The Role of Mitochondria in Reactive Oxygen Species Generation and Its Implications for Neurodegenerative Diseases
Abstract
:1. Introduction
2. The Structure and Functions of Mitochondria
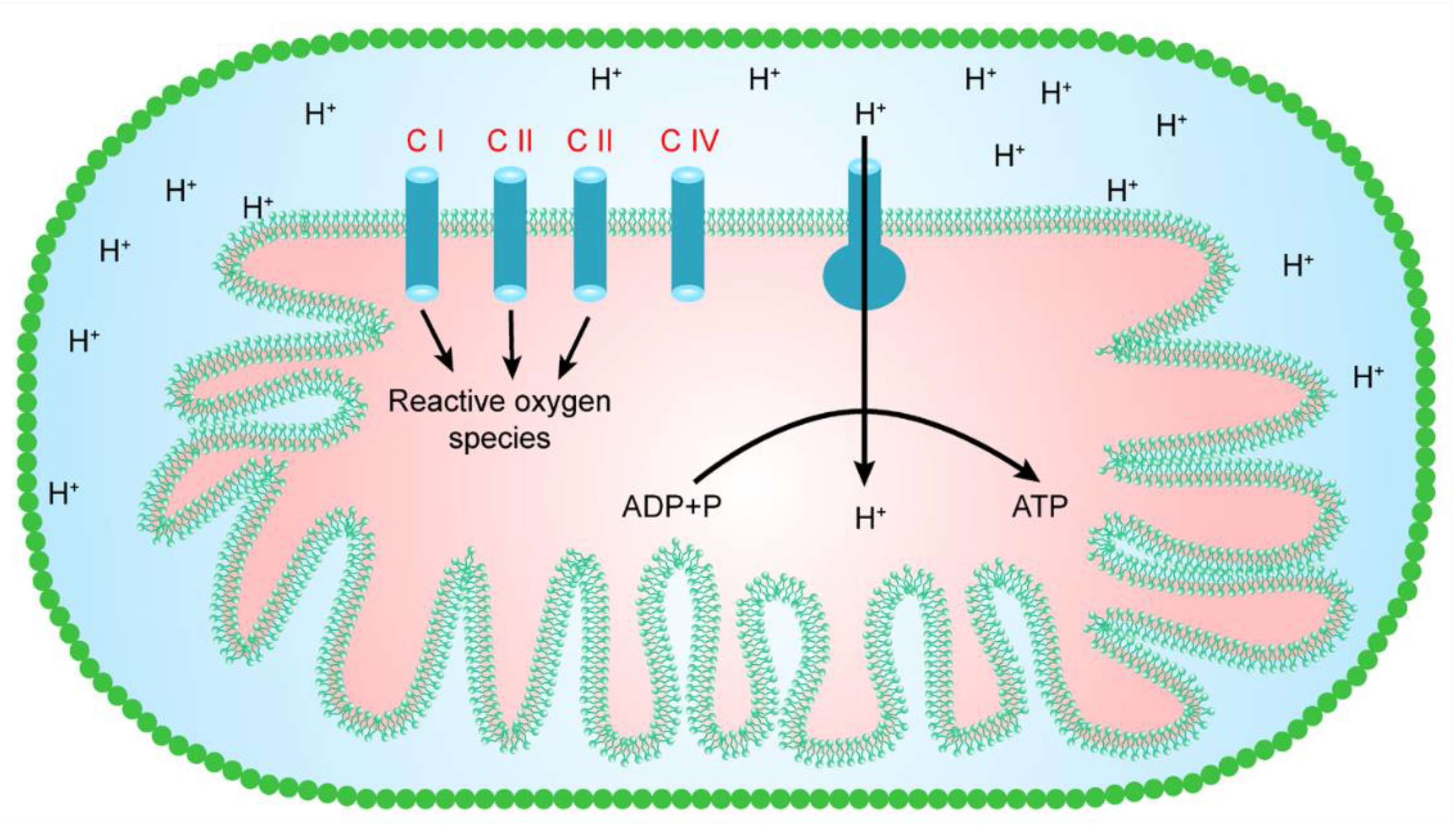
3. Mitochondrial Sites for the Generation of Free Radicals
4. Complex I and the Generation of Free Radicals
5. Complex II and the Generation of Free Radicals
6. Complex III and the Generation of Free Radicals
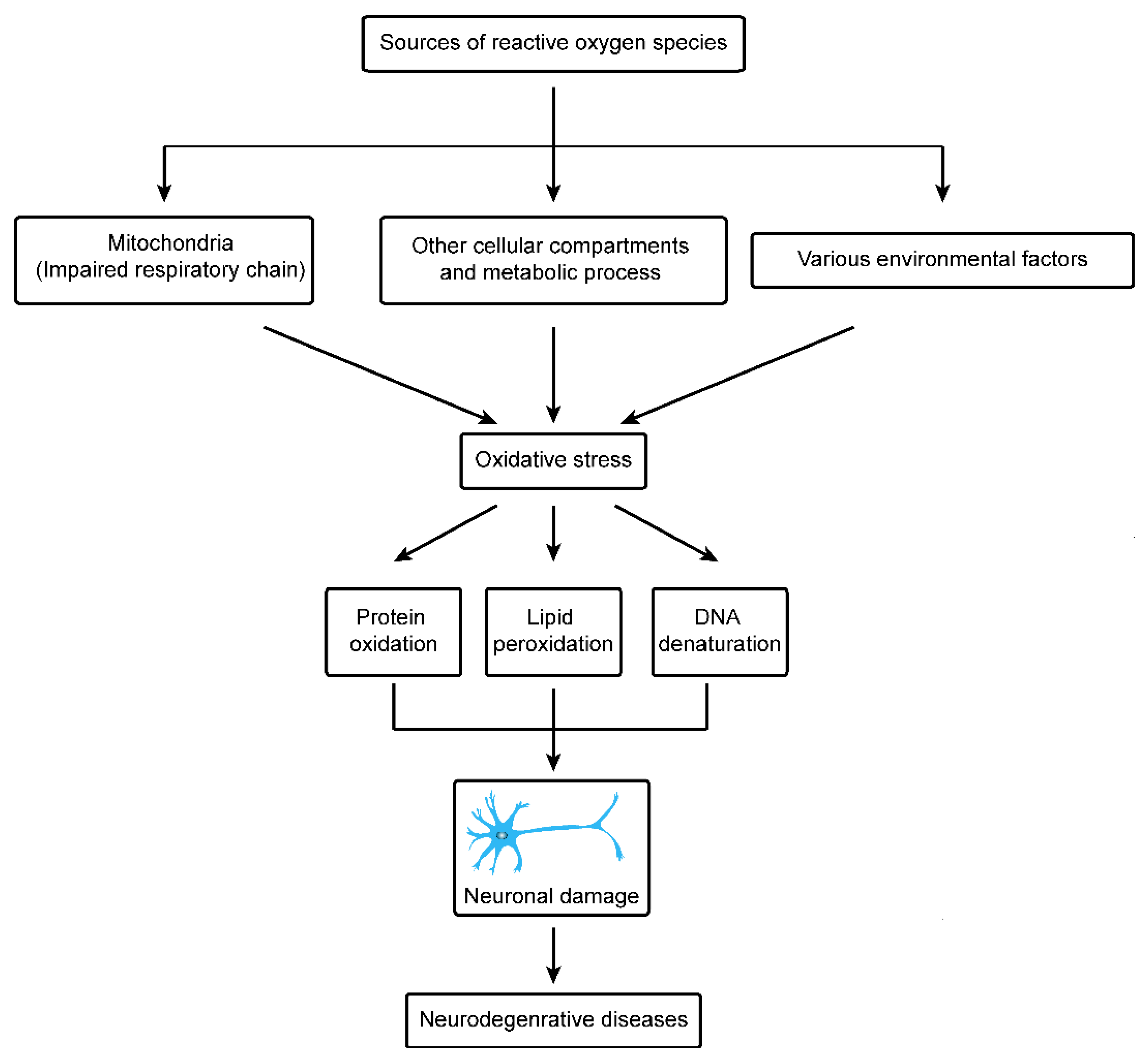
7. Other Cellular Components and Free Radicals
8. Environmental Sources of Oxidative Stress and Neurodegenerative Disorder
9. Physiological Functions of Mitochondria in Neurodegenerative Disease
10. Mitochondria and Huntington’s Disease
11. Mitochondria and Parkinson’s Disease
12. Oxidative Stress and Parkinson’s Disease
13. Oxidative Stress and Huntington’s Disease
14. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chan, D.C. Mitochondria: Dynamic organelles in disease, aging and development. Cell 2000, 125, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.X.; Wang, L.; Wei, G.Q.; Qian, C.; Dai, L.S.; Sun, Y.; Abbas, M.N.; Zhu, B.J.; Liu, C.L. Characterization of the complete mitochondrial genome of Leucoma salicis (Lepidoptera: Lymantriidae) and comparison with other lepidopteran insects. Sci. Rep. 2016, 6, 39153. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Taivassalo, T.; Gouspillou, G.; Hepple, R.T. Mitochondria: Isolation, structure and function. J. Physiol. 2011, 589, 4413–4421. [Google Scholar] [CrossRef] [PubMed]
- Duchen, M.R. Mitochondria in health and disease: Perspectives on a new mitochondrial biology. Mol. Aspects Med. 2004, 25, 365–451. [Google Scholar] [CrossRef] [PubMed]
- Pryde, K.R.; Hirst, J. Superoxide is produced by the reduced flavin in mitochondrial complex I: A single, unified mechanism that applies during both forward and reverse electron transfer. J. Biol. Chem. 2011, 286, 18056–18065. [Google Scholar] [CrossRef] [PubMed]
- Starkov, A.A.; Fiskum, G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J. Neurochem. 2003, 86, 1101–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarewicz, M.; Borek, A.; Cieluch, E.; Swierczek, M.; Osyczka, A. Discrimination between two possible reaction sequences that create potential risk of generation of deleterious radicals by cytochrome bc 1. Implications for the mechanism of superoxide production. Biochim. Biophys. Acta 2010, 1797, 1820–1827. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, E.; Davies, K.J.A. Mitochondrial free radical generation, oxidative stress and aging. Free Rad. Biol. Med. 2000, 29, 222–230. [Google Scholar] [CrossRef]
- Migliorea, L.; Coppedè, F. Environmental-induced oxidative stress in neurodegenerative disorders and aging. Toxicol. Environ. Mutagen. 2009, 674, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Trushina, E.; McMurray, C.T. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience 2007, 145, 1233–1248. [Google Scholar] [CrossRef] [PubMed]
- Beal, M.F. Mitochondrial dysfunction in neurodegenerative diseases. Biochim. Biophys. Acta 1998, 1366, 211–223. [Google Scholar] [CrossRef]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Bayrhuber, M.; Meins, T.; Habeck, M.; Becker, S.; Giller, K.; Villinger, S.; Vonrhein, C.; Griesinger, C.; Zweckstetter, M.; Zeth, K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA 2008, 105, 15370–15375. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.S.; Zhou, X.D.; Kausar, S.; Abbas, M.N.; Wu, L.; Zhou, H.L. Mitochondrial genome of Diaphania indica (saunders) (Lepidoptera: Pyraloidea) and implications for its phylogeny. Intl. J. Biol. Macromol. 2018, 108, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Kühlbrandt, W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Hoppins, S.; Collins, S.R.; Cassidy-Stone, A.; Hummel, E.; DeVay, R.M.; Lackner, L.L.; Westermann, B.; Schuldiner, M.; Weissman, J.S.; Nunnari, J. A mitochondrialfocused genetic interaction map reveals a scaffoldlike complex required for inner membrane organization in mitochondria. J. Cell Biol. 2011, 195, 323–340. [Google Scholar] [CrossRef] [PubMed]
