Transcriptional Regulation of Acyl-CoA:Glycerol-sn-3-Phosphate Acyltransferases
Abstract
:1. Introduction
2. Characteristics of GPAT Isoforms
2.1. Classification of GPAT Isoforms
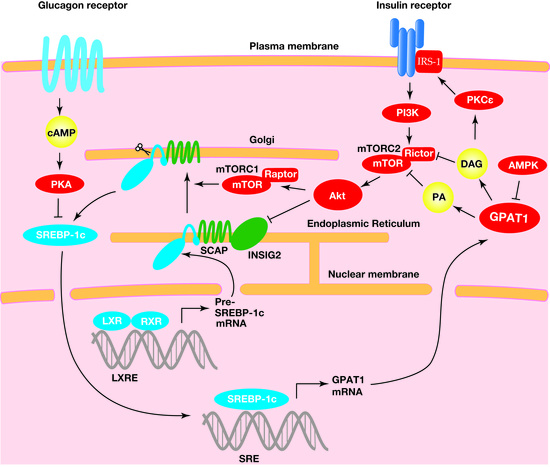
2.2. GPAT1
2.3. GPAT2
2.4. GPAT3
2.5. GPAT4
3. Regulation of GPAT Isoforms
3.1. Transcriptional Regulation
3.2. Post-Translational Regulation
4. Association of the GPAT Expression and Activity with Pathological and Physiological Functions
4.1. Insulin Resistance
4.2. Obesity
4.3. Tumorigenesis
4.4. Spermatogenesis
5. Conclusion and Future Directions
Funding
Conflicts of Interest
References
- Coleman, R.A.; Lee, D.P. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004, 43, 134–176. [Google Scholar] [CrossRef]
- Wendel, A.A.; Lewin, T.M.; Coleman, R.A. Glycerol-3-phosphate acyltransferases: Rate limiting enzymes of triacylglycerol biosynthesis. Biochim. Biophys. Acta 2009, 6, 501–506. [Google Scholar] [CrossRef]
- Takeuchi, K.; Reue, K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1195–E1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Loh, K.; Song, Z.Y.; Yang, H.Q.; Zhang, Y.; Lin, S. Update on glycerol-3-phosphate acyltransferases: The role in the development of insulin resistance. Nutr. Diabetes 2018, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Hayashi, Y.; Matsumoto, N.; Nemoto-Sasaki, Y.; Oka, S.; Tanikawa, T.; Sugiura, T. Glycerophosphate/acylglycerophosphate acyltransferases. Biology 2014, 3, 801–830. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Hayashi, Y.; Nemoto-Sasaki, Y.; Ito, M.; Oka, S.; Tanikawa, T.; Waku, K.; Sugiura, T. Acyltransferase and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 2014, 53, 18–81. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Baro, M.R.; Granger, D.A.; Coleman, R.A. Mitochondrial glycerol phosphate acyltransferase contains two transmembrane domains with the active site in the N-terminal domain facing the cytosol. J. Biol. Chem. 2001, 276, 43182–43188. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Harada, N.; Miyamoto, A.; Kawanishi, Y.; Yoshida, M.; Shono, M.; Mawatari, K.; Takahashi, A.; Skaue, H.; Nakaya, Y. Membrane topology of murine glycerol-3-phosphate acyltransferase 2. Biochem. Biophys. Res. Commun. 2012, 418, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Paulauskis, J.D.; Sul, H.S. Cloning and expression of mouse fatty acid synthase and other specific mRNAs. Developmental and hormonal regulation in 3T3-L1 cells. J. Biol. Chem. 1988, 263, 7049–7054. [Google Scholar]
- Yet, S.F.; Moon, Y.K.; Sul, H.S. Purification and reconstitution of murine mitochondrial glycerol-3-phosphate acyltransferase. Biochemistry 1995, 34, 7303–7310. [Google Scholar] [CrossRef] [PubMed]
- Vancura, A.; Haldar, D. Purification and characterization of glycerophosphate acyltransferase from rat liver mitochondria. J. Biol. Chem. 1994, 269, 27209–27215. [Google Scholar] [PubMed]
- Gonzalez-Baro, M.R.; Coleman, R.M. Mitochondrial acyltransferases and glycerophospholipid metabolism. Biochim. Biophys. Acta 2017, 1862, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Lewin, T.M.; Schwerbrock, N.M.; Lee, D.P.; Coleman, R.A. Identification of a new glycerol-3-phosphate acyltransferase isoenzyme, mtGAT2, in mitochondria. J. Biol. Chem. 2004, 279, 13488–13495. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lee, D.P.; Gong, N.; Schwerbrock, N.M.; Mashek, D.G.; Gonzalez-Baró, M.R.; Stapleton, C.; Li, L.O.; Lewin, T.M.; Coleman, R.A. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2). Arch. Biochem. Biophys. 2007, 465, 347–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattaneo, E.R.; Pellon-Maison, M.; Rabassa, M.E.; Lacunza, E.; Coleman, R.A. glycerol-3-phsophate acyltransferase-2 is expressed in spermatic germ cells and incorporates arachidonic acid into triacylglycerols. PLoS ONE 2012, 7, e42986. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Barnes, R.I.; Garg, A.; Agarwal, A.K. Functional characterization of the human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 10/glycerol-3-phosphate acyltransferase isoform 3. J. Mol. Endocrinol. 2009, 42, 469–478. [Google Scholar] [CrossRef]
