1. Introduction
Multiple sclerosis (MS) is a chronic and complex demyelinating disease of the central nervous system (CNS) with an occurrence of about 0.1% in Caucasian young adults [
1]. The average age of onset of MS is between 20 and 40, and it is twice as common in women [
2]. It is widely believed that the development of MS is attributed mainly to genetic susceptibility combined with environmental factors [
3,
4]. The most common genetic variants that are known to date belong to HLA class II, are located in chromosome 6p21 and are in linkage disequilibrium, while a variant of HLA-A (class I) is known to confer protection from MS [
5]. According to a number of genome wide association studies (GWAS), at least 29 additional genomic regions are suggested to be associated with MS susceptibility [
5] (and references therein).
Of particular note,
CD24, although not detected in GWAS, is a gene that has been investigated thoroughly in regard to its association with autoimmune diseases, including MS, with promising results. The
CD24 gene is located in the chromosomal region 6q21. Interestingly, the particular region (6q) has previously been suggested to be in linkage with MS [
6,
7]. The CD24 protein is a GPI-anchored cell surface glycoprotein abundantly expressed in a variety of hematopoietic cells such as T and B cells, macrophages, neutrophiles, eosinophils and dendritic cells [
8,
9,
10].
CD24 is also expressed in cells of the CNS such as neural and gaglion cells, astrocytes and microglia [
8,
9,
10,
11], cells involved in the pathogenesis of MS. When expressed in T-cells of the nervous system,
CD24 expression is required for T-cell homeostatic proliferation [
10]. It has also been shown that CD24 is responsible for the local expansion of T cells after migration to the CNS and the development of experimental autoimmune encephalomyelitis (EAE) in mouse models [
12,
13].
Four polymorphisms of the
CD24 gene have been found to be implicated in the etiology of MS and various degenerative diseases [
10]; these polymorphisms are: (a) a C-to-T substitution at nucleotide 226 resulting in a Ala57Val substitution; (b) a TG dinucleotide deletion at positions 1527–1528; (c) an A-to-G substitution at nucleotide 1056; and (d) an A-to-G substitution at nucleotide 1626. The later three polymorphisms are located in exon 2 in the 3ʹ UTR, a region that confers mRNA stability [
14,
15]. rs numbers have been assigned to these polymorphisms (rs52812045, rs3838646, rs1058818 and rs1058881 respectively); however, all correspond to the intronless
CD24 pseudogene, which is located in the chromosomal region Yq11. A number of case-control studies have been performed to investigate the putative association of
CD24 gene polymorphisms with MS development and progression, although the results are controversial. In the present study, we performed a complete meta-analysis of the currently available bibliographic data in order to decipher these associations and add statistical support to them.
3. Discussion
CD24 is a GPI-anchored cell membrane protein, with a molecular mass of 20 to 70 kD, and varying glycosylation. CD24 is a multifaceted protein implicated in inflammation, immunity—both adaptive and autoimmunity—and cancer [
10]. Many experimental studies have focused on investigating the associations of
CD24 polymorphisms with autoimmune diseases such as MS, systemic lupus erythematosus (SLE), rheumatoid arthritis or giant cell arthritis [
10] (and references therein). Although
CD24 was not detected in any GWASs, earlier classical linkage analyses [
6] together with case control studies carried out in 2003 [
11] suggested that
CD24 is a very promising biomarker for MS development. The fact that
CD24 was not identified in GWASs does not necessarily underestimate our study since GWASs often capture only a part of the phenotypic variance, due to intrinsic methodological limitations. The stringent criteria used for statistical significance in GWAS result, in many circumstances, in decreased power to identify a moderate effect. This happens because in the GWAS context, we usually work in an agnostic manner and the primary concern is to reduce the risk of spurious findings and false positive results arising from multiple testing. On the contrary, in the current context, we followed a hypothesis-driven approach. That is, the
CD24 gene had been chosen and studied in many independent studies due to some preliminary genetic evidence (
i.e., as mentioned, the linkage studies concerning the 6q region), as well as due to the presumed biological plausibility that might implicate CD24 antigen in the disease formation. Moreover, the present study includes populations from Iran and Argentina that were not included in previous GWASs. In support of the hypothesis that
CD24 plays a pivotal role in the development of autoimmune diseases, Li
et al. (2006) [
26] found that T cells undergo uncontrolled extensive proliferation in
CD24-deficient mice. Thus, the homeostatic proliferation, which should be slow-paced, loses its self-limiting capacity, and this results in the rapid death of the recipient
CD24−/− mice.