- Von der Malsburg, K.; Müller, J.M.; Bohnert, M.; Oeljeklaus, S.; Kwiatkowska, P.; Becker, T.; Loniewska-Lwowska, A.; Wiese, S.; Rao, S.; Milenkovic, D.; et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 2011, 21, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.A.; Ellisman, M.H.; Fox, D.A. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: Bioenergetic and functional implications. Mol. Vision 2003, 9, 60–73. [Google Scholar]
- Cogliati, S.; Enriquez, J.A.; Scorrano, L. Mitochondrial cristae: Where beauty meets functionality. Trends Biochem. Sci. 2016, 41, 3. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.F.; Lin, S.S.; Chen, J.M.; Tsai, H.Z.; Hsieh, T.S.; Fu, C.Y. Electron tomographic analysis reveals ultrastructural features of mitochondrial cristae architecture which reflect energetic state and aging. Sci. Rep. 2017, 7, 45474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, L.S.; Kausar, S.; Abbas, M.N.; Wang, T.T. Complete sequence and characterization of the Ectropis oblique mitochondrial genome and its phylogenetic implications. Int. J. Biol. Macromol. 2018, 107, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Sun, Y.X.; Yang, L.L.; Wu, L.; Abbas, M.N.; Chen, C.; Gao, J.; Li, X.K.; Liu, C.L.; Dai, L.S. Characterization of the complete mitochondrial genome of Biston marginata (Lepidoptera: Geometridae) and phylogenetic analysis among lepidopteran insects. Int. J. Biol. Macromol. 2018, 113, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Lenaz, G. The mitochondrial production of reactive oxygen species: Mechanisms and implications in human pathology. IUBMB Life 2001, 52, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010, 45, 466–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolisetty, S.; Jaimes, E.A. Mitochondria and reactive oxygen species: Physiol. Pathophysiol. Int. J. Mol. Sci. 2013, 14, 6306–6344. [Google Scholar] [CrossRef] [PubMed]
- Warnaua, J.; Sharmac, V.; Gamiz-Hernandez, A.P.; Luca, A.D.; Haapanen, O.; Vattulainen, I.; Wikström, M.; Hummer, G.; Kaila, V.R.I. Redox-coupled quinone dynamics in the respiratory complex I. Proc. Natl. Acad. Sci. USA 2018, 115, E8413–E8420. [Google Scholar] [CrossRef] [PubMed]
- Dominiak, K.; Koziel, A.; Jarmuszkiewicz, W. The interplay between mitochondrial reactive oxygen species formation and the coenzyme Q reduction level. Redox Biol. 2018, 18, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Lenaz, G. Role of mitochondria in oxidative stress and aging. Biochim. Biophys. Acta 1998, 1366, 53–67. [Google Scholar] [CrossRef]
- Galkin, A.; Brandt, U. Superoxide radical formation by pure Complex I (NADH:Ubiquinone Oxidoreductase) from Yarrowia lipolytica. J. Biol. Chem. 2005, 280, 30129–30135. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.J.; Doroshow, J.H. Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J. Biol. Chem. 1986, 261, 3060–3067. [Google Scholar] [PubMed]
- Degli, E.M.; Ngo, A.; Ghelli, A.; Benelli, B.; Carelli, V.; McLennan, H.; Lirnnane, A.W. The interaction of Q analogs, particularly hydroxydecyl benzoquinone (idebenone), with the respiratory complexes of heart mitochondria. Arch. Biochem. Biophys. 1996, 330, 395–400. [Google Scholar]
- Takeshige, K.; Minakami, S. NADH and NADPH-dependent formation of superoxide anions by bovine heart submitochondrial particles and NADH-ubiquinone reductase preparation. Biochem. J. 1979, 180, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Barja, G. Mitochondrial oxygen radical generation and leak: Sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J. Bioenerg. Biomembr. 1999, 31, 347–366. [Google Scholar] [CrossRef] [PubMed]