- Cao, J.; Li, J.L.; Li, D.; Tobin, J.F.; Gimeno, R.E. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19695–19700. [Google Scholar] [CrossRef]
- Coleman, R.A.; Reed, B.C.; Mackall, J.C.; Student, A.K.; Lane, M.D.; Bell, R.M. Selective changes in microsomal enzymes of triacylglycerol, phosphatidylcholine, and phosphatidylethanolamine biosynthesis during differentiation of 3T3-L1 preadipocytes. J. Biol. Chem. 1978, 253, 7256–7261. [Google Scholar]
- Cao, J.; Perez, S.; Goodwin, B.; Lin, Q.; Peng, H.; Qadri, A.; Zhou, Y.; Clark, R.W.; Perreault, M.; Tobin, J.F.; et al. Mice deleted for GPAT3 have reduced GPAT activity in white adipose tissue and altered energy and cholesterol homeostasis in diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1176–E1187. [Google Scholar] [CrossRef] [Green Version]
- Wilfling, F.; Wang, H.; Haas, J.T.; Krahmer, N.; Gould, T.J.; Uchida, A.; Cheng, J.X.; Graham, M.; Christiano, R.; Fröhlich, F.; et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 2013, 24, 384–399. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Kuo, M.S.; Li, S.; Bui, H.H.; Peake, D.A.; Sanders, P.E.; Thibodeaux, S.J.; Chu, S.; Qian, Y.W.; Zhao, Y.; et al. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. J. Biol. Chem. 2008, 283, 10048–10057. [Google Scholar] [CrossRef] [PubMed]
- Nagel, C.A.; Vergnes, L.; DeJong, H.; Wang, S.; Lewin, T.M.; Reue, K.; Coleman, R.A. Identification of a novel sn-glycero-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J. Lipid Res. 2008, 49, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Beigneux, A.P.; Vergnes, L.; Qiao, X.; Quatela, S.; Davis, R.; Watkins, S.M.; Coleman, R.A.; Walzem, R.L.; Philips, M.; Reue, K.; et al. AGPAT6-a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J. Lipid Res. 2006, 47, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. The SREBP Pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef]
- Amemiya-Kudo, M.; Shimano, H.; Hasty, A.H.; Yahagi, N.; Yoshikawa, T.; Matsuzaka, T.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Ohashi, K.; et al. Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promotors of lipogenic and cholesterogenic genes. J. Lipid Res. 2002, 43, 1220–1235. [Google Scholar]
- Shimano, H.; Yahagi, N.; Amemiya-Kudo, M.; Hasty, A.H.; Osuga, J.; Tamura, Y.; Shinoiri, F.; Iizuka, Y.; Ohashi, K.; Harada, K.; et al. Sterol regulatory element-binding protein-1 as a key transcription for nutritional induction of lipogenic enzyme genes. J. Biol. Chem. 1999, 274, 35832–35839. [Google Scholar] [CrossRef]
- Repa, J.J.; Liang, G.; Ou, J.; Bashmakov, Y.; Lobaccaro, J.M.A.; Shimomura, I.; Shan, B.; Brown, M.S.; Goldstein, J.L.; Mangelsdorf, D.J. Regulation of mouse sterol regulatory element-binding protein-1-c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000, 14, 2819–2830. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Shimano, H.; Amemiya-Kudo, M.; Yahagi, N.; Hasty, A.H.; Matsuzaka, T.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Ohashi, K.; et al. Identification of liver X-receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell Biol. 2001, 21, 2991–3000. [Google Scholar] [CrossRef]
- Shin, D.H.; Paulauskis, J.D.; Moustaïd, N.; Sul, H.S. Transcriptional regulation of p90 with sequence homology to Escherichia coli glycerol-3-phosphate acyltransferase. J. Biol. Chem. 1991, 266, 23834–23839. [Google Scholar]