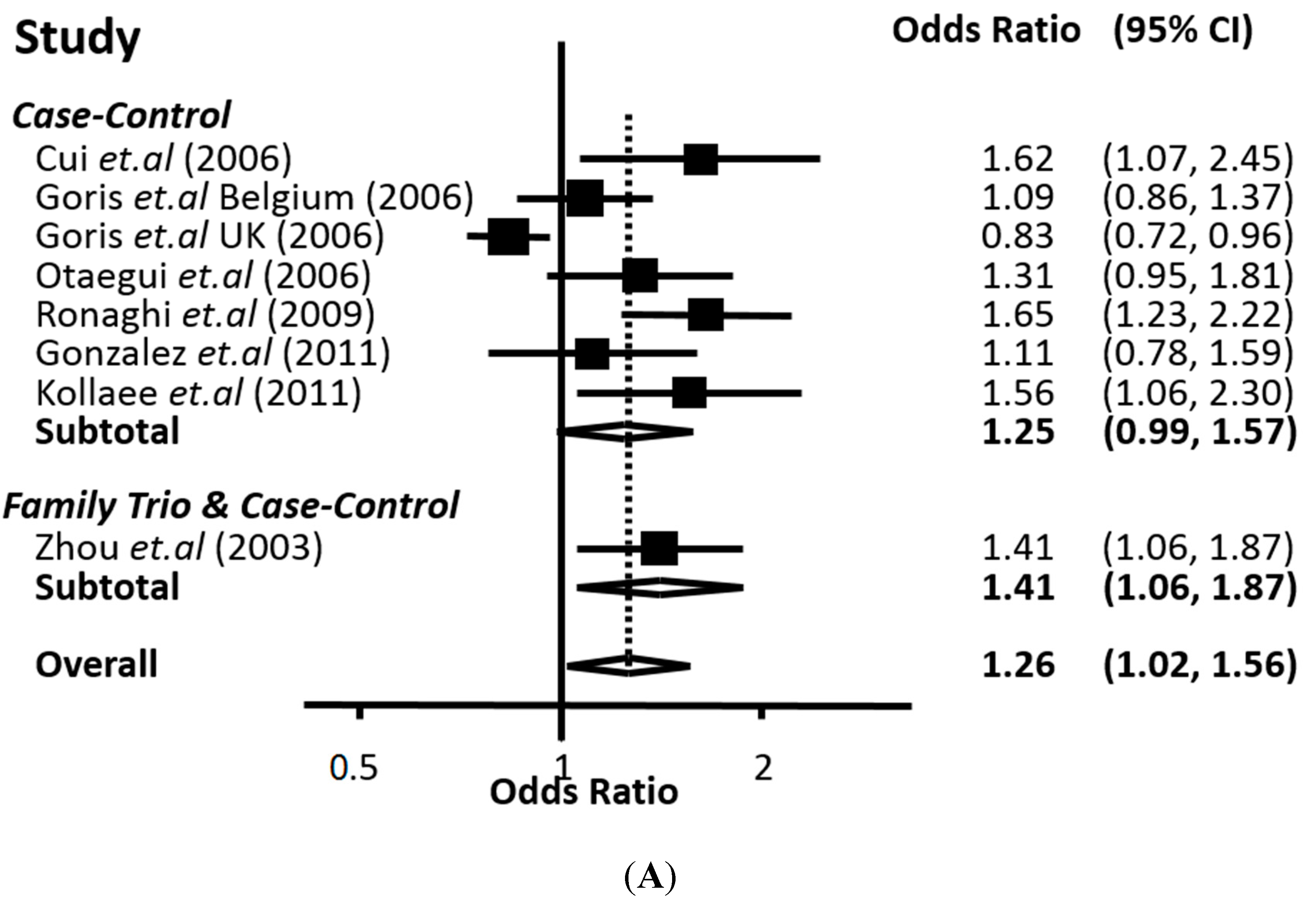
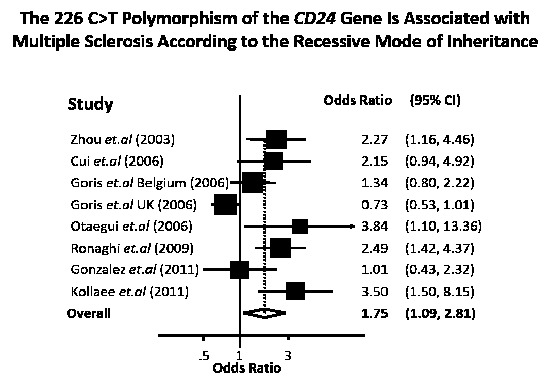
The meta-analysis performed in this work, based on all the available data from the biomedical literature, clearly demonstrates a genetic association between the 226 C>T polymorphism of
CD24 with MS. Our conclusion is supported by two lines of evidence. First, with univariate meta-analysis, significant association was identified with all modes of inheritance (co-dominant, recessive and dominant). Second, from multivariate meta-analysis, which can indicate the mode of inheritance, the significant association was strengthened and the recessive mode appeared to be the most prominent. In any case, the association of the
CD24 226 C>T polymorphism with MS proved to be quite robust according to the overall estimate of our analysis. The findings of our study are in accordance with previous case-control studies showing that the 226 C>T polymorphism of
CD24 is associated with another autoimmune disease, the SLE [
27,
28].