- 35 Robb, E.L.; Hall, A.R.; Prime, T.A.; Eaton, S.; Szibor, M.; Viscomi, C.; James, A.M.; Murphy, M.P. Control of mitochondrial superoxide production by reverse electron transport at Complex I. The J. Biol. Chem. 2018, 293, 9869–9879. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Barja, G. Localization of the site of oxygen radical generation inside the Complex I of heart and nonsynaptic brain mammalian mitochondria. J. Bioenerg. Biomembr. 2000, 32, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Treberg, J.R.; Quinlan, C.L.; Brand, M.D. Evidence for two sites of superoxide production by mitochondrial NADH-ubiquinone oxidoreductase (complex I). J. Biol. Chem. 2011, 286, 27103–27110. [Google Scholar] [CrossRef] [PubMed]
- McLennan, H.R.; Degli, E.M. The contribution of mitochondrial respiratory complexes to the production of reactive oxygen species. J. Bioenerg. Biomembr. 2000, 32, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kussmaul, L.; Hirst, J. The mechanism of superoxide production by NADH: Ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. PNAS 2006, 103, 7607–7612. [Google Scholar] [CrossRef] [PubMed]
- Cortopassi, G.; Wang, E. Modelling the effects of age-related mtDNA mutation accumulation: Complex I defciency, superoxide and cell death. Biochim. Biophys. Acta 1995, 1271, 171–176. [Google Scholar] [CrossRef]
- Votyakova, T.V.; Reynolds, I.J. ΔΨ-dependent and independent production of reactive oxygen species by rat brain mitochondria. J. Neurochem. 2001, 79, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, C.L.; Gerencser, A.A.; Treberg, J.R.; Brand, M.D. The mechanism of superoxide production by the antimycin-inhibited mitochondrial Q-cycle. J. Biol. Chem. 2011, 286, 31361–31372. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, C.L.; Orr, A.L.; Perevoshchikova, I.V.; Treberg, J.R.; Ackrell, B.A.; Brand, M.D. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 2012, 287, 27255–27264. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Sanchez, R.; Hernandez-Esquivel, L.; Rivero-Segura, N.A.; Marin-Hernandez, A.; Neuzil, J.; Ralph, S.J.; Rodriguez-Enriquez, S. Reactive oxygen species are generated by the respiratory complex II – evidence for lack of contribution of the reverse electron flow in Complex I. FEBS J. 2013, 280, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Ralph, S.J.; Moreno-Sanchez, R.; Neuzil, J.; Rodriguez-Enriquez, S. Inhibitors of succinate: Quinone reductase/complex II regulate production of mitochondrial reactive oxygen species and protect normal cells from ischemic damage but induce specific cancer cell death. Pharm. Res. 2011, 28, 2695–2730. [Google Scholar] [CrossRef] [PubMed]
- Guzy, R.D.; Sharma, B.; Bell, E.; Chandel, N.S.; Schumacker, P.T. Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell Biol. 2008, 28, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Miyazawa, M.; Onodera, A.; Yasuda, K.; Kawabe, N.; Kirinashizawa, M.; Yoshimura, S.; Maruyama, N.; Hartman, P.S.; Ishii, N. Mitochondrial reactive oxygen species generation by the SDHC V69E mutation causes low birth weight and neonatal growth retardation. Mitochondrion 2011, 11, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Owens, K.M.; Aykin-Burns, N.; Dayal, D.; Coleman, M.C.; Domann, F.E.; Spitz, D.R. Genomic instability induced by mutant succinate dehydrogenase subunit D (SDHD) is mediated by O2 − degrees and H2O2. Free Rad. Biol. Med. 2012, 52, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Ishii, N.; Fujii, M.; Hartman, P.S.; Tsuda, M.; Yasuda, K.; Senoo-Matsuda, N.; Yanase, S.; Ayusawa, D.; Suzuki, K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature 1998, 394, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Senoo-Matsuda, N.; Yasuda, K.; Tsuda, M.; Ohkubo, T.; Yoshimura, S.; Nakazawa, H.; Hartman, P.S.; Ishii, N. A defect in the cytochrome b large subunit in complex II causes both superoxide anion over production and abnormal energy metabolism in Caenorhabditis elegans. J. Biol. Chem. 2001, 276, 41553–41558. [Google Scholar] [CrossRef] [PubMed]
