Cathepsin B Is Upregulated and Mediates ECM Degradation in Colon Adenocarcinoma HT29 Cells Overexpressing Snail
Abstract
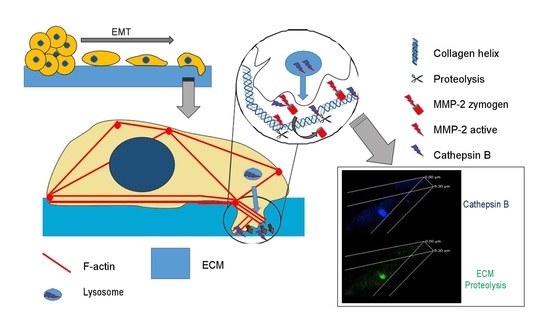
:1. Introduction
2. Methods
2.1. Cell Culture and Reagents
2.2. Western Immunoblotting
2.3. Fluorescent Dequenching (DQ) Gelatine Assay
2.4. Proteome Analysis of Cancer Tissue Samples
2.5. Transwell Migration Assay
2.6. Viability Assay
2.7. Microscopy
2.8. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hüsemann, Y.; Klein, C.A. The analysis of metastasis in transgenic mouse models. Transgenic Res. 2009, 18, 1–5. [Google Scholar] [CrossRef]
- Kubeček, O.; Laco, J.; Špaček, J.; Petera, J.; Kopecký, J.; Kubečková, A.; Filip, S. The pathogenesis, diagnosis, and management of metastatic tumors to the ovary: A comprehensive review. Clin. Exp. Metastasis. 2017, 34, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Kryczka, J.; Stasiak, M.; Dziki, L.; Mik, M.; Dziki, A.; Cierniewski, C.S. Matrix metalloproteinase-2 cleavage of the β1 integrin ectodomain facilitates colon cancer cell motility. J. Biol. Chem. 2012, 287, 36556–36566. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Nosrati, R.; Salmaninejad, A.; Dehghani, S.; Shahryari, A.; Saberi, A. Organ-specific metastasis of breast cancer: Molecular and cellular mechanisms underlying lung metastasis. Cell. Oncol. 2018, 41, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A. The metastasis cascade. Science 2008, 321, 1785–1787. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Schindeler, A.; Kolind, M.; Little, D.G. Cellular Transitions and Tissue Engineering. Cell. Reprogram. 2013, 15, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Micalizzi, D.S.; Farabaugh, S.M.; Ford, H.L. Epithelial-Mesenchymal Transition in Cancer: Parallels Between Normal Development and Tumor Progression. J. Mammary Gland Biol. Neoplasia 2010, 15, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Przygodzka, P.; Papiewska-Pajak, I.; Bogusz, H.; Kryczka, J.; Sobierajska, K.; Kowalska, M.A.; Boncela, J. Neuromedin U is upregulated by Snail at early stages of EMT in HT29 colon cancer cells. Biochim. Biophys. Acta 2016, 1860, 2445–2453. [Google Scholar] [CrossRef]
- Qiao, B.; Johnson, N.W.; Gao, J. Epithelial-mesenchymal transition in oral squamous cell carcinoma triggered by transforming growth factor-beta1 is Snail family-dependent and correlates with matrix metalloproteinase-2 and -9 expressions. Int. J. Oncol. 2010, 37, 663–668. [Google Scholar]
- Kryczka, J.; Przygodzka, P.; Bogusz, H.; Boncela, J. HMEC-1 adopt the mixed amoeboid-mesenchymal migration type during EndMT. Eur. J. Cell Biol. 2017, 96, 289–300. [Google Scholar] [CrossRef]
- Bear, J.E.; Haugh, J.M. Directed migration of mesenchymal cells: Where signaling and the cytoskeleton meet. Curr. Opin. Cell Biol. 2014, 30, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, J.; Henriet, E.; Ezzoukhry, Z.; Goetz, J.G.; Moreau, V.; Saltel, F. The microenvironment controls invadosome plasticity. J. Cell Sci. 2016, 129, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Rottiers, P.; Saltel, F.; Daubon, T.; Chaigne-Delalande, B.; Tridon, V.; Billottet, C.; Reuzeau, E.; Génot, E. TGFbeta-induced endothelial podosomes mediate basement membrane collagen degradation in arterial vessels. J. Cell Sci. 2009, 122, 4311–4318. [Google Scholar] [CrossRef]
