tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA
Abstract
:1. Introduction
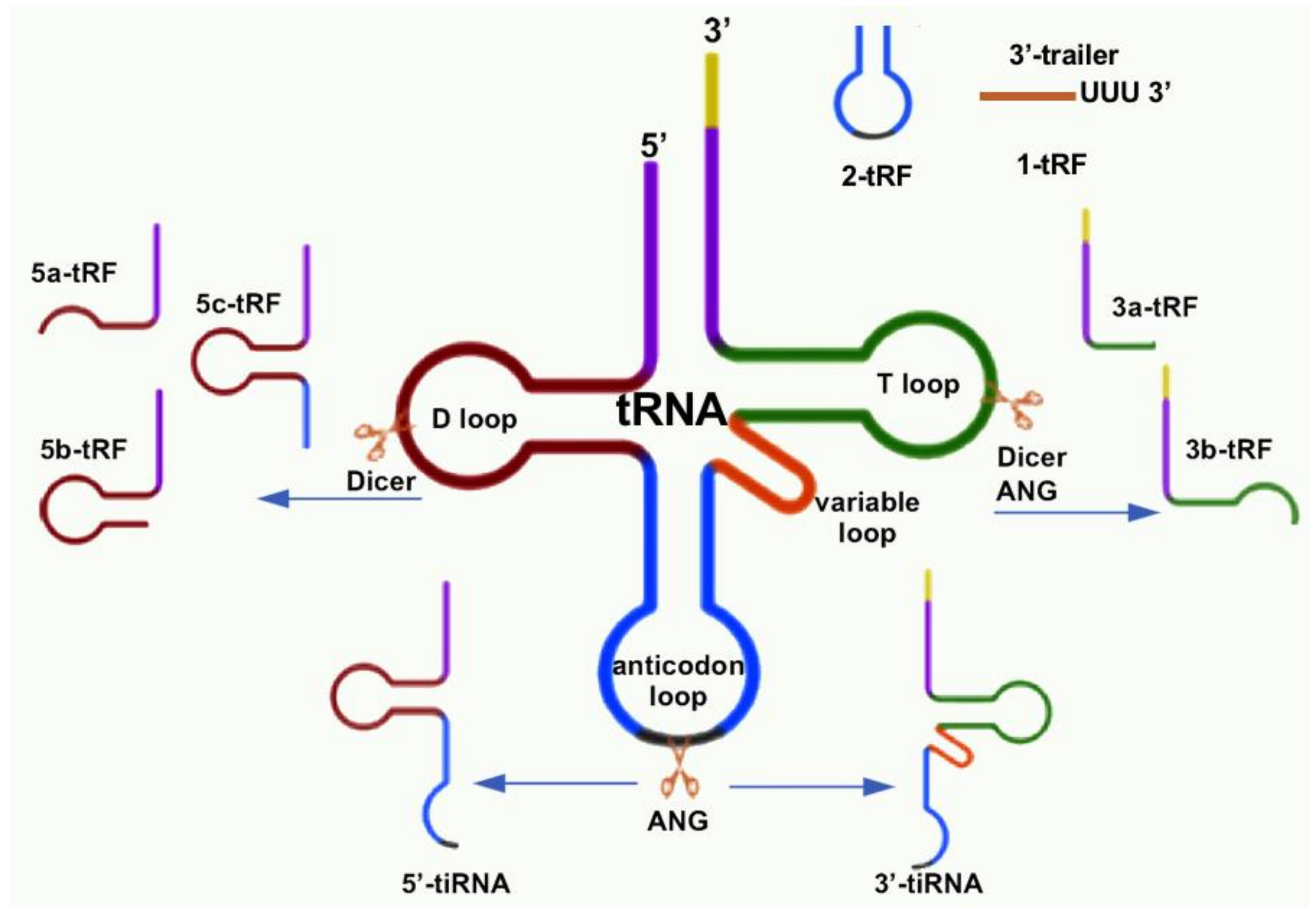
2. tRNA-Derived Small RNA Classification and Biogenesis
2.1. tRNA-Derived Stress-Induced RNA
2.2. tRNA-Derived Fragment
2.3. Effects of Transfer RNA Modification and Structure on tRNA-Derived Small RNA Biogenesis
3. Biological Roles and Mechanisms of Action of tRNA-Derived Small RNAs
3.1. Regulating mRNA Stability
3.2. Inhibiting Translation Initiation and Elongation
3.3. Regulating Ribosome Biogenesis
3.4. Functioning as a Novel Epigenetic Factor
3.5. Regulating RNA Reverse Transcription as a Guide RNA
3.6. Preventing Apoptosis by Binding to Cytochrome C
3.7. Immune Regulation
3.8. Other Possible Roles and Mechanisms of Action
4. Roles of tRNA-Derived Small RNA in Diseases
4.1. tRNA-Derived Small RNA and Cancer
4.2. tRNA-Derived Small RNA and Neurodegenerative Diseases
4.3. tRNA-Derived Small RNA and Acquired Metabolic Diseases
4.4. tRNA-Derived Small RNA and Infectious Diseases
5. Conclusions and Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Chen, S.; Dare, K.; Gilreath, M.; Praetorius-Ibba, M.; Raina, M.; Reynolds, N.M.; Rogers, T.; Roy, H.; Yadavalli, S.S.; et al. tRNAs: Cellular barcodes for amino acids. FEBS Lett. 2010, 584, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P. The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018, 19, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Simos, G.; Hurt, E. Transfer RNA biogenesis: A visa to leave the nucleus. Curr. Biol. 1999, 9, R238–R241. [Google Scholar] [CrossRef]
- Rich, A.; RajBhandary, U.L. Transfer RNA: Molecular structure, sequence, and properties. Annu. Rev. Biochem. 1976, 45, 805–860. [Google Scholar] [CrossRef] [PubMed]
- Borek, E.; Baliga, B.S.; Gehrke, C.W.; Kuo, C.W.; Belman, S.; Troll, W.; Waalkes, T.P. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977, 37, 3362–3366. [Google Scholar] [PubMed]
- Speer, J.; Gehrke, C.W.; Kuo, K.C.; Waalkes, T.P.; Borek, E. tRNA breakdown products as markers for cancer. Cancer 1979, 44, 2120–2123. [Google Scholar] [CrossRef]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. The RNAse Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.R.; Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005, 280, 42744–42749. [Google Scholar] [CrossRef] [PubMed]
- Jochl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.L.; Schrettl, M.; Haas, H.; Huttenhofer, A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, J.; Zhou, H.; Liao, J.Y.; Ma, L.M.; Chen, Y.Q.; Qu, L.H. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008, 36, 6048–6055. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA Pub. RNA Soc. 2008, 14, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, L.C.; Lin, S.I.; Shih, A.C.; Chen, J.W.; Lin, W.Y.; Tseng, C.Y.; Li, W.H.; Chiou, T.J. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Loher, P.; Shigematsu, M.; Palazzo, J.P.; Suzuki, R.; Imoto, I.; Rigoutsos, I.; Kirino, Y. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. USA 2015, 112, E3816–E3825. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Yamakawa, A.; Boffelli, D.; Mote, P.; Martin, D.I.K. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013, 14, 298. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. TRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015, 43, D141–D145. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Ender, C.; Meister, G.; Moore, P.S.; Chang, Y.; John, B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012, 40, 6787–6799. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Lamichhane, T.N. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip. Rev. RNA 2011, 2, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.M.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.Y.; Ma, L.M.; Guo, Y.H.; Zhang, Y.C.; Zhou, H.; Shao, P.; Chen, Y.Q.; Qu, L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE 2010, 5, e10563. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Liu, X.H.; Nguyen, H.C.B.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with Ago proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef] [PubMed]
- Reifur, L.; Garcia-Silva, M.R.; Poubel, S.B.; Alves, L.R.; Arauco, P.; Buiar, D.K.; Goldenberg, S.; Cayota, A.; Dallagiovanna, B. Distinct subcellular localization of tRNA-derived fragments in the infective metacyclic forms of Trypanosoma cruzi. Mem. Inst. Oswaldo Cruz. 2012, 107, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Goktas, C.; Yigit, H.; Cosacak, M.I.; Akgul, B. Differentially expressed tRNA-derived small RNAs co-sediment primarily with non-polysomal fractions in Drosophila. Genes 2017, 8, E333. [Google Scholar] [CrossRef] [PubMed]
- Hori, H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and Nsun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. Nsun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Mishima, E.; Inoue, C.; Saigusa, D.; Inoue, R.; Ito, K.; Suzuki, Y.; Jinno, D.; Tsukui, Y.; Akamatsu, Y.; Araki, M.; et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J. Am. Soc. Nephrol. 2014, 25, 2316–2326. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Sun, Q.; Liu, X.; Wang, P.; Wu, R.; Ma, Z. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem. Biol. Drug Des. 2017, 90, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; He, F.; Luo, J.; Dou, S.; Wang, Y.; Guo, A.; Lu, J. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. MicroRNAs associated with the different human argonaute proteins. Nucleic acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Krokowski, D.; Guan, B.J.; Ivanov, P.; Parisien, M.; Hu, G.F.; Anderson, P.; Pan, T.; Hatzoglou, M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 2012, 287, 42708–42725. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; O’Day, E.; Emara, M.M.; Wagner, G.; Lieberman, J.; Anderson, P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA 2014, 111, 18201–18206. [Google Scholar] [CrossRef] [PubMed]
- Lyons, S.M.; Achorn, C.; Kedersha, N.L.; Anderson, P.J.; Ivanov, P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 2016, 44, 6949–6960. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, N.; Ciesla, M.; Ngoc, P.C.T.; Lang, S.; Arora, S.; Dimitriou, M.; Pimkova, K.; Sommarin, M.N.E.; Munita, R.; Lubas, M.; et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Yim, D.; Krishna, S.; Lakshmanan, V.; Koh, J.; Park, J.E.; Cheong, J.K.; Low, J.L.; Lim, M.; Ip, J.; Nah, J.M.; et al. Dynamic expression of tRNA-derived small RNAs define cellular states. BioRxiv 2017. [Google Scholar] [CrossRef]