- Borek, A.; Sarewicz, M.; Osyczka, A. Movement of the iron–sulfur head domain of cytochrome bc1 transiently opens the catalytic Qo site for reaction with oxygen. Biochemistry 2008, 47, 12365–12370. [Google Scholar] [CrossRef] [PubMed]
- Gurung, B.; Yu, L.; Yu, C.A. Stigmatellin induces reduction of iron–sulfur protein in the oxidized cytochrome bc1 complex. J. Biol. Chem. 2008, 283, 28087–28094. [Google Scholar] [CrossRef] [PubMed]
- Loschen, G.; Azzi, A.; Flohe, L. Mitochondrial H2O2 formation: Relationship with energy conservation. FEBS Lett. 1973, 33, 84–87. [Google Scholar] [CrossRef]
- Starkov, A.A.; Fiskum, G. Myxothiazol induces H2O2 production from mitochondrial respiratory chain. Biochem. Biophys. Res. Commun. 2001, 281, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, K.D.; Guzy, R.D.; Pan, Y.; Young, R.M.; Cash, T.P.; Schumacker, P.T.; Simon, M.C. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005, 1, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.L.; Klimova, T.A.; Eisenbart, J.; Moraes, C.T.; Murphy, M.P.; Budinger, G.R.; Chandel, N.S. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 2007, 177, 1029–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, F.; Crofts, A.R.; Kramer, D.M. Multiple Q-cycle bypass reactions at the Qo site of the cytochrome bc 1 complex. Biochemistry 2002, 41, 7866–7874. [Google Scholar] [CrossRef] [PubMed]
- Osyczka, A.; Moser, C.C.; Daldal, F.; Dutton, P.L. Reversible redox energy coupling in electron transfer chains. Nature 2004, 427, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Orrenius, S.; Gogvadze, V.; Zhivotovsky, B. Mitochondrial oxidative stress: Implications for cell death. Ann. Rev. Pharmacol. Toxicol. 2007, 47, 143–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Ischemic defects in the electron transport chain increase the production of reactive oxygen species from isolated rat heart mitochondria. AJP Cell. Physiol. 2008, 294, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Korge, P.; Calmettes, G.; John, S.A.; Weiss, J.N. Reactive oxygen species production induced by pore opening in cardiac mitochondria: The role of Complex III. J. Biol. Chem. 2017, 292, 9882–9895. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.L.; Roberts, A.G.; Bowman, M.K.; Kramer, D.M. Architecture of the Qo site of the cytochrome bc1 complex probed by superoxide production. Biochemistry 2003, 42, 6493–6499. [Google Scholar] [CrossRef] [PubMed]
- Dröse, S.; Brandt, U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J. Biol. Chem. 2008, 283, 21649–21654. [Google Scholar] [CrossRef] [PubMed]
- Dröse, S.; Brandt, U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv. Exp. Med. Biol. 2012, 748, 145–169. [Google Scholar] [PubMed]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Weng, M.; Xie, X.; Liu, C.; Lim, K.L.; Zhang, C.W.; Li, L. The sources of reactive oxygen species and its possible role in the pathogenesis of Parkinson’s disease. Parkinson’s Dis. 2018, 9163040, 9. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Cross, C.E. Oxygen-derived species: Their relation to human disease and environmental stress. Environ. Health Perspect. 1994, 102, 5–12. [Google Scholar] [PubMed]
- Jodeiri, F.M.; Ghaedi, K. Huntington’s disease and mitochondria. Neurotox. Res. 2017, 32, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Bates, G.P.; Dorsey, R.; Gusella, J.F.; Hayden, M.R.; Kay, C.; Leavitt, B.R.; Nance, M.; Ross, C.A.; Scahill, R.I.; Wetzel, R.; et al. Huntington disease. Nat. Rev. Dis. Prim. 2015, 1, 15005. [Google Scholar] [CrossRef] [PubMed]