- Horton, J.D.; Bashimakov, Y.; Shimomura, I.; Shimano, H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. USA 1998, 95, 5987–5992. [Google Scholar] [CrossRef] [Green Version]
- Shimomura, I.; Bashmakov, Y.; Ikemoto, S.; Horton, J.D.; Brown, M.S.; Goldstein, J.L. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 1999, 96, 13656–13661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Repa, J.J.; Mangelsdorf, D.J. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu. Rev. Cell Dev. Biol. 2000, 16, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.Y.; Repa, J.J. The liver X receptor (LXR) and hepatic lipogenesis: The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 2007, 282, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Jerkins, A.A.; Liu, W.R.; Lee, S.; Sul, H.S. Characterization of the murine mitochondrial glycerol-3-phosphate acyltransferase promoter. J. Biol. Chem. 1995, 270, 1416–1421. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, J.; Jackson, S.M.; Kim, J.B.; Spiegelman, B.M.; Edwards, P.A. Identification of glycerol-3-phosphate acyltransferase as an adipocyte determination and differentiation factor 1- and sterol regulatory element-binding protein-responsive gene. J. Biol. Chem. 1997, 272, 7298–7305. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Harada, N.; Yamamoto, H.; Taketani, Y.; Nakagawa, T.; Yin, Y.; Hattori, A.; Zenitani, T.; Hara, S.; Yonemoto, H. Identification of cis-acting promoter sequences required for expression of the glycerol-3-phosphate acyltransferase 1 gene in mice. Biochim. Biophys. Acta 2009, 1791, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Aneja, K.K.; Guha, P.; Shilpi, R.Y.; Chakraborty, S.; Schramm, L.M.; Haldar, D. The presence of distal and proximal promoters for rat mitochondrial glycerol-3-phosphate acyltransferase. Arch. Biochem. Biophys. 2008, 470, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Guha, P.; Aneja, K.K.; Shilpi, R.Y.; Haldar, D. Transcriptional regulation of mitochondrial Glycerophosphate acyltransferase is mediated by distal promoter via ChREBP and SREBP-1. Arch. Biochem. Biophys. 2009, 490, 85–95. [Google Scholar] [CrossRef]
- Harada, N.; Fujimoto, E.; Okuyama, M.; Sakaue, H.; Nakaya, Y. Identification and functional characterization of human glycerol-3-phophate acyltransferase 1 gene promoters. Biochem. Biophys. Res. Commun. 2012, 423, 128–133. [Google Scholar] [CrossRef]
- Moya, M.; Benet, M.; Guzmán, C.; Tolosa, L.; Carcia-Monzón, C.; Pareja, E.; Castell, J.V.; Jover, R. Foxa1 induces lipid accumulation in human hepatocytes and is down-regulated in nonalcoholic fatty liver. PLoS ONE 2012, 7, e30014. [Google Scholar] [CrossRef]
- Hughes, D.E.; Stolz, D.B.; Yu, S.; Tan, Y.; Reddy, J.K.; Watkins, S.C.; Diehl, A.M.; Costa, R.H. Elevated hepatocyte levels of the Forkhead box A2 (HNF-3β) transcription factor cause postnatal steatosis and mitochondria damage. Hepatology 2003, 37, 1414–1424. [Google Scholar] [CrossRef] [PubMed]
- Ehara, T.; Kamei, Y.; Takahashi, M.; Yuan, X.; Kanai, S.; Tamura, E.; Tanaka, M.; Yamazaki, T.; Miura, S.; Ezaki, O.; et al. Role of DNA-methylation in the regulation of lipogenic glycerol-3-phosphate acyltransferase 1 gene expression in the mouse neonatal liver. Diabetes 2012, 61, 2442–2450. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fabiani, M.B.; Montanaro, M.A.; Lacunza, E.; Cattaneo, E.R.; Coleman, R.A.; Pellon-Maison, M.; Conzalez- Baró, M.R. Methylation of the Gpat2 promoter regulates transient expression during mouse spermatohenesis. Biochem. J. 2015, 471, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.; Spiegelman, B.M. PPARγ: A nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001, 41, 37731–37734. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Jiang, Y.J.; Kim, P.; Moser, A.; Elias, P.M.; Grunfeld, C.; Feingold, K.R. Expression and regulation of GPAT isoforms in cultured human keratinocytes and rodent epidermis. J. Lipid Res. 2010, 51, 3207–3216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, C.; Zhu, Y.; Reddy, J.K. Peroxisome proliferator-activated receptors, coactivators, and downstream targets. Cell Biochem. Biophys. 2003, 32, 187–204. [Google Scholar] [CrossRef]