- Sobala, A.; Hutvagner, G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013, 10, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Gebetsberger, J.; Wyss, L.; Mleczko, A.M.; Reuther, J.; Polacek, N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017, 14, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Bounova, G.; Purdom, E.; Speed, T.P.; Collins, K. A Tetrahymena Piwi bound to mature trna 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell 2012, 48, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-Chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell 2017, 170, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Choudury, S.G.; Slotkin, R.K. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017, 45, 5142–5152. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shi, J.; Zhang, Y.; Zhang, H.; Liao, S.; Li, W.; Lei, L.; Han, C.; Ning, L.; Cao, Y.; et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012, 22, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, K.; Guffanti, A.; Corradin, A.; Sharma, V.K.; De Bellis, G.; Corti, G.; Grassi, A.; Zanovello, P.; Bronte, V.; Ciminale, V.; et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: A role for a tRNA fragment as a primer for reverse transcriptase. J. Virol. 2014, 88, 3612–3622. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.F.; Ptashkin, R.N.; Chen, Y.; Cheng, Z.; Liu, G.L.; Phan, T.; Deng, X.L.; Zhou, J.H.; Lee, I.; Lee, Y.S.; et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 2015, 23, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.R.; Lee, I.; Ren, J.P.; Ajay, S.S.; Lee, Y.S.; Bao, X.Y. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013, 21, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, S.; Chen, Y.; Fu, Y.; Silver, A.J.; Hill, M.S.; Lee, I.; Lee, Y.S.; Bao, X. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J. Gen. Virol. 2017, 98, 1600–1610. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.D.; Yong, J.; Liu, H.T.; Shi, Y.G.; Meinkoth, J.; Dreyfuss, G.; Yang, X.L. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 2010, 37, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Jobava, R.; Parisien, M.; Putnam, A.; Krokowski, D.; Gao, X.H.; Guan, B.J.; Yuan, Y.; Jankowsky, E.; Feng, Z.; et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol. 2014, 34, 2450–2463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.; Shi, J.; Zhang, H.; Cao, Z.; Gao, X.; Ren, W.; Ning, Y.; Ning, L.; Cao, Y.; et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell. Biol. 2014, 6, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, X.; Liu, C.; Liu, J.; Hu, Q.; Pan, T.; Duan, X.; Liu, B.; Zhang, Y.; Chen, J.; et al. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J. Immunol. 2016, 196, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Xiang, L.; Shao, J.J.; Yuan, Z.H. The 3′ CCACCA sequence of tRNAAla(UGC) is the motif that is important in inducing Th1-like immune response, and this motif can be recognized by Toll-like receptor 3. Clin. Vaccine Immunol. 2006, 13, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Schopman, N.C.; Heynen, S.; Haasnoot, J.; Berkhout, B. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 2010, 7, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by par-clip. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.; Simon, M.C. Hypoxia-inducible factors, stem cells, and cancer. Cell 2007, 129, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Tolkach, Y.; Schmidt, D.; Kristiansen, G.; Muller, S.C.; Ellinger, J. 5′-tRNA halves are dysregulated in clear cell renal cell carcinoma. J. Urol. 2018, 199, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Magee, R.G.; Telonis, A.G.; Loher, P.; Londin, E.; Rigoutsos, I. Profiles of miRNA isoforms and tRNA fragments in prostate cancer. Sci. Rep. 2018, 8, 5314. [Google Scholar] [CrossRef] [PubMed]