- Franco-Iborra, S.; Vila, M.; Perier, C. Mitochondrial quality control in neurodegenerative diseases: Focus on Parkinson’s disease and Huntington’s disease. Front. Neurosci. 2018, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Szlachcic, W.J.; Switonski, P.M.; Krzyzosiak, W.J.; Figlerowicz, M.; Figiel, M. Huntington disease iPSCs show early molecular changes in intracellular signaling, the expression of oxidative stress proteins and the p53 pathway. Dis. Model. Mech. 2015, 8, 1047–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manoharan, S.; Guillemin, G.J.; Abiramasundari, R.S.; Essa, M.M.; Akbar, M.; Akbar, M.D. The role of reactive oxygen species in the pathogenesis of Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease: A Mini Review. Oxid. Med. Cell. Longev. 2016, 8590578, 15. [Google Scholar] [CrossRef] [PubMed]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: A clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Panov, A.V.; Gutekunst, C.A.; Leavitt, B.R.; Hayden, M.R.; Burke, J.R.; Strittmatter, W.J.; Greenamyre, J.T. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 2002, 5, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Gleichmann, M.; Cheng, A. Mitochondria in neuroplasticity and neurological disorder. Cell 2008, 60, 748–766. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Jeong, H.; Borovecki, F.; Parkhurst, C.N.; Tanese, N.; Krainc, D. Transcriptional repression of PGC-1 α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 2006, 127, 59–69. [Google Scholar] [CrossRef] [PubMed]
- De Maagd, G.; Philip, A. Parkinson’s disease and its management: Part 1: Disease entity, risk factors, pathophysiology, clinical presentation, and diagnosis. Pharm. Ther. 2015, 40, 504–532. [Google Scholar]
- Massano, J.; Bhatia, K.P. Clinical approach to Parkinson’s disease: Features, diagnosis, and principles of management. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Wang, G. Mitochondrial dysfunction in Parkinson’s disease. Transl. Neurodegener. 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Langston, J.W.; Ballard, P.; Tetrud, J.W.; Irwin, I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 1983, 219, 979–980. [Google Scholar] [CrossRef] [PubMed]
- Carreras, M.; Franco, M.C.; Peralta, J.G.; Poderoso, J.J. Nitric oxide, complex I, and the modulation of mitochondrial reactive species in biology and disease. Mol. Aspects Med. 2004, 25, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Haas, R.H.; Nasirian, F.; Nakano, K.; Ward, D.; Pay, M.; Hill, R.; Clifford, W.; Shults, M.D. Low platelet mitochondrial complex I and complex II/III activity in early untreated Parkinson’s disease. Ann. Neurol. 1995, 37, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.D.; Parks, J.K.; Swerdlow, R.H.; Swerdlow, R.H. Complex I deficiency in Parkinson’s disease frontal cortex. Brain Res. 2008, 1189, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, R.H.; Parks, J.K.; Miller, S.W.; Davis, R.E.; Tuttle, J.B.; Trimmer, P.A.; Jason, P.; Sheehan, B.S. Origin and functional consequences of the Complex I defect in Parkinson’s disease. Ann. Neurol. 1996, 40, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Betarbet, R.; Sherer, T.B.; MacKenzie, G.; Garcia-Osuna, M.; Panov, A.V.; Greenamyre, J.T. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat. Neurosci. 2000, 3, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Guardia-Laguarta, C.; Area-Gomez, E.; Rüb, C.; Liu, Y.; Magrané, J.; Becker, D.; Voos, W.; Schon, E.A.; Przedborski, S. α-Synuclein is localized to mitochondria-associated ER membranes. J. Neurosci. 2014, 34, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Area-Gomez, E.; Castillo, M.D.C.L.; Tambini, M.D.; Guardia-Laguarta, C.; de Groof, A.J.C.; Madra, M.; Ikenouchi, J.; Umeda, M.; Bird, T.D.; Sturley, S.L.; et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012, 31, 4106–4123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Saif, A.; Al-Mohanna, F.; Bohlega, S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann. Neurol. 