- Azzouzi, K.E.; Wiesner, C.; Linder, S. Metalloproteinase MT1-MMP islets act as memory devices for podosome reemergence. J. Cell Biol. 2016, 213, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.G.; Tan, K.W.; Osa-Andrews, B.; Iram, S.H. High-content screening of clinically tested anticancer drugs identifies novel inhibitors of human MRP1 (ABCC1). Pharmacol. Res. 2017, 119, 313–326. [Google Scholar] [CrossRef]
- Linder, S.; Aepfelbacher, M. Podosomes: Adhesion hot-spots of invasive cells. Trends Cell Biol. 2003, 13, 376–385. [Google Scholar] [CrossRef]
- Starnes, T.W.; Cortesio, C.L.; Huttenlocher, A. Imaging podosome dynamics and matrix degradation. Methods Mol. Biol. 2011, 769, 111–136. [Google Scholar] [CrossRef]
- Alblazi, K.M.O.; Siar, C.H. Cellular protrusions--lamellipodia, filopodia, invadopodia and podosomes--and their roles in progression of orofacial tumours: Current understanding. Asian Pac. J. Cancer Prev. 2015, 16, 2187–2191. [Google Scholar] [CrossRef] [PubMed]
- Overall, C.M.; Kleifeld, O. Tumour microenvironment-opinion: Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 2006, 6, 227–239. [Google Scholar] [CrossRef]
- Wilkinson, R.D.A.; Williams, R.; Scott, C.J.; Burden, R.E. Cathepsin S: Therapeutic, diagnostic, and prognostic potential. Biol. Chem. 2015, 396, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Kryczka, J.; Boncela, J. Proteases Revisited: Roles and Therapeutic Implications in Fibrosis. Mediat. Inflamm. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Sloane, B.F.; Dunn, J.R.; Honn, K.V. Lysosomal cathepsin B: Correlation with metastatic potential. Science 1981, 212, 1151–1153. [Google Scholar] [CrossRef] [PubMed]
- Gocheva, V.; Zeng, W.; Ke, D.; Klimstra, D.; Reinheckel, T.; Peters, C.; Hanahan, D.; Joyce, J.A. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006, 20, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Bakst, R.L.; Xiong, H.; Chen, C.-H.; Deborde, S.; Lyubchik, A.; Zhou, Y.; He, S.; McNamara, W.; Lee, S.-Y.; Olson, O.C.; et al. Inflammatory Monocytes Promote Perineural Invasion via CCL2-Mediated Recruitment and Cathepsin B Expression. Cancer Res. 2017, 77, 6400–6414. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, R.; Astorgues-Xerri, L.; Bekradda, M.; Gormley, J.; Buick, R.; Kerr, P.; Cvitkovic, E.; Raymond, E.; D’Incalci, M.; Frapolli, R.; et al. Fsn0503h antibody-mediated blockade of cathepsin S as a potential therapeutic strategy for the treatment of solid tumors. Biochimie 2015, 108, 101–107. [Google Scholar] [CrossRef]
- Caculitan, N.G.; Dela Cruz Chuh, J.; Ma, Y.; Zhang, D.; Kozak, K.R.; Liu, Y.; Pillow, T.H.; Sadowsky, J.; Cheung, T.K.; Phung, Q.; et al. Cathepsin B Is Dispensable for Cellular Processing of Cathepsin B-Cleavable Antibody-Drug Conjugates. Cancer Res. 2017, 77, 7027–7037. [Google Scholar] [CrossRef] [PubMed]
- Jevnikar, Z.; Mirković, B.; Fonović, U.P.; Zidar, N.; Švajger, U.; Kos, J. Three-dimensional invasion of macrophages is mediated by cysteine cathepsins in protrusive podosomes. Eur. J. Immunol. 2012, 42, 3429–3441. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Ortega-Cava, C.F.; Chen, G.; Fernandes, N.D.; Cavallo-Medved, D.; Sloane, B.F.; Band, V.; Band, H. Lysosomal cathepsin B participates in the podosome-mediated extracellular matrix degradation and invasion via secreted lysosomes in v-Src fibroblasts. Cancer Res. 2008, 68, 9147–9156. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.-Q.; Xu, M.; Yuan, Y.; Li, F.-F.; Yang, Z.; Liu, Y.; Zhou, M.-Q.; Bian, Z.-Y.; Deng, W.; Gao, L.; et al. Cathepsin B deficiency attenuates cardiac remodeling in response to pressure overload via TNF-α/ASK1/JNK pathway. Am. J. Physiol. Heart Circ. Physiol. 2015, 308, H1143–H1154. [Google Scholar] [CrossRef] [PubMed]