- Yeri, A.; Courtright, A.; Reiman, R.; Carlson, E.; Beecroft, T.; Janss, A.; Siniard, A.; Richholt, R.; Balak, C.; Rozowsky, J.; et al. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci. Rep. 2017, 7, 44061. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Martin, D.I. Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and yRNA fragment expression associated with breast cancer. Biomark. Cancer 2014, 6, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.L.; Xu, W.L.; Liu, S.; Sun, W.J.; Li, J.H.; Wu, J.; Yang, J.H.; Qu, L.H. tRF2Cancer: A web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016, 44, W185–W193. [Google Scholar] [CrossRef] [PubMed]
- Balatti, V.; Rizzotto, L.; Miller, C.; Palamarchuk, A.; Fadda, P.; Pandolfo, R.; Rassenti, L.Z.; Hertlein, E.; Ruppert, A.S.; Lozanski, A.; et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2015, 112, 2169–2174. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Balatti, V.; Palamarchuk, A.; Rizzotto, L.; Veneziano, D.; Nigita, G.; Rassenti, L.Z.; Pass, H.I.; Kipps, T.J.; Liu, C.G.; et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 5071–5076. [Google Scholar] [CrossRef] [PubMed]
- Greenway, M.J.; Alexander, M.D.; Ennis, S.; Traynor, B.J.; Corr, B.; Frost, E.; Green, A.; Hardiman, O. A novel candidate region for ALS on chromosome 14q11.2. Neurology 2004, 63, 1936–1938. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, A.E.; Eggens, V.R.; Caglayan, A.O.; Reuter, M.S.; Scott, E.; Coufal, N.G.; Silhavy, J.L.; Xue, Y.; Kayserili, H.; Yasuno, K.; et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 2014, 157, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, Y.; Sun, D.; Bai, R.; Gao, X.; Yang, Y.; Sheng, J.; Xu, Z. Angiogenin prevents progranulin A9D mutation-induced neuronal-like cell apoptosis through cleaving tRNAs into tiRNAs. Mol. Neurobiol. 2018, 55, 1338–1351. [Google Scholar] [CrossRef] [PubMed]
- Huypens, P.; Sass, S.; Wu, M.; Dyckhoff, D.; Tschop, M.; Theis, F.; Marschall, S.; Hrabe de Angelis, M.; Beckers, J. Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nat. Genet. 2016, 48, 497–499. [Google Scholar] [CrossRef] [PubMed]
- De Castro Barbosa, T.; Ingerslev, L.R.; Alm, P.S.; Versteyhe, S.; Massart, J.; Rasmussen, M.; Donkin, I.; Sjogren, R.; Mudry, J.M.; Vetterli, L.; et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol. Metab. 2016, 5, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Schuster, A.; Skinner, M.K.; Yan, W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ. Epigenet. 2016, 2, dvw001. [Google Scholar] [CrossRef] [PubMed]
