Matefin/SUN-1 Phosphorylation on Serine 43 Is Mediated by CDK-1 and Required for Its Localization to Centrosomes and Normal Mitosis in C. elegans Embryos
Abstract
:1. Introduction
2. Experimental Section
2.1. Strains
2.2. RNAi Experiments
2.3. Fecundity Experiments
2.4. Indirect Immunofluorescence
2.4.1. Embryo Staining
2.4.2. Gonad Staining
2.5. Molecular Cloning and Strain Creation
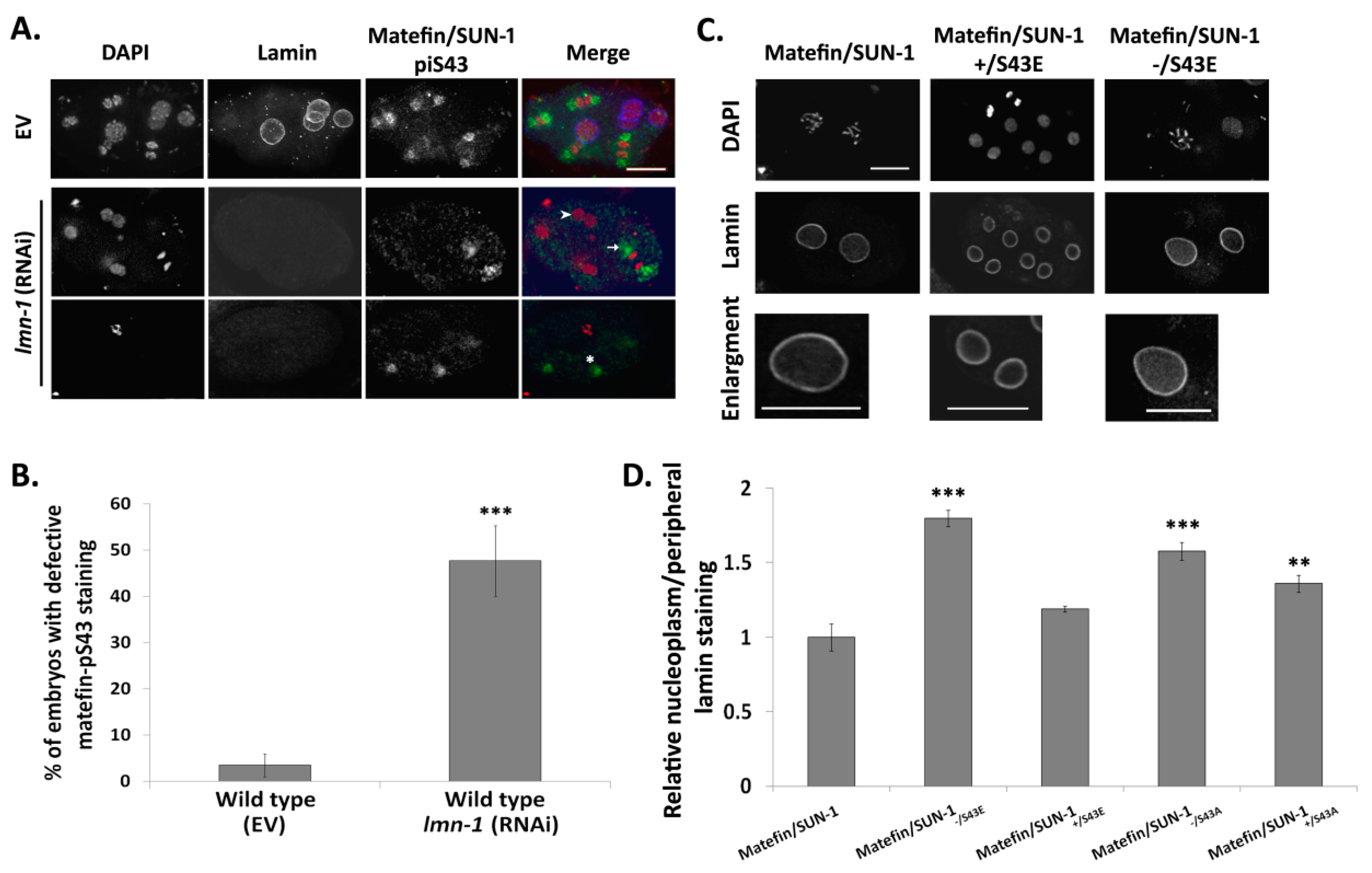
2.6. Analysis of Relative Nucleoplasmic Peripheral Staining of Lamin
3. Results and Discussion
3.1. Results
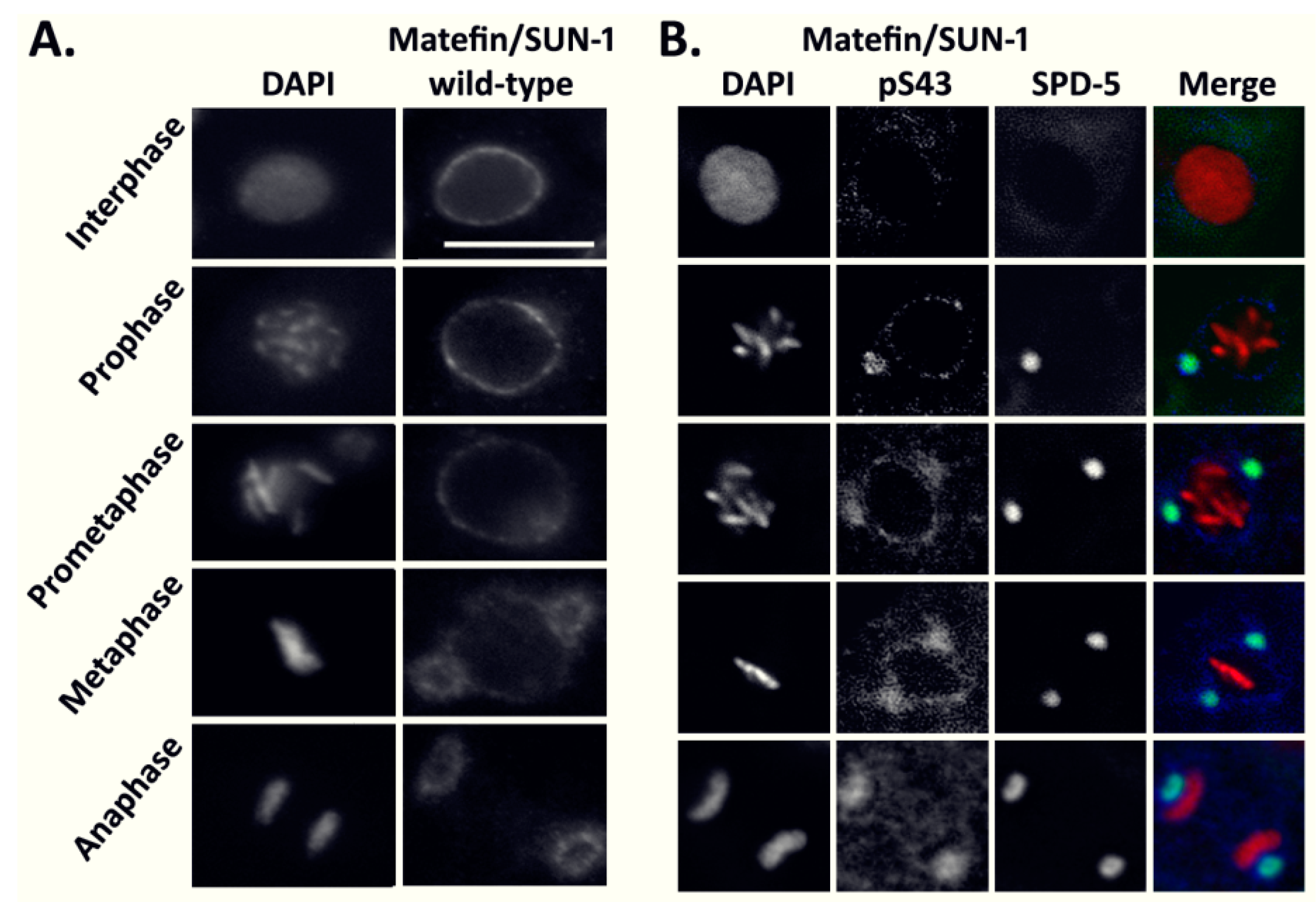