- Bronnikov, G.E.; Aboulaich, N.; Vener, A.V.; Strålfors, P. Acute effects of insulin on the activity of mitochondrial GPAT1 in primary adipocytes. Biochem. Biophys. Res. Commun. 2008, 367, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.E.; Mitchelhill, K.I.; Stapleton, D.; Michell, B.J.; Chen, Z.P.; Witters, L.A. Dealing with energy demand: The AMP-activated protein kinase. Trens Biochem. Sci. 1999, 24, 22–25. [Google Scholar] [CrossRef]
- Muoio, D.M.; Seefeld, K.; Witters, L.A.; Coleman, R.A. AMP-activated kinase reciprocally regulates triacylglycerol synthesis and fatty acid oxidation in liver and muscle: Evidence that sn-glycero-3-phosphate acyltransferase is a novel target. Biochem. J. 1999, 338, 783–791. [Google Scholar] [CrossRef]
- Merrill, G.F.; Kurth, E.J.; Hardie, D.G.; Winder, W.W. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation and glucose uptake in rat muscle. Am. J. Physiol. 1997, 273, E1107–E1112. [Google Scholar] [CrossRef]
- McGarry, J.D.; Leatherman, G.F.; Foster, D.W. Carnitine palmitoyltransferase 1: The site of inhibition of hepatic fatty acid oxidation by malonyl-CoA. J. Biol. Chem. 1978, 253, 4128–4236. [Google Scholar] [PubMed]
- Henriksen, B.S.; Curtis, M.E.; Fillmore, N.; Cardon, B.R.; Thomson, D.M.; Hancock, C.R. The effects of chronic AMPK activation on hepatic triglyceride accumulation and glycerol-3-phsophate acyltransferase activity with high fat feeding. Diabetol. Metab. Syndr. 2013, 5, 29. [Google Scholar] [CrossRef] [PubMed]
- Hocking, S.; Samocha-Bonet, D.; Milner, K.L.; Greenfield, J.R.; Chisholm, D.J. Adiposity and insulin resistance in humans: The role of the different tissue and cellular lipid depots. Endocr. Rev. 2013, 34, 463–500. [Google Scholar] [CrossRef] [PubMed]
- Nagel, C.A.; An, J.; Shiota, M.; Torres, T.P.; Cline, G.W.; Liu, Z.X.; Wang, S.; Catlin, R.L.; Shulman, G.I.; Newgard, C.B.; et al. Hepatic overexpression of glycerol-sn-3-phsphate acyltransferase 1 in rats causes insulin resistance. J. Biol. Chem. 2007, 282, 14807–14815. [Google Scholar] [CrossRef] [PubMed]
- Neschen, S.; Morino, K.; Hammond, L.E.; Zhang, D.; Liu, Z.X.; Romanelli, A.J.; Cline, G.W.; Pongratz, R.L.; Zhang, X.M.; Choi, C.S.; et al. Prevention of hepatic steatosis and hepatic insulin resistance in mitochondrial acyl-CoA:glycerol-sn-3-phsphate acyltransferase 1 knockout mice. Cell Metab. 2005, 2, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Samuel, V.T.; Liu, Z.X.; Qu, X.; Elder, B.D.; Bilz, S.; Befroy, D.; Rommanelli, A.J.; Shulman, G.I. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J. Biol. Chem. 2004, 279, 32345–32353. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A.A.; Li, L.O.; Li, Y.; Cline, G.W.; Shulman, G.I.; Coleman, R.A. Glycerol-3-phosphate acyltransferase 1 deficiency in ob/ob mice diminishes hepatic steatosis but does not protect against insulin resistance or obesity. Diabetes 2010, 59, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wendel, A.A.; Keogh, M.R.; Harris, T.E.; Chen, J.; Coleman, R.A. Glycerolipid signals alter mTOR complex 2 (mTORC2) to diminish insulin signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 1667–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Cooper, D.E.; Grevengoed, T.J.; Li, L.O.; Klett, E.L.; Eaton, J.M.; Harris, T.E.; Coleman, R.A. Glycerol-3-phosphate acyltransferase-4-deficient mice are protected from diet-induced insulin resistance by the enhanced association of mTOR and rictor. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E305–E315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopelman, P.G. Obesity as a medical problem. Nature 2004, 404, 635–643. [Google Scholar] [CrossRef]