2011, 70, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Sherer, T.; Betarbet, R.; Stout, A.K.; Lund, S.; Baptista, M.; Panov, A.V.; Cookson, M.R.; Greenamyre, J.T. An in vitro model of Parkinson’s disease: Linking mitochondrial impairment to altered alpha-synuclein metabolism and oxidative damage. J. Neurosci. 2002, 22, 7006–70015. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.S.; Hawrylycz, M.; Miller, J.A.; Snyder, A.Z.; Raichle, M.E. Aerobic glycolysis in the human brain is associated with development and neotenous gene expression. Cell Metab. 2014, 19, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Magistretti, P.J.; Allaman, I. A cellular perspective on brain energy metabolism and functional imaging. Neuron 2015, 86, 883–901. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.L.; Brannock, C. Invited review free radicals and the pathobiology of brain dopamine systems. Neurochem. Int. 1998, 32, 117–131. [Google Scholar] [CrossRef]
- Gao, L.; Laude, K.; Cai, H. Mitochondrial pathophysiology, reactive oxygen species, and cardiovascular diseases. Vet. Clin. North Am. Small Anim. Pr. 2008, 38, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Zecca, L.; Shima, T.; Stroppolo, A.; Goj, C.; Battiston, G.A.; Gerbasi, R.; Sarna, T.; Swartz, H.M. Interaction of neuromelanin and iron in substantia nigra and other areas of human brain. Neuroscience 1996, 73, 407–415. [Google Scholar] [CrossRef]
- Perier, C.; Vila, M. Mitochondrial biology and Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 4, a009332. [Google Scholar] [CrossRef] [PubMed]
- Nagatsu, T.; Sawada, M. Molecular mechanism of the relation of monoamine oxidase inhibitors to Parkinson’s disease: Possible implications of glial cells. J. Neural. Transm. 2006, 71, 53–65. [Google Scholar]
- Meiser, J.; Weind, D.; Hiller, K. Complexity of dopamine metabolism. Cell Commun. Signal. 2013, 11, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochizuki, H.; Yasuda, T. Iron accumulation in Parkinson’s disease. J. Neural Transm. 2012, 119, 1511–1514. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, Y.; Carrasco, C.M.; Campos, J.D.; Aguirre, P.; Núñez, M.T. Parkinson’s disease: The mitochondria-iron link. Parkinson’s Dis. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Valko, M. Advances in metal-induced oxidative stress and human disease. Toxicology 2011, 283, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Nunez, M.T.; Urrutia, P.; Mena, N.; Aguirre, P.; Tapia, V.; Salazar, J. Iron toxicity in neurodegeneration. Biometals 2012, 25, 761–776. [Google Scholar] [CrossRef] [PubMed]
- Atasoy, H.T.; Nuyan, O.; Tunc, T.; Yorubulut, M.; Unal, A.E.; Inan, L.E. T2-weighted MRI in Parkinson’s disease; substantia nigra pars compacta hypointensity correlates with the clinical scores. Neurol. India 2004, 52, 332–337. [Google Scholar] [PubMed]
- Shichir, M. The role of lipid peroxidation in neurological disorders. J. Clin. Biochem. Nutr. 2014, 54, 151–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faucheux, B.A.; Martin, M.E.; Beaumont, C.; Hauw, J.J.; Agid, Y.; Hirsch, E.C. Neuromelanin associated redox-active iron is increased in the substantia nigra of patients with Parkinson’s disease. J. Neurochem. 2003, 86, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Zucca, F.A.; Segura-Aguilar, J.; Ferrari, E.; Muñoz, P.; Paris, I.; Sulzerd, D.; Sarna, T.; Casella, L.; Zecca, L. Interactions of iron, dopamine and neuromelanin pathways in brain aging and Parkinson’s disease. Prog. Neurobiol. 2017, 155, 96–119. [Google Scholar] [CrossRef] [PubMed]
