Copy Number Analysis of 24 Oncogenes: MDM4 Identified as a Putative Marker for Low Recurrence Risk in Non Muscle Invasive Bladder Cancer
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
| Gene | Overall Series (43 Cases) | Non Recurrent Tumors (20 Cases) | Recurrent Tumors (23 Cases) | p Value * |
|---|---|---|---|---|
| MDM4 | 20/43 | 13/20 | 7/23 | |
| 0.46 | 0.65 | 0.3 | 0.023 | |
| MET | 15/43 | 10/20 | 5/23 | |
| 0.35 | 0.5 | 0.22 | 0.052 | |
| CDK4 | 12/43 | 7/20 | 5/23 | NS |
| 0.28 | 0.35 | 0.22 | ||
| MYC | 12/43 | 8/20 | 4/23 | NS |
| 0.28 | 0.4 | 0.17 | ||
| TOP2A | 12/43 | 8/20 | 4/23 | NS |
| 0.27 | 0.4 | 0.17 | ||
| AURKA | 11/43 | 5/20 | 6/23 | NS |
| 0.26 | 0.25 | 0.26 | ||
| ERBB2 | 9/43 | 2/20 | 7/23 | NS |
| 0.21 | 0.10 | 0.30 | ||
| MDM2 | 8/43 | 6/20 | 2/23 | NS |
| 0.19 | 0.3 | 0.09 | ||
| CCND1 | 6/43 | 4/20 | 2/23 | NS |
| 0.14 | 0.2 | 0.09 | ||
| BRAF | 6/43 | 3/20 | 3/23 | NS |
| 0.14 | 0.15 | 0.13 | ||
| DHFR | 5/43 | 3/20 | 2/23 | NS |
| 0.12 | 0.15 | 0.09 | ||
| FGFR | 4/43 | 2/20 | 2/23 | NS |
| 0.09 | 0.01 | 0.09 | ||
| CCND2 | 4/43 | 1/20 | 3/23 | NS |
| 0.09 | 0.05 | 0.13 | ||
| KIT | 3/43 | 2/20 | 1/23 | NS |
| 0.07 | 0.1 | 0.04 | ||
| KDR | 1/43 | 0/20 | 1/23 | NS |
| 0.02 | 0 | 0.04 | ||
| MYCN | 0/43 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 | ||
| ALK | 0/43 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 | ||
| PDGFR | 0/43 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 | ||
| EGFR | 0/40 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 | ||
| ABL | 0/43 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 | ||
| RET | 0/43 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 | ||
| AURKB | 0/43 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 | ||
| SMO | 0/43 | 0/20 | 0/23 | NS |
| 0 | 0 | 0 |
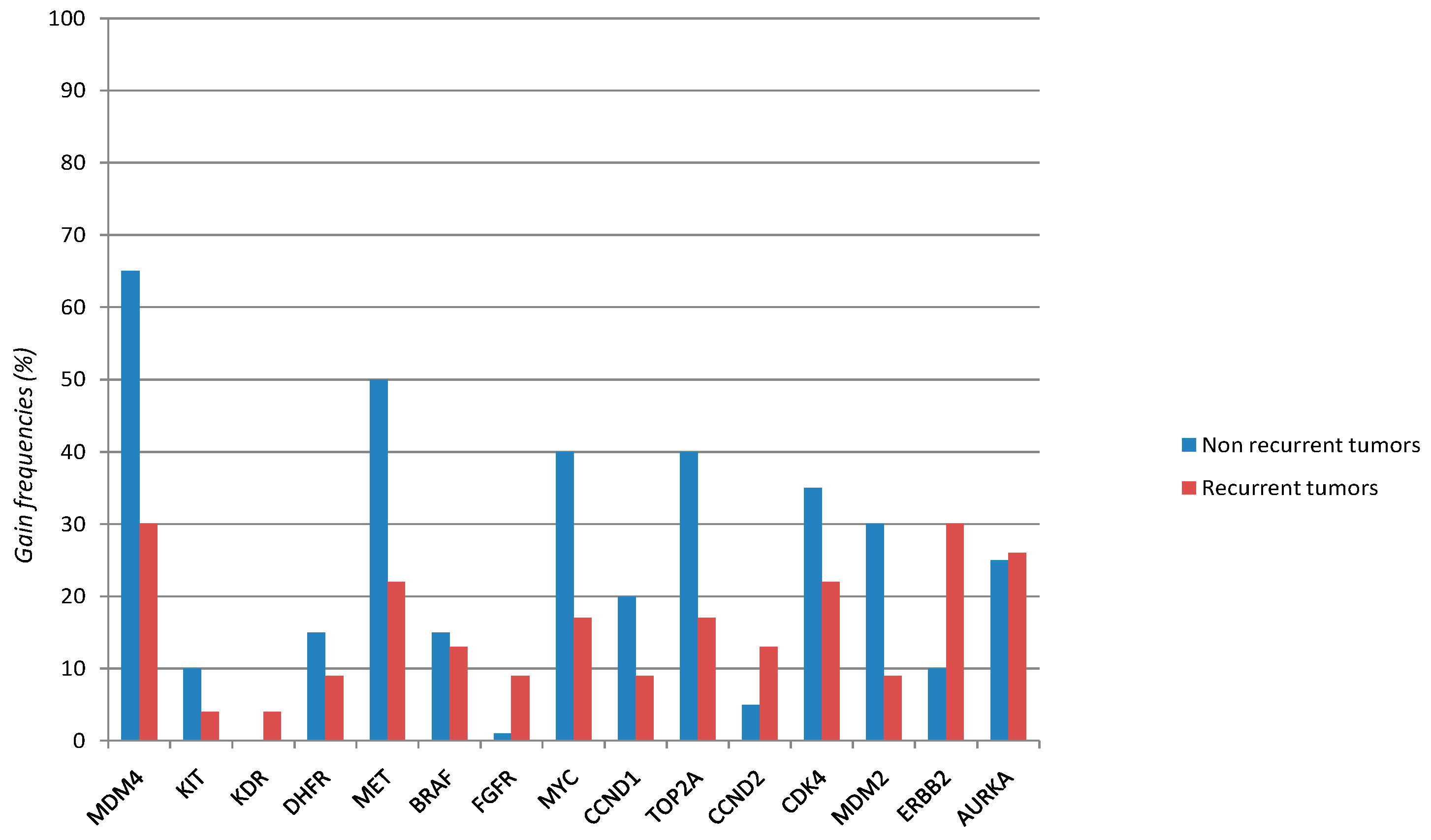

| Variables | Categories | Raw HR (95% CI) | p | Adjusted HR (95% CI) | p |
|---|---|---|---|---|---|
| Age, years | <70 vs. ≥70 | 3.023 (1.301–7.026) | 0.010 | 3.318 (1.420–7.753) | 0.006 |
| Stage | T1 vs. Ta | 1.472 (0.605–3.580) | 0.394 | – | – |
| Grade | High vs. Low | 1.369 (0.573–3.269) | 0.479 | – | – |
| MDM4 | N vs. G | 2.447 (1.000–5.987) | 0.05 | 2.745 (1.114–6.766) | 0.028 |
2.2. Discussion
3. Experimental Section
3.1. Case Series (Retrospective Cohort Study)
| Clinical and Pathological Characteristics | Patients (n = 43) | |
|---|---|---|
| Non Relapsed | Relapsed | |
| Sex | ||
| Male | 18 | 21 |
| Female | 2 | 2 |
| Age, years | ||
| <70 | 4 | 14 |
| ≥70 | 16 | 9 |
| Grade | ||
| High | 5 | 8 |
| Low | 14 | 14 |
| Stage | ||
| Ta | 16 | 16 |
| T1 | 4 | 7 |
| Number of tumors | ||
| Single | 14 | 13 |
| Multiple | 7 | 9 |