- Masoodi, M.; Kuda, O.; Rossmeisl, M.; Flachs, P.; Kopecky, J. Lipid signaling in adipose tissue: Commenting inflammation and metabolism. Biochim. Biophys. Acta 2015, 1851, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Hammond, L.E.; Gallagher, P.A.; Wang, S.; Hiller, S.; Kluckman, K.D.; Posey-Marcos, E.L.; Maeda, N.; Coleman, R.A. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol. Cell Biol. 2002, 22, 8204–8214. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wilcox, D.; Nguyen, P.; Voorbach, M.; Suhar, T.; Morgan, S.J.; An, W.F.; Ge, L.; Green, J.; Wu, Z.; et al. Hepatic knockdown of mitochondrial GPAT1 in ob/ob mice improves metabolic profile. Biochem. Biophys. Res. Commun. 2006, 349, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Lindén, D.; William-Olsson, L.; Rhedin, M.; Asztély, A.K.; Clapham, J.C.; Schreyer, S. Overexpression of mitochondrial GPAT in rat hepatocytes leads to decreased fatty acid oxidation and increased glycerolipids biosynthesis. J. Lipid Res. 2004, 45, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Morgan-Bathke, M.; Chen, L.; Oberschneider, E.; Harteneck, D.; Jensen, M.D. Sex and depot differences in ex vivo adipose tissue fatty acid storage and glycerol-3-phosphat acyltransferase activity. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E830–E846. [Google Scholar] [CrossRef] [PubMed]
- Vergnes, L.; Beigneux, A.P.; Davis, R.; Watkins, S.M.; Young, S.G.; Reue, K. Agpat6 deficiency causes subdermal lipodystrophy and resistance to obesity. J. Lipid Res. 2006, 47, 745–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massoud, O.; Charlton, M. Nonalcoholic fatty liver disease/nonalcoholic steatohepatitis and hepatocellular carcinoma. Clin. Liver Dis. 2018, 22, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Lee, J.H.; Yu, G.Y.; He, G.; Ali, S.R.; Holzer, R.G.; Österreicher, C.H.; Takahashi, H.; Karin, M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010, 140, 197–208. [Google Scholar] [CrossRef]
- Ellis, J.M.; Paul, D.S.; Depetrillo, N.A.; Singh, B.P.; Malarkey, D.E.; Coleman, R.A. Mice deficient in glycerol-3-phosphate acyltransferase-1 have a reduced susceptibility to liver cancer. Toxicol. Pathol. 2012, 40, 513–521. [Google Scholar] [CrossRef]
- Stewart, J.D.; Marchan, R.; Lesjak, M.S.; Lambert, J.; Hergenroeder, R.; Ellis, J.K.; Lau, C.H.; Keun, H.C.; Schmitz, G.; Schiller, J.; et al. Choline-releasing glycerophosphodiesterase ED13 drives tumor cell migration and metastasis. Proc. Natl. Acad. Sci. USA 2012, 109, 8155–8160. [Google Scholar] [CrossRef]
- Marchan, R.; Büttner, B.; Lambert, J.; Edlund, K.; Glaeser, I.; Blaszkewicz, M.; Leohardt, G.; Marienhoff, L.; Kaszta, D.; Anft, M.; et al. Glycerol-3-phosphate acyltransferase 1 promotes tumor cell migration and poor survival in ovarian carcinoma. Cancer Res. 2017, 77, 4589–4601. [Google Scholar] [CrossRef] [PubMed]
- Moolenaar, W.H.; van Meeteren, L.A.; Giepmans, B.N. The ins and outs of lysophosphatidic acid signaling. Bioessays 2004, 26, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Lysophospholipid signaling in the epithelial ovarian cancer tumor microenvironment. Cancers 2018, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Scanlan, M.J.; Simpson, A.J.; Old, L.J. The cancer/testis genes: Review, standardization, and commentary. Cancer Immun. 2004, 4, 1. [Google Scholar] [PubMed]