- Floor, E.; Wetzel, M.G. Increased protein oxidation in human substantia nigra pars compacta in comparison with basal ganglia and prefrontal cortex measured with an improved dinitrophenylhydrazine assay. J. Neurochem. 1998, 70, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-Hydroxy-2-Nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Huang, Y.; Przedborski, S. Oxidative stress in Parkinson’s disease: A mechanism of pathogenic and therapeutic significance. Ann. N. Y. Acad. Sci. 2008, 1147, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Danielson, S.R.; Andersen, J.K. Oxidative and nitrative protein modifications in Parkinson’s disease. Free Radic. Biol. Med. 2008, 44, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.; Soundararajan, C.C.; Vivekanandhan, S.; Behari, M. Erythrocyte antioxidant enzymes in Parkinson’s disease. Indian J. Med. Res. 2005, 121, 111–115. [Google Scholar] [PubMed]
- Beal, M.F. Experimental models of Parkinson’s disease. Nat. Rev. Neurosci. 2001, 2, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Surendran, S.; Rajasankar, S. Parkinson’s disease: Oxidative stress and therapeutic approaches. Neurol. Sci. 2010, 31, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.W. Linking selective vulnerability to cell death mechanisms in Parkinson’s disease. Am. J. Pathol. 2007, 170, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Liu, J.L.; Wu, Y.R.; Chen, Y.C.; Cheng, H.S.; Cheng, M.L.; Chiu, D.T.Y. Increased oxidative damage in peripheral blood correlates with severity of Parkinson’s disease. Neurobiol. Dis. 2009, 33, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Filograna, R.; Beltramini, M.; Bubacco, L.; Bisaglia, M. Anti-Oxidants in Parkinson’s disease therapy: A critical point of view. Curr. Neuropharmacol. 2016, 14, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Dias, V.; Junn, E.; Mouradian, M.M. The role of oxidative stress in Parkinson’s disease. J. Parkinson’s Dis. 2013, 3, 461–491. [Google Scholar]
- Stack, E.C.; Matson, W.R.; Ferrante, R.J. Evidence of oxidant damage in Huntington’s disease: Translational strategies using antioxidants. Ann. N. Y. Acad. Sci. 2008, 1147, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Tasset, I.; Sanchez, F.; Tunez, I. The molecular bases of Huntington’s disease: The role played by oxidative stress. Rev. Neurol. 2009, 49, 424–429. [Google Scholar] [PubMed]
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef]
- Gil-Mohapel, J.; Brocardo, P.S.; Christie, B.R. The role of oxidative stress in Huntington’s disease: Are antioxidants good therapeutic candidates? Curr. Drug Targets 2014, 15, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Rotblat, B.; Southwell, A.L.; Ehrnhoefer, D.E.; Skotte, N.H.; Metzlerc, M.; Franciosi, S.; Leprivier, G.; Somasekharan, S.P.; Barokas, A.; Deng, Y.; et al. HACE1 reduces oxidative stress and mutant Huntingtin toxicity by promoting the NRF2 response. Proc. Natl. Acad. Sci. USA 2014, 111, 3032–3037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Q.; Huang, B.; Cheng, J.; Seefelder, M.; Engler, T.; Pfeifer, G.; Oeckl, P.; Otto, M.; Moser, F.; Maurer, M.; et al. The cryo-electron microscopy structure of huntingtin. Nature 2018, 555, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Shirendeb, U.P.; Calkins, M.J.; Manczak, M.; Anekonda, V.; Dufour, B.; McBride, J.L.; Mao, P.; Reddy, P.H. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Human Mol. Genet. 2012, 21, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Johri, A.; Beal, M.F. Antioxidants in Huntington’s disease. Biochim. Biophys. Acta 2012, 1822, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Deckel, A.W.; Tang, V.; Nuttal, D.; Gary, K.; Elder, R. Altered neuronal nitric oxide synthase expression contributes to disease progression in Huntington’s disease transgenic mice. Brain Res. 2002, 939, 7686. [Google Scholar] [CrossRef]
- Perez-Severiano, F.; Escalante, B.; Vergara, P.; Rios, C.; Segovia, J. Age-dependent changes in nitric oxide synthase activity and protein expression in striata of mice transgenic for the Huntington’s disease mutation. Brain Res. 2002, 951, 36–42. [Google Scholar] [CrossRef]