- Donkin, I.; Versteyhe, S.; Ingerslev, L.R.; Qian, K.; Mechta, M.; Nordkap, L.; Mortensen, B.; Appel, E.V.; Jorgensen, N.; Kristiansen, V.B.; et al. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab. 2016, 23, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomita, K.; Ogawa, T.; Uozumi, T.; Watanabe, K.; Masaki, H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl. Acad. Sci. USA 2000, 97, 8278–8283. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Czech, B.; Crunk, A.; Wallace, A.; Mitreva, M.; Hannon, G.J.; Davis, R.E. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011, 21, 1462–1477. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Frugier, M.; Tosar, J.P.; Correa-Dominguez, A.; Ronalte-Alves, L.; Parodi-Talice, A.; Rovira, C.; Robello, C.; Goldenberg, S.; Cayota, A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2010, 171, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Calero, T.; Garcia-Silva, R.; Pena, A.; Robello, C.; Persson, H.; Rovira, C.; Naya, H.; Cayota, A. Profiling of small RNA cargo of extracellular vesicles shed by Trypanosoma cruzi reveals a specific extracellular signature. Mol. Biochem. Parasitol. 2015, 199, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012, 40, 10937–10949. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Güida, M.C.; Cayota, A. Novel aspects of tRNA-derived small RNAs with potential impact in infectious diseases. Adv. Biosci. Biotechnol. 2013, 4, 17–25. [Google Scholar] [CrossRef]
- Gebetsberger, J.; Zywicki, M.; Kunzi, A.; Polacek, N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 2012, 260909. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M. 5′ trna halves: The next generation of immune signaling molecules. Front. Immunol. 2015, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Haiser, H.J.; Karginov, F.V.; Hannon, G.J.; Elliot, M.A. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008, 36, 732–741. [Google Scholar] [CrossRef] [PubMed]


| tsRNA Type | Subtype | Published Name | Suggested Rename | Biogenesis | Refs. |
|---|---|---|---|---|---|
| tRF | 1-tRF | tRF-1001 | 1-tRFSer-TGA | pre-tRNASer-TGA | [20] |
| Cand45 | 1-tRFSer-TGA | pre-tRNASer-TGA | [28] | ||
| 2-tRF | tRFAsp, tRFGlu, tRFGly | 2-tRFAsp-GTC, 2-tRFGlu-YTC, 2-tRFGly-TCC | tRNAAsp-GTC tRNAGlu-YTC tRNAGly-TCC | [25] | |
| 3-tRF | tRF-3019 | 3a-tRFPro | tRNAPro | [57] | |
| miR-1280 | 3a-tRFLeu | tRNALeu | [66] | ||
| miR-720 | 3a-tRFThr | tRNAThr | [66] | ||
| miR-1274a/b | 3a-tRFLys5, 3a-tRFLys3 | tRNALys | [66] | ||
| CU1276 | 3b-tRFGly-GCC | tRNAGly-GCC | [37] | ||
| 5-tRF | miR-1308 | 5a-tRFGly | tRNAGly | [66] | |
| miR-886-5P | 5a-tRFAla | tRNAAla | [66] | ||
| Val-tRF | 5b-tRFVal-GAC | tRNAVal-GAC | [89] | ||
| td-piR(Glu) | 5c-tRFGlu | tRNAGlu | [64] | ||
| tiRNA | 5′-tiRNA 3′-tiRNA | tRF5-GluCTC | 5′tiRNAGlu-CTC | tRNAGlu-CTC | [58,59] |
| tsRNA | e.g., 5′-tiRNAGlu-CTC, 3′-tiRNAGlu-CTC | tRNA | [55] | ||
| SHOT-RNA | e.g., 5′-tiRNAGlu-CTC, 3′-tiRNAGlu-CTC | tRNA | [16] | ||
| tRNA halve | e.g., 5′-tiRNAGlu-CTC, 3′-tiRNAGlu-CTC | tRNA | [12,90,91] | ||
| tiRNA | e.g., 5′-tiRNAGlu-CTC, 3′-tiRNAGlu-CTC | tRNA | [42,43] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Xu, Z.; Sheng, J. tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes 2018, 9, 246. https://doi.org/10.3390/genes9050246
Li S, Xu Z, Sheng J. tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes. 2018; 9(5):246. https://doi.org/10.3390/genes9050246
Chicago/Turabian StyleLi, Siqi, Zhengping Xu, and Jinghao Sheng. 2018. "tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA" Genes 9, no. 5: 246. https://doi.org/10.3390/genes9050246