3.2. Macrodissection and DNA Isolation
3.3. Multiplex Ligation Probe Amplification (MLPA) Analysis
| Gene | Function | Chromosomal Localization | Exons |
|---|---|---|---|
| MDM4 | Cell cycle and apoptosis | 1q32.1 | 2,8 |
| MYCN | Transcription factor | 2p.24 | 3 |
| ALK | Receptor tyrosine kinase | 2p23.2 | 4,6 |
| PDGFRA | Cell proliferation and survival | 4q12 | 3,22 |
| KIT | Cell proliferation and survival | 4q12 | 2,20 |
| KDR | Regulation of angiogenesis | 4q12 | 14,19 |
| DHFR | Cell growth and proliferation | 5q14.1 | 1,2 |
| EGFR | Cell proliferation | 7P11.2 | 8,22 |
| MET | Tumor growth, angiogenesis and metastasis | 7q31 | 4,10 |
| SMO | Maintenance of tissue homeostasis | 7q32.1 | 4,12 |
| FGFR | Cell proliferation | 8p11.2 | 8,14 |
| MYC | Regulation of gene transcription | 8q24.1 | 3 |
| ABL | Cell growth and survival | 9q34.1 | 1,12 |
| RET | Cell proliferation, cell migration and cell differentiation | 10q11.2 | 8,14 |
| CCND1 | Cell-cycle regulation | 11q13 | 2,3,5 |
| TOP2A | DNA replication, transcription and repair | 17q21.2 | 7,33 |
| CCND2 | Cell-cycle regulation | 12p13.3 | 1,5 |
| CDK4 | Cell-cycle regulation | 12q14 | 3,8 |
| MDM2 | Cell cycle and apoptosis | 12q14.3 | 4,5 |
| ERBB2 | Transcriptional regulation | 17q12 | 7,29 |
| AURKB | Regulator of mitosis | 17p13.1 | 4,5 |
| AURKA | Regulator of mitosis | 20q13.2 | 8,10 |
| AR | Androgen receptor | Xq12 | 9 |
3.4. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef]
- Van Rhijn, B.W.; Burger, M.; Lotan, Y.; Solsona, E.; Stief, C.G.; Sylvester, R.J.; Witjes, J.A.; Zlotta, A.R. Recurrence and progression of disease in non-muscle-invasive bladder cancer: From epidemiology to treatment strategy. Eur. Urol. 2009, 56, 430–442. [Google Scholar] [CrossRef]
- Bryan, R.T.; Collins, S.I.; Daykin, M.C.; Zeegers, M.P.; Cheng, K.K.; Wallace, D.M.; Sole, G.M. Mechanisms of recurrence of Ta/T1 bladder cancer. Ann. R. Coll. Surg. Engl. 2010, 92, 519–524. [Google Scholar] [CrossRef]
- Casadio, V.; Molinari, C.; Calistri, D.; Tebaldi, M.; Gunelli, R.; Serra, L.; Falcini, F.; Zingaretti, C.; Silvestrini, R.; Amadori, D.; et al. DNA methylation profiles as predictors of recurrence in non muscle invasive bladder cancer: An MS-MLPA approach. J. Exp. Clin. Cancer Res. 2013, 32, 94. [Google Scholar] [CrossRef]
- Lindgren, D.; Gudjonsson, S.; Jee, K.J.; Liedberg, F.; Aits, S.; Andersson, A.; Chebil, G.; Borg, A.; Knuutila, S.; Fioretos, T.; et al. Recurrent and multiple bladder tumors show conserved expression profiles. BMC Cancer 2008, 8, 183. [Google Scholar] [CrossRef]