- Perez-Severiano, F.; Santamaria, A.; Pedraza-Chaverri, J.; Medina-Campos, O.N.; Rios, C.; Segovia, J. Increased formation of reactive oxygen species, but no changes in glutathione peroxidase activity, in striata of mice transgenic for the Huntington’s disease mutation. Neurochem. Res. 2004, 29, 729–733. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kosaras, B.; Del Signore, S.J.; Cormier, K.; McKee, A.; Ratan, R.R.; Kowall, N.W.; Ryu, H. Modulation of lipid peroxidation and mitochondrial function improves neuropathology in Huntington’s disease mice. Acta Neuropathol. 2011, 121, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Pitts, A.; Dailey, K.; Newington, J.T.; Chien, A.; Arseneault, R.; Cann, T.; Thompson, L.M.; Cumming, R.C. Dithiol-based compounds maintain expression of antioxidant protein peroxiredoxin 1 that counteracts toxicity of mutant huntingtin. J. Biol. Chem. 2012, 287, 22717–22729. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.H.; Dara, K.B.; Anees, S.; Zargar, M.A.; Masood, A.; Sofi, M.A.; Ganie, S.A. Oxidative stress, mitochondrial dysfunction and neurodegenerative diseases; a mechanistic insight. Biomed. Pharmacother. 2015, 74, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Sayre, L.M.; Perry, G.; Smith, M.A. Oxidative stress and neurotoxicity. Chem. Res. Toxicol. 2008, 21, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.M.; Wu, Y.R.; Chengetal, M.L. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington’s disease patients. Biochem. Biophysi. Res. Commun. 2007, 359, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Gash, M.T.; Mann, V.M.; Javoy-Agid, F.; Cooper, J.M.; Schapira, A.H. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann. Neurol. 1996, 39, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Kumar, P.; Malik, J. Plants and phytochemicals for Huntington’s disease. Pharmacog. Rev. 2013, 7, 81–91. [Google Scholar]
- Klepac, N.; Relja, M.; Klepac, R.; Hecimovic, S.; Babic, T.; Trkulja, V. Oxidative stress parameters in plasma of Huntington’s disease patients, asymptomatic Huntington’s disease gene carriers and healthy subjects: A cross-sectional study. J. Neurol. 2007, 254, 1676–1683. [Google Scholar] [CrossRef] [PubMed]
- Goula, A.V.; Berquist, B.R.; Wilson, D.M., III; Wheeler, V.C.; Trottier, Y.; Merienne, K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009, 5, e1000749. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.; Rosenstock, T.R.; Cunha-Oliveira, T.; Ferreira, I.L.; Oliveira, C.R.; Rego, A.C. Glutathione redox cycle dysregulation in Huntington’s disease knock-in striatal cells. Free Radcal. Biol. Med. 2012, 53, 1857–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoy, N.; Mackay, G.M.; Forrest, C.M.; Christofides, J.; Egerton, M.; Stone, T.W.; Darlington, L.G. Tryptophan metabolism and oxidative stress in patients with Huntington’s disease. J. Neurochem. 2005, 93, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.; Barrero, F.J.; Morales, B.; Luna, J.D.; Ramirez, M.; Vives, F. Oxidative stress and plasma amino peptidase activity in Huntington’s disease. J. Neural Transm. 2010, 117, 325–332. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kausar, S.; Wang, F.; Cui, H. The Role of Mitochondria in Reactive Oxygen Species Generation and Its Implications for Neurodegenerative Diseases. Cells 2018, 7, 274. https://doi.org/10.3390/cells7120274
Kausar S, Wang F, Cui H. The Role of Mitochondria in Reactive Oxygen Species Generation and Its Implications for Neurodegenerative Diseases. Cells. 2018; 7(12):274. https://doi.org/10.3390/cells7120274
Chicago/Turabian StyleKausar, Saima, Feng Wang, and Hongjuan Cui. 2018. "The Role of Mitochondria in Reactive Oxygen Species Generation and Its Implications for Neurodegenerative Diseases" Cells 7, no. 12: 274. https://doi.org/10.3390/cells7120274