3.1.1. The Phosphorylated Form of Serine 43 in Matefin/SUN-1 Co-Localizes with Centrosomes
3.1.2. Phosphorylation of Serine 43 in Matefin/SUN-1 Requires CDK-1 Activity
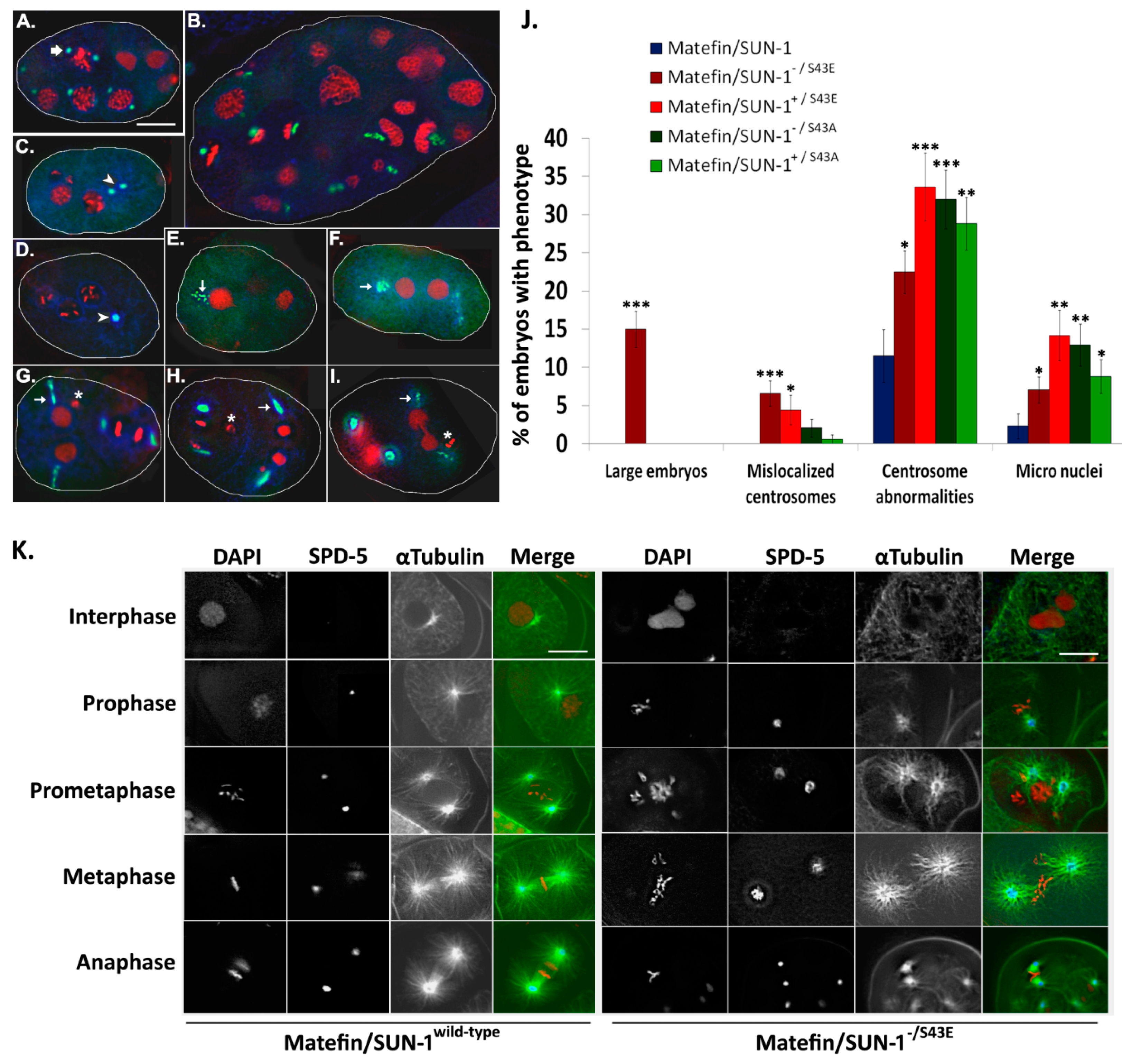
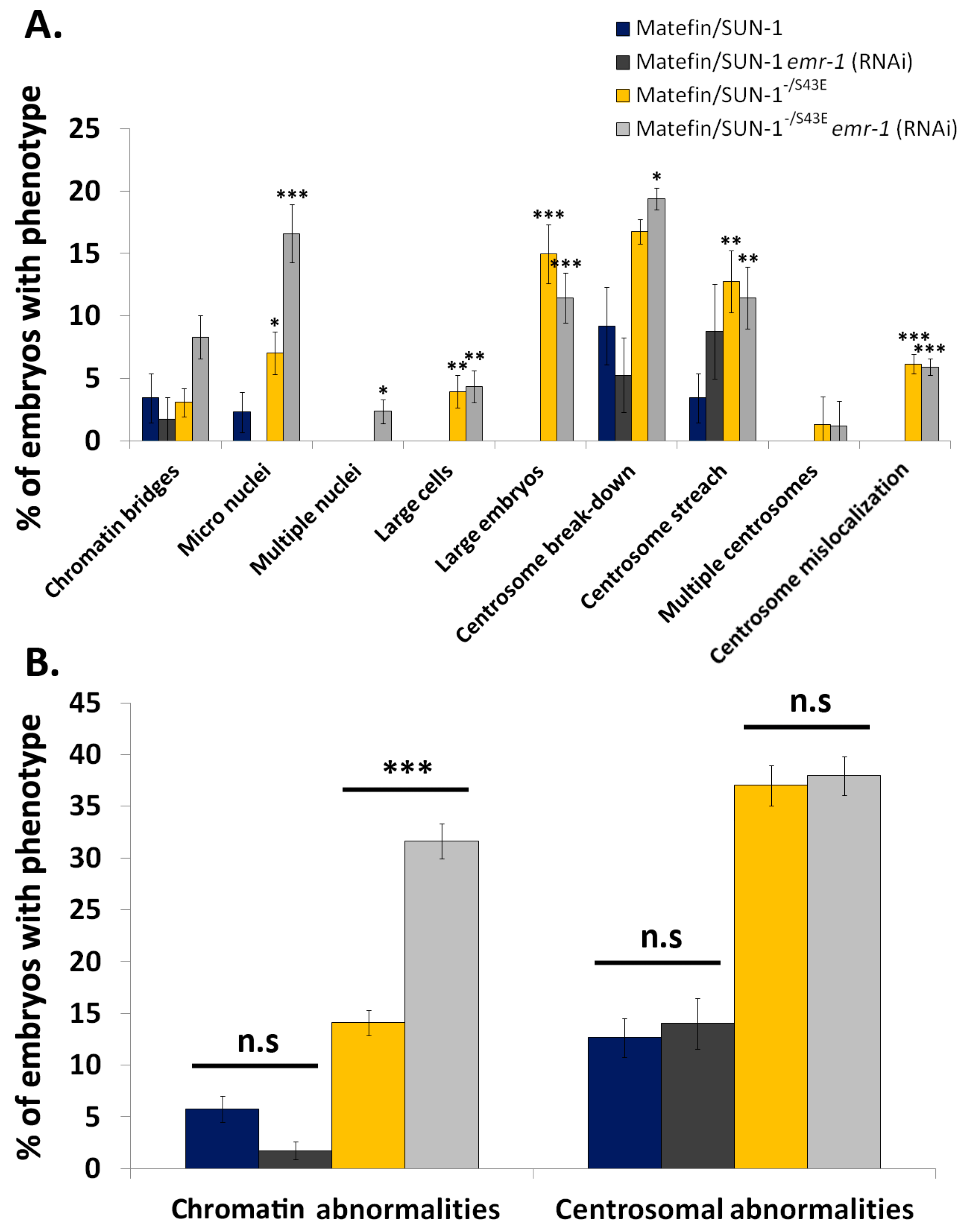
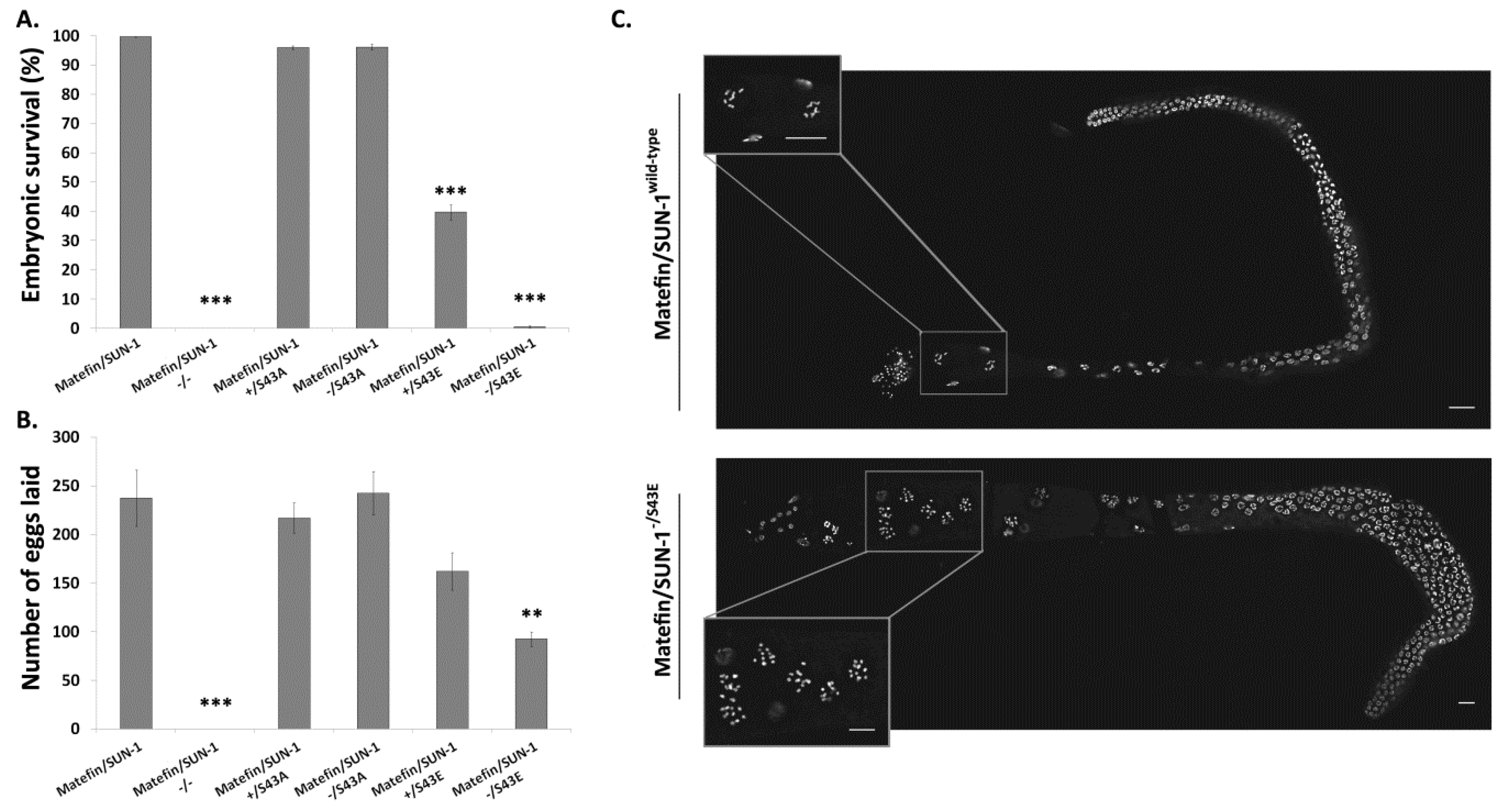
3.1.3. Phosphorylation of Serine 43 in Matefin/SUN-1 Is Required to Maintain Centrosome Shape and Function