- Lindgren, D.; Sjödahl, G.; Lauss, M.; Staaf, J.; Chebil, G.; Lövgren, K.; Gudjonsson, S.; Liedberg, F.; Patschan, O.; Månsson, W.; et al. Integrated genomic and gene expression profiling identifies two majorgenomic circuits in urothelial carcinoma. PLoS One 2012, 7, e38863. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Matsuyama, H.; Kawauchi, S.; Furuya, T.; Liu, X.P.; Ikemoto, K.; Oga, A.; Naito, K.; Sasaki, K. Biological characteristics in bladder cancer depend on the type of genetic instability. Clin. Cancer Res. 2006, 12, 2752–2758. [Google Scholar] [CrossRef]
- Hurst, C.D.; Platt, F.M.; Taylor, C.F.; Knowles, M.A. Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clin. Cancer Res. 2012, 18, 5865–5877. [Google Scholar] [CrossRef]
- Höglund, M.; Säll, T.; Heim, S.; Mitelman, F.; Mandahl, N.; Fadl-Elmula, I. Identification of cytogenetic subgroups and karyotypic pathways in transitional cell carcinoma. Cancer Res. 2001, 61, 8241–8246. [Google Scholar]
- Watters, A.D.; Latif, Z.; Forsyth, A.; Dunn, I.; Underwood, M.A.; Grigor, K.M.; Bartlett, J.M. Genetic aberrations of c-myc and CCND1 in the development of invasive bladder cancer. Br. J. Cancer 2002, 87, 654–658. [Google Scholar] [CrossRef]
- Veerakumarasivam, A.; Scott, H.E.; Chin, S.F.; Warren, A.; Wallard, M.J.; Grimmer, D.; Ichimura, K.; Caldas, C.; Collins, V.P.; Neal, D.E.; et al. High-resolution array-based comparative genomic hybridization of bladder cancers identifies mouse double minute 4 (MDM4) as an amplification target exclusive of MDM2 and TP53. Clin. Cancer Res. 2008, 14, 2527–2534. [Google Scholar] [CrossRef]
- Simon, R.; Struckmann, K.; Schraml, P.; Wagner, U.; Forster, T.; Moch, H.; Fijan, A.; Bruderer, J.; Wilber, K.; Mihatsch, M.J.; et al. Amplification pattern of 12q13-q15 genes (MDM2, CDK4, GLI) in urinary bladder cancer. Oncogene 2002, 21, 2476–2483. [Google Scholar] [CrossRef]
- Park, H.S.; Park, W.S.; Bondaruk, J.; Tanaka, N.; Katayama, H.; Lee, S.; Spiess, P.E.; Steinberg, J.R.; Wang, Z.; Katz, R.L.; et al. Quantitation of aurora kinase a gene copy number in urine sediments and bladder cancer detection. J. Natl. Cancer Inst. 2008, 100, 1401–1411. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [CrossRef]
- Schouten, J.P.; McElgunn, C.J.; Waaijer, R.; Zwijnenburg, D.; Diepvens, F.; Pals, G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids. Res. 2002, 30, e57. [Google Scholar] [CrossRef]
- Toledo, F.; Wahl, G.M. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int. J. Biochem. Cell. Biol. 2007, 39, 1476–1482. [Google Scholar] [CrossRef]
- Mancini, F.; di Conza, G.; Moretti, F. MDM4 (MDMX) and its transcript variants. Curr. Genomics 2009, 10, 42–50. [Google Scholar] [CrossRef]