- Brisson, L.; Reshkin, S.J.; Goré, J.; Roger, S. pH regulators in invadosomal functioning: Proton delivery for matrix tasting. Eur. J. Cell Biol. 2012, 91, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Brahimi-Horn, M.C.; Pouysségur, J. Hypoxia in cancer cell metabolism and pH regulation. Essays Biochem. 2007, 43, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Attanasio, F.; Caldieri, G.; Giacchetti, G.; van Horssen, R.; Wieringa, B.; Buccione, R. Novel invadopodia components revealed by differential proteomic analysis. Eur. J. Cell Biol. 2011, 90, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, M.; Boncela, J.; Perreau, C.; Karamanou, K.; Chatron-Colliet, A.; Proult, I.; Przygodzka, P.; Chakravarti, S.; Maquart, F.-X.; Kowalska, M.A.; et al. Lumican Inhibits SNAIL-Induced Melanoma Cell Migration Specifically by Blocking MMP-14 Activity. PLoS ONE 2016, 11, e0150226. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.K.; Park, S.J.; Shin, J.H.; Kim, Y.; Hwang, J.J.; Cho, D.-H.; Kim, J.C. ARP101, a selective MMP-2 inhibitor, induces autophagy-associated cell death in cancer cells. Biochem. Biophys. Res. Commun. 2011, 404, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. Ponten, A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Blal, H.A.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A Human Protein Atlas for Normal and Cancer Tissues Based on Antibody Proteomics. Mol. Cell Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Foucquier, J.; Guedj, M. Analysis of drug combinations: Current methodological landscape. Pharmacol. Res. Perspect. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Cosen-Binker, L.I.; Kapus, A. Cortactin: The gray eminence of the cytoskeleton. Physiology 2006, 21, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Kampik, D.; Grehn, F.; Schlunck, G. TGF-β2-Induced Invadosomes in Human Trabecular Meshwork Cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; Zhu, X.; Kaverina, I.; Weaver, A.M. Cortactin controls cell motility and lamellipodial dynamics by regulating ECM secretion. Curr. Biol. 2011, 21, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Bernadzki, K.M.; Rojek, K.O.; Prószyński, T.J. Podosomes in muscle cells and their role in the remodeling of neuromuscular postsynaptic machinery. Eur. J. Cell Biol. 2014, 93, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Giubellino, A.; Burke, T.R.; Bottaro, D.P. Grb2 signaling in cell motility and cancer. Expert Opin. Ther. Targets 2008, 12, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.C.; Popp, J.L.; Tang, L.K.; Gibbs, H.C.; Schmitt, E.; Chaki, S.P.; Bywaters, B.C.; Yeh, A.T.; Porter, W.W.; Burghardt, R.C.; et al. Nck deficiency is associated with delayed breast carcinoma progression and reduced metastasis. Mol. Biol. Cell 2017, 28, 3500–3516. [Google Scholar] [CrossRef] [PubMed]
