; Galbraith, J.A. Polymerizing Actin Fibers Position Integrins Primed to Probe for Adhesion Sites. Science 2007, 315, 992–995. [Google Scholar] [CrossRef] [Green Version]
- Diz-Muñoz, A.; Romanczuk, P.; Yu, W.; Bergert, M.; Ivanovitch, K.; Salbreux, G.; Heisenberg, C.-P.; Paluch, E.K. Steering cell migration by alternating blebs and actin-rich protrusions. BMC Boil. 2016, 14, 523. [Google Scholar] [CrossRef]
- Fackler, O.T.; Grosse, R. Cell motility through plasma membrane blebbing. J. Cell Boil. 2008, 181, 879–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casadevall, A.; Pirofski, L. Host-Pathogen Interactions: The Attributes of Virulence. J. Infect. Dis. 2001, 184, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Hurst, C.; Alexander, J.; Dolan, B.; Jia, L.; Bartholomew, J. Outcome of within-host competition demonstrates that parasite virulence doesn’t equal success in a myxozoan model system. Int. J. Parasitol. Parasites Wildl. 2019, 9, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Bunnell, T.M.; Burbach, B.J.; Shimizu, Y.; Ervasti, J.M. β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Boil. Cell 2011, 22, 4047–4058. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Goode, B.L. Coronin: The Double-Edged Sword of Actin Dynamics. In The Coronin Family of Proteins. Subcellular Biochemistry vol 48; Clemen, C.S., Eichinger, L., Rybakin, V., Eds.; Springer: New York, NY, USA, 2008. [Google Scholar]
- Hou, X.; Katahira, T.; Ohashi, K.; Mizuno, K.; Sugiyama, S.; Nakamura, H. Coactosin accelerates cell dynamism by promoting actin polymerization. Dev. Boil. 2013, 379, 53–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrie, R.J.; Gavara, N.; Chadwick, R.S.; Yamada, K.M. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Boil. 2012, 197, 439–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K.; Petri, W.A. Molecular biology research to benefit patients with Entamoeba histolytica infection. Mol. Microbiol. 2015, 98, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Rikitake, Y.; Takai, Y. Chapter three—Directional Cell Migration: Regulation by Small G Proteins, Nectin-like Molecule-5, and Afadin. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 287, pp. 97–143. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPase signalling in cell migration. Curr. Opin. Cell Boil. 2015, 36, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Lo, C.M.; Buxton, D.B.; Chua, G.C.; Dembo, M.; Adelstein, R.S.; Wang, Y.L. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Boil. Cell 2004, 15, 982–989. [Google Scholar] [CrossRef]
- Fenix, A.M.; Burnette, D.T. A small part of myosin IIB takes on a big role in cell polarity. J. Cell Boil. 2015, 209, 11–12. [Google Scholar] [CrossRef]
- Tovy, A.; Hertz, R.; Siman-Tov, R.; Syan, S.; Faust, D.; Guillén, N.; Ankri, S. Glucose Starvation Boosts Entamoeba histolytica Virulence. PLoS Negl. Trop. Dis. 2011, 5, e1247. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.A.; O’Byrne, S.; Keir, M.E.; Butcher, E.C. Gut-Selective Integrin-Targeted Therapies for Inflammatory Bowel Disease. J. Crohn Colitis 2018, 12, S653–S668. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alama-Bermejo, G.; Holzer, A.S.; Bartholomew, J.L. Myxozoan Adhesion and Virulence: Ceratonova shasta on the Move. Microorganisms 2019, 7, 397. https://doi.org/10.3390/microorganisms7100397
Alama-Bermejo G, Holzer AS, Bartholomew JL. Myxozoan Adhesion and Virulence: Ceratonova shasta on the Move. Microorganisms. 2019; 7(10):397. https://doi.org/10.3390/microorganisms7100397
Chicago/Turabian StyleAlama-Bermejo, Gema, Astrid S. Holzer, and Jerri L. Bartholomew. 2019. "Myxozoan Adhesion and Virulence: Ceratonova shasta on the Move" Microorganisms 7, no. 10: 397. https://doi.org/10.3390/microorganisms7100397