- Bakkar, A.A.; Wallerand, H.; Radvanyi, F.; Lahaye, J.B.; Pissard, S.; Lecerf, L.; Kouyoumdjian, J.C.; Abbou, C.C.; Pairon, J.C.; Jaurand, M.C.; et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003, 63, 8108–8112. [Google Scholar]
- Wang, L.; Feng, C.; Ding, G.; Ding, Q.; Zhou, Z.; Jiang, H.; Wu, Z. Ki67 and TP53 expressions predict recurrence of non-muscle-invasive bladder cancer. Tumour Biol. 2014, 35, 2989–2995. [Google Scholar] [CrossRef]
- Papadogianni, D.; Soulitzis, N.; Delakas, D.; Spandidos, D.A. Expression of p53 family genes in urinary bladder cancer: Correlation with disease aggressiveness and recurrence. Tumour Biol. 2014, 35, 2481–2489. [Google Scholar] [CrossRef]
- George, B.; Datar, R.H.; Wu, L.; Cai, J.; Patten, N.; Beil, S.J.; Groshen, S.; Stein, J.; Skinner, D.; Jones, P.A.; et al. P53 gene and protein status: The role of p53 alterations in predicting outcome in patients with bladder cancer. J. Clin. Oncol. 2007, 25, 5352–5358. [Google Scholar] [CrossRef]
- Marín, A.P.; Arranz, E.E.; Sánchez, A.R.; Auñón, P.Z.; Barón, M.G. Role of anti-Her-2 therapy in bladder carcinoma. J. Cancer Res. Clin. Oncol. 2010, 136, 1915–1920. [Google Scholar] [CrossRef]
- Sharma, N.; Adjei, A.A. In the clinic: Ongoing clinical trials evaluating c-MET-inhibiting drugs. Ther. Adv. Med. Oncol. 2011, 3, S37–S50. [Google Scholar] [CrossRef]
- Iyer, G.; Al-Ahmadie, H.; Schultz, N.; Hanrahan, A.J.; Ostrovnaya, I.; Balar, A.V.; Kim, P.H.; Lin, O.; Weinhold, N.; Sander, C.; et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J. Clin. Oncol. 2013, 31, 3133–3140. [Google Scholar] [CrossRef]
- MRC-Holland. Available online: https://www.mlpa.com (accessed on 20 March 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Salvi, S.; Calistri, D.; Gurioli, G.; Carretta, E.; Serra, L.; Gunelli, R.; Zoli, W.; Casadio, V. Copy Number Analysis of 24 Oncogenes: MDM4 Identified as a Putative Marker for Low Recurrence Risk in Non Muscle Invasive Bladder Cancer. Int. J. Mol. Sci. 2014, 15, 12458-12468. https://doi.org/10.3390/ijms150712458
Salvi S, Calistri D, Gurioli G, Carretta E, Serra L, Gunelli R, Zoli W, Casadio V. Copy Number Analysis of 24 Oncogenes: MDM4 Identified as a Putative Marker for Low Recurrence Risk in Non Muscle Invasive Bladder Cancer. International Journal of Molecular Sciences. 2014; 15(7):12458-12468. https://doi.org/10.3390/ijms150712458
Chicago/Turabian StyleSalvi, Samanta, Daniele Calistri, Giorgia Gurioli, Elisa Carretta, Luigi Serra, Roberta Gunelli, Wainer Zoli, and Valentina Casadio. 2014. "Copy Number Analysis of 24 Oncogenes: MDM4 Identified as a Putative Marker for Low Recurrence Risk in Non Muscle Invasive Bladder Cancer" International Journal of Molecular Sciences 15, no. 7: 12458-12468. https://doi.org/10.3390/ijms150712458