The idea that polymorphisms located in regulatory regions, can account for MS susceptibility has recently gained interest [
29]. To this end, the 1527–1528 TG>del, 1056 A>G and 1626 A>G polymorphisms located in the 3′ UTR of the
CD24 gene were investigated. Our analysis showed that the 1527–1528 TG>del polymorphism has a protective role against the occurrence of MS. Significant associations were found in the allele and the del/del + TG/del
vs. TG/TG genotype contrasts which both had zero heterogeneity. Multivariate meta-analysis further confirmed the above association and reinforced the dominant mode of inheritance. The same TG>del polymorphism has been shown in a case-control study to confer protection against SLE, as well [
15]. Our findings are further corroborated by previous studies showing that this region [
14] and especially the TG dinucleotide deletion confers instability to the
CD24 transcript [
15], thus granting protection against MS and SLE onset and progression.
Conversely, based on meta-analyses we performed for the 1056 A>G and 1626 A>G
CD24 gene polymorphisms, no association was found under any contrast. Noticeably, all SNPs are in close chromosomal proximity to each other and one would expect Linkage Disequilibrium (LD) between them. The two associated polymorphisms with MS have a distance of about 1400 bp. Nevertheless, no definite LD has been observed between 226 C>T and 1527–1528 TG>del in the literature, neither for MS and SLE nor for other diseases such as Chron’s disease [
15,
30]. It is postulated that the
CD24 gene may have a recombination hotspot between these two polymorphisms [
15]. Moreover, the fact that the other two SNPs, though located very close to the associated ones, are not associated with MS may suggest that the 226 C>T and the 1527–1528 TG>del polymorphisms play a causal role in MS, either promoting or preventive, respectively. A haplotype analysis, that would shed more light to this hypothesis, could not be performed in the present study due to lack of data in the included studies. Hence, it is clear that more studies (or collaborative meta-analyses of the current studies) investigating
CD24 haplotypes and their associations with MS are greatly needed to clarify the causative polymorphisms.
Despite the robustness of most of our results, the present study has some limitations. First, the number of studies included is relatively small, while the populations’ ethnic diversity is limited to Asians and Caucasians of European, USA, Argentinean and Iranian ancestry. Hence, additional studies with more participants from diverse ethnic backgrounds would be of great value, especially for the meta-analyses concerning the three polymorphisms located in the 3′ UTR. Second, significant heterogeneity was detected in three out of four analyzed polymorphisms. Regarding the 226 C>T polymorphism, heterogeneity was detected in the allele and the recessive mode contrasts. However, when the analyses were restricted to studies with populations in HWE, the heterogeneity was decreased. As far as the analysis of the 1527–1528 TG>del polymorphism is concerned, heterogeneity was present only in the recessive contrast, which yields non-significant results. Importantly, in the other contrasts, and particularly in the dominant mode contrast (the prevailing one) the heterogeneity was almost zero, reinforcing the robustness of the association. It is of particular interest that high heterogeneity appeared in all contrasts for the 1056 A>G polymorphism while zero heterogeneity was present in the contrasts of 1626 A>G polymorphism. Heterogeneity could be due mainly to small sample size, but also to the different criteria used to diagnose MS. Third, meta-analysis is subjected to methodological weaknesses of the original studies.
Overall, our study has several important advantages. First, a thorough and meticulous search was performed, including both a computer-aided and manual search, to identify all possible eligible studies included in published literature. Second, we did not impose any quality restriction and we did not exclude any study based on the design, the language or any other criteria, since most recent guidelines advocate against this. Third, we used modern statistical techniques in order to include all available studies (i.e., family-based and population-based ones) and to detect the mode of inheritance. The multivariate method in particular is capable not only of identifying the mode of inheritance but also preserving the nominal type I error rate and protecting from multiple comparisons. Although the method identified the most plausible mode of inheritance, we chose to report all the available contrasts only for completeness. Finally, we took any available precaution in order to minimize different sources of bias, which is the main source of concern in this type of studies. Along these lines, we investigated the sources of heterogeneity, we searched for influential studies, and we performed all the available tests for detecting publication bias and time-trend bias.
In summary, we have conducted meta-analyses to investigate associations between
CD24 polymorphisms and MS using a method that combines data from case-control studies with family-based data [
31]. We found that
CD24 226 C>T polymorphism constitutes a risk of MS development while the dinucleotide TG deletion at 1527–1528 confers protection against the disease, and they can be used as prognostic biomarkers for MS onset. Any associations of 1056 A>G and 1626 A>G polymorphisms with MS could not be identified due to the small number of studies. We also estimated the number of the additional participants needed in order to reach significant results, using statistical methods. Given that these numbers can be attained relatively easily, biomedical researchers should be encouraged to investigate the association of the three 3′ UTR polymorphisms with MS and possibly with other autoimmune diseases. Furthermore, additional studies investigating linkage disequilibrium between these
CD24 polymorphisms, gene–gene interactions, or even gene-environment interactions would be helpful in better understanding the role of the
CD24 gene in MS onset and development.