- Condomines, M.; Hose, D.; Rème, T.; Requirand, G.; Hundemer, M.; Schoenhals, M.; Goldschmidt, H.; Klein, B. Gene expression profiling and real-time PCR analyses identify novel potential cancer-testis antigens in multiple myeloma. J. Immunol. 2009, 183, 832–840. [Google Scholar] [CrossRef] [PubMed]
- Pellon-Maison, M.; Montanaro, M.A.; Lacunza, E.; Garcia-Fabiani, M.B.; Soler-Gerino, M.C.; Cattaneo, E.R.; Quiroga, I.Y.; Abba, M.C.; Coleman, R.A.; Conzalez-Baró, M.R. Glycerol-3-phosphate acyltransferase-2 behaves as a cancer testis gene and promotes growth and tumorigenicity of the breast cancer MDA-MB-231 cell line. PLoS ONE 2014, 9, e100896. [Google Scholar] [CrossRef]
- Cattaneo, E.R.; Prieto, E.D.; Garcia-Fabiani, M.B.; Montanaro, M.A.; Guillou, H.; Gonzalez-Baro, M.R. Glycero-3-phosphate acyltransferase-2 expression modulates cell roughness and membrane permeability: An atomic force microscopy study. PLoS ONE 2017, 12, e0189031. [Google Scholar] [CrossRef]
- Shiromoto, Y.; Kuramochi-Miyagawa, S.; Daiba, A.; Chuma, S.; Katanaya, A.; Katsumata, A.; Nishimura, K.; Ohtaka, M.; Nakanishi, M.; Nakamura, T.; et al. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA 2013, 19, 803–810. [Google Scholar] [CrossRef] [Green Version]
- Janic, A.; Mendizabal, L.; Llamazares, S.; Rossell, D.; Gonzalez, C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 2010, 330, 1824–1827. [Google Scholar] [CrossRef]
- Cheng, J.; Guo, J.M.; Xiao, B.X.; Miao, Y.; Jiang, Z.; Li, Q.N. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta 2011, 412, 1621–1625. [Google Scholar] [CrossRef]
- Lacunza, E.; Montanaro, M.A.; Salvati, A.; Memoli, D.; Rizzo, F.; Henning, M.F.; Quiroga, I.Y.; Guillou, H.; Abba, M.C.; Gonzalez-Baró, M.R.; et al. Small non-coding RNA landscape is modified by GPAT2 silencing in MDA-MB-231 cells. Oncotarget 2018, 9, 28141–28154. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Sachidanandam, R.; Girard, A.; Fejes-Toth, K.; Hannon, G.J. Develompentally regulated piRNA clusters implicate MILI1 in transposon control. Science 2007, 316, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fabiani, M.B.; Montanaro, M.A.; Stringa, P.; Lacunza, E.; Cattaneo, E.R.; Santana, M.; Pellon-Maison, M.; Conzalez-Baró, M.R. Glycerol-3-phsphate acyltransferase 2 is essential for normal spermatogenesis. Biochem. J. 2017, 474, 3093–3107. [Google Scholar] [CrossRef] [PubMed]
- Gale, S.E.; Frolov, A.; Han, X.; Bickel, P.E.; Cao, L.; Bowcock, A. A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem. 2006, 281, 11082–11089. [Google Scholar] [CrossRef] [PubMed]
- Outlaw, V.K.; Wydysh, E.A.; Vadlamudi, A.; Medghalchi, S.; Tawnsend, C.A. Design, synthesis, and evaluation of 4- and 5-substituted o-(octanedulfonamide) benzoic acids as inhibitors of glycerol-3-phosphate acyltransferase. Medchemcomm 2014, 5, 826–830. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karasawa, K.; Tanigawa, K.; Harada, A.; Yamashita, A. Transcriptional Regulation of Acyl-CoA:Glycerol-sn-3-Phosphate Acyltransferases. Int. J. Mol. Sci. 2019, 20, 964. https://doi.org/10.3390/ijms20040964
Karasawa K, Tanigawa K, Harada A, Yamashita A. Transcriptional Regulation of Acyl-CoA:Glycerol-sn-3-Phosphate Acyltransferases. International Journal of Molecular Sciences. 2019; 20(4):964. https://doi.org/10.3390/ijms20040964
Chicago/Turabian StyleKarasawa, Ken, Kazunari Tanigawa, Ayako Harada, and Atsushi Yamashita. 2019. "Transcriptional Regulation of Acyl-CoA:Glycerol-sn-3-Phosphate Acyltransferases" International Journal of Molecular Sciences 20, no. 4: 964. https://doi.org/10.3390/ijms20040964