- Sariahmetoglu, M.; Crawford, B.D.; Leon, H.; Sawicka, J.; Li, L.; Ballermann, B.J.; Holmes, C.; Berthiaume, L.G.; Holt, A.; Sawicki, G.; et al. Regulation of matrix metalloproteinase-2 (MMP-2) activity by phosphorylation. FASEB J. 2007, 21, 2486–2495. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Kinoshita, T.; Takino, T.; Nakayama, K.; Seiki, M. Activation of a recombinant membrane type 1-matrix metalloproteinase (MT1-MMP) by furin and its interaction with tissue inhibitor of metalloproteinases (TIMP)-2. FEBS Lett. 1996, 393, 101–104. [Google Scholar] [CrossRef]
- Stawowy, P.; Meyborg, H.; Stibenz, D.; Borges, N. Pereira Stawowy, M. Roser, U. Thanabalasingam, J.P. Veinot, M. Chrétien, N.G. Seidah, E. Fleck, K. Graf, Furin-like proprotein convertases are central regulators of the membrane type matrix metalloproteinase-pro-matrix metalloproteinase-2 proteolytic cascade in atherosclerosis. Circulation 2005, 111, 2820–2827. [Google Scholar] [CrossRef] [PubMed]
- Cepeda, C.L.; Evered, J.J.H.; Pelling, S. Damjanovski, Inhibition of MT1-MMP proteolytic function and ERK1/2 signalling influences cell migration and invasion through changes in MMP-2 and MMP-9 levels. J. Cell Commun. Signal. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tuccinardi, T.; Martinelli, A.; Nuti, E.; Carelli, P.; Balzano, F.; Uccello-Barretta, G.; Murphy, G.; Rossello, A. Amber force field implementation, molecular modelling study, synthesis and MMP-1/MMP-2 inhibition profile of (R)- and (S)-N-hydroxy-2-(N-isopropoxybiphenyl-4-ylsulfonamido)-3-methylbutanamides. Bioorg. Med. Chem. 2006, 14, 4260–4276. [Google Scholar] [CrossRef] [PubMed]
- Victor, B.C.; Anbalagan, A.; Mohamed, M.M.; Sloane, B.F.; Cavallo-Medved, D. Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion. Breast Cancer Res. 2011, 13, R115. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.-J.; Peng, Z.-K.; Lu, J.-P.; Tang, F.-Q. Cathepsins mediate tumor metastasis. World J. Biol. Chem. 2013, 4, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fu, X.-H.; Yu, Y.; Shui, R.-H.; Li, C.; Zeng, H.-Y.; Qiao, Y.-L.; Ni, L.-Y.; Wang, Q. Treatment with CA-074Me, a Cathepsin B inhibitor, reduces lung interstitial inflammation and fibrosis in a rat model of polymyositis. Lab. Investig. 2015, 95, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Bliss, C.I. The Toxicity of Poisons Applied Jointly1. Ann. Appl. Biol. 1939, 26, 585–615. [Google Scholar] [CrossRef]
- Berenbaum, M.C. The expected effect of a combination of agents: The general solution. J. Theor. Biol. 1985, 114, 413–431. [Google Scholar] [CrossRef]
- McCarthy, J.B.; El-Ashry, D.; Turley, E.A. Hyaluronan, Cancer-Associated Fibroblasts and the Tumor Microenvironment in Malignant Progression. Front. Cell Dev. Biol. 2018, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Sisto, M.; Lisi, S.; Ribatti, D. The role of the epithelial-to-mesenchymal transition (EMT) in diseases of the salivary glands. Histochem. Cell Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69. [Google Scholar] [CrossRef] [PubMed]
- Simeone, P.; Trerotola, M.; Franck, J.; Cardon, T.; Marchisio, M.; Fournier, I.; Salzet, M.; Maffia, M.; Vergara, D. The multiverse nature of epithelial to mesenchymal transition. Semin. Cancer Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Liao, T.-T.; Yang, M.-H. Revisiting epithelial-mesenchymal transition in cancer metastasis: The connection between epithelial plasticity and stemness. Mol. Oncol. 2017, 11, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Kupai, K.; Szucs, G.; Cseh, S.; Hajdu, I.; Csonka, C.; Csont, T.; Ferdinandy, P. Matrix metalloproteinase activity assays: Importance of zymography. J. Pharmacol. Toxicol. Methods 2010, 61, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Takkunen, M.; Hukkanen, M.; Liljeström, M.; Grenman, R.; Virtanen, I. Podosome-like structures of non-invasive carcinoma cells are replaced in epithelial-mesenchymal transition by actin comet-embedded invadopodia. J. Cell. Mol. Med. 2010, 14, 1569–1593. [Google Scholar] [CrossRef] [PubMed]
- Van Hinsbergh, V.W.M.; Engelse, M.A.; Quax, P.H.A. Pericellular proteases in angiogenesis and vasculogenesis. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Oser, M.; Dovas, A.; Cox, D.; Condeelis, J. Nck1 and Grb2 localization patterns can distinguish invadopodia from podosomes. Eur. J. Cell Biol. 2011, 90, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Aga, M.; Bradley, J.M.; Keller, K.E.; Kelley, M.J.; Acott, T.S. Specialized podosome- or invadopodia-like structures (PILS) for focal trabecular meshwork extracellular matrix turnover. Invest. Ophthalmol. Vis. Sci. 2008, 49, 5353–5365. [Google Scholar] [CrossRef] [PubMed]
- Gawden-Bone, C.; Zhou, Z.; King, E.; Prescott, A.; Watts, C.; Lucocq, J. Dendritic cell podosomes are protrusive and invade the extracellular matrix using metalloproteinase MMP-14. J. Cell Sci. 2010, 123, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- McNiven, M.A. Breaking away: Matrix remodeling from the leading edge. Trends Cell Biol. 2013, 23, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Foxall, E.; Pipili, A.; Jones, G.E.; Wells, C.M. Significance of kinase activity in the dynamic invadosome. Eur. J. Cell Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Albiges-Rizo, C.; Destaing, O.; Fourcade, B.; Planus, E.; Block, M.R. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J Cell Sci. 2009, 122, 3037–3049. [Google Scholar] [CrossRef] [PubMed]
- Paterson, E.K.; Courtneidge, S.A. Invadosomes are coming: New insights into function and disease relevance. FEBS J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Strongin, A.Y.; Collier, I.; Bannikov, G.; Marmer, B.L.; Grant, G.A.; Goldberg, G.I. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 1995, 270, 5331–5338. [Google Scholar] [CrossRef] [PubMed]
- Jacob, A.; Jing, J.; Lee, J.; Schedin, P.; Gilbert, S.M.; Peden, A.A.; Junutula, J.R.; Prekeris, R. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J. Cell Sci. 2013, 126, 4647–4658. [Google Scholar] [CrossRef] [PubMed]
- Coppola, J.M.; Bhojani, M.S.; Ross, B.D.; Rehemtulla, A. A Small-Molecule Furin Inhibitor Inhibits Cancer Cell Motility and Invasiveness. Neoplasia 2008, 10, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Rosenberg, G.A. Matrix metalloproteinases as therapeutic targets for stroke. Brain Res. 2015, 1623, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Takino, T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci. 2010, 101, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Hannocks, M.-J.; Zhang, X.; Gerwien, H.; Chashchina, A.; Burmeister, M.; Korpos, E.; Song, J.; Sorokin, L. The gelatinases, MMP-2 and MMP-9, as fine tuners of neuroinflammatory processes. Matrix Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, T.; Yap, A.S. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 2015, 17, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Dinicola, S.; Pasqualato, A.; Proietti, S.; Masiello, M.G.; Palombo, A.; Coluccia, P.; Canipari, R.; Catizone, A.; Ricci, G.; Harrath, A.H.; et al. Bizzarri, Paradoxical E-cadherin increase in 5FU-resistant colon cancer is unaffected during mesenchymal-epithelial reversion induced by γ-secretase inhibition. Life Sci. 2016, 145, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Gondi, C.S.; Rao, J.S. Cathepsin B as a cancer target. Expert Opin. Ther. Targets 2013, 17, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Ward, R.; Gavrilovic, J.; Atkinson, S. Physiological mechanisms for metalloproteinase activation. Matrix Suppl. 1992, 1, 224–230. [Google Scholar] [PubMed]
- Hofschröer, V.; Koch, K.A.; Ludwig, F.T.; Friedl, P.; Oberleithner, H.; Stock, C.; Schwab, A. Extracellular protonation modulates cell-cell interaction mechanics and tissue invasion in human melanoma cells. Sci. Rep. 2017, 7, 42369. [Google Scholar] [CrossRef] [PubMed]
- Stüwe, L.; Müller, M.; Fabian, A.; Waning, J.; Mally, S.; Noël, J.; Schwab, A.; Stock, C. pH dependence of melanoma cell migration: Protons extruded by NHE1 dominate protons of the bulk solution. J. Physiol. 2007, 585, 351–360. [Google Scholar] [CrossRef] [PubMed]
- McKerrow, J.H.; Bhargava, V.; Hansell, E.; Huling, S.; Kuwahara, T.; Matley, M.; Coussens, L.; Warren, R. A functional proteomics screen of proteases in colorectal carcinoma. Mol. Med. 2000, 6, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, H.F. Tumors: Wounds That Do Not Heal. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Donthireddy, L.; Marvel, D.; Condamine, T.; Wang, F.; Lavilla-Alonso, S.; Hashimoto, A.; Vonteddu, P.; Behera, R.; Goins, M.A.; et al. Cancer-Associated Fibroblasts Neutralize the Anti-tumor Effect of CSF1 Receptor Blockade by Inducing PMN-MDSC Infiltration of Tumors. Cancer Cell. 2017, 32, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; He, M.-Y.; Zhu, L.-F.; Yang, C.-C.; Zhou, M.-L.; Wang, Q.; Zhang, W.; Zheng, Y.-Y.; Wang, D.-M.; Xu, Z.-Q.; et al. Tumor-associated macrophages correlate with the clinicopathological features and poor outcomes via inducing epithelial to mesenchymal transition in oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 12. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Mazo, I.; Leung, H.; Engelke, K.; von Andrian, U.H.; Deryugina, E.I.; Strongin, A.Y.; Bröcker, E.-B.; Friedl, P. Compensation mechanism in tumor cell migration. J. Cell Biol. 2003, 160, 267–277. [Google Scholar] [CrossRef] [PubMed]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kryczka, J.; Papiewska-Pajak, I.; Kowalska, M.A.; Boncela, J. Cathepsin B Is Upregulated and Mediates ECM Degradation in Colon Adenocarcinoma HT29 Cells Overexpressing Snail. Cells 2019, 8, 203. https://doi.org/10.3390/cells8030203
Kryczka J, Papiewska-Pajak I, Kowalska MA, Boncela J. Cathepsin B Is Upregulated and Mediates ECM Degradation in Colon Adenocarcinoma HT29 Cells Overexpressing Snail. Cells. 2019; 8(3):203. https://doi.org/10.3390/cells8030203
Chicago/Turabian StyleKryczka, Jakub, Izabela Papiewska-Pajak, M. Anna Kowalska, and Joanna Boncela. 2019. "Cathepsin B Is Upregulated and Mediates ECM Degradation in Colon Adenocarcinoma HT29 Cells Overexpressing Snail" Cells 8, no. 3: 203. https://doi.org/10.3390/cells8030203