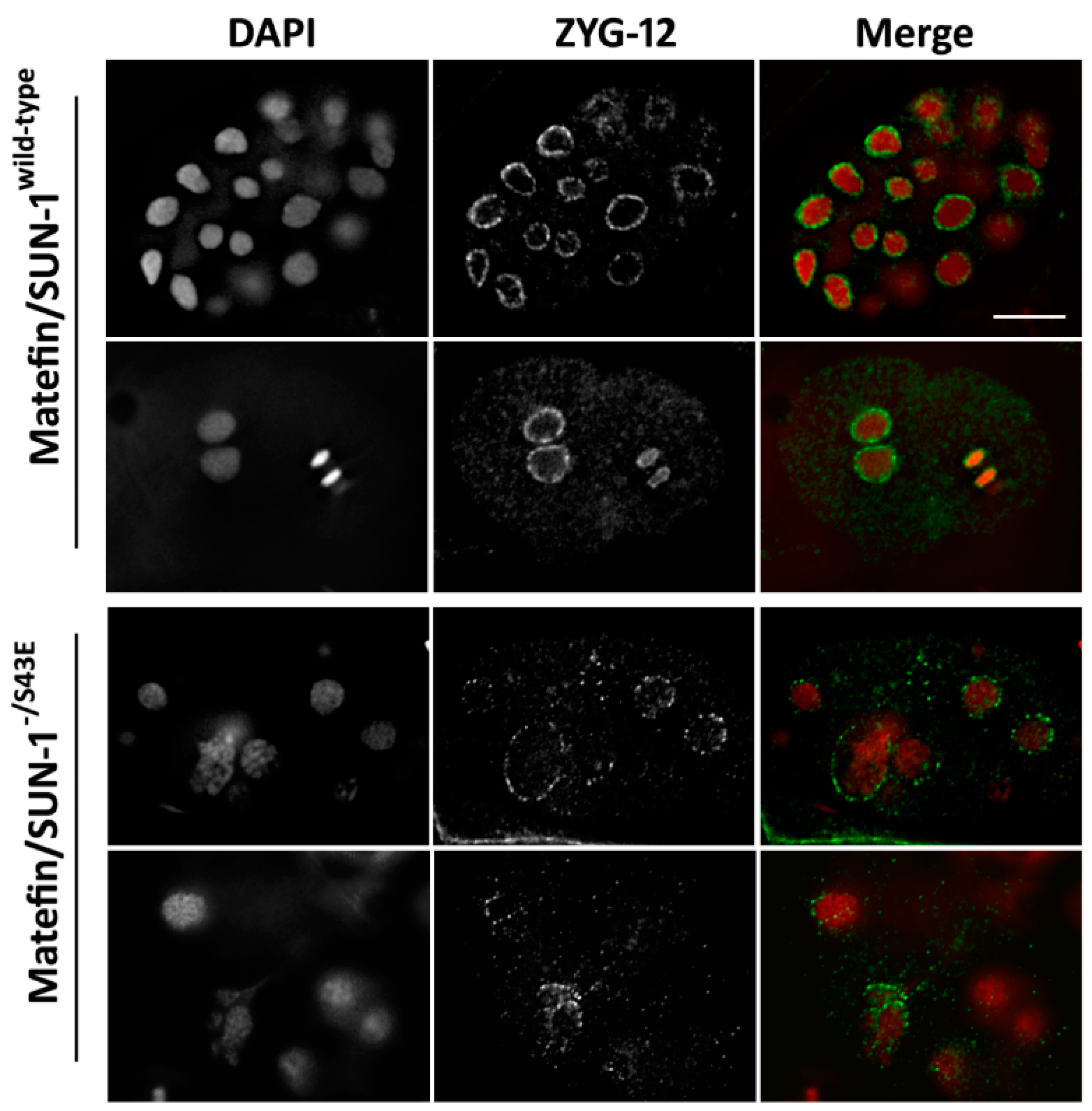
3.1.4. Lamin Is Required for the Centrosome Localization of pS43.
3.1.5. Fertility Is Reduced in Animals Expressing S43E Matefin/SUN-1
3.2. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015, in press. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, E.C.; Florens, L.; Guan, T.; Yates, J.R.; Gerace, L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003, 531, 1380–1382. [Google Scholar] [CrossRef] [PubMed]
- De Las Heras, J.I.; Meinke, P.; Batrakou, D.G.; Srsen, V.; Zuleger, N.; Kerr, A.R.; Schirmer, E.C. Tissue specificity in the nuclear envelope supports its functional complexity. Nucleus 2013, 4, 460–477. [Google Scholar] [CrossRef] [PubMed]
- Bronshtein, I.; Kepten, E.; Kanter, I.; Berezin, S.; Lindner, M.; Redwood, A.B.; Mai, S.; Gonzalo, S.; Foisner, R.; Shav-Tal, Y.; et al. Loss of lamin a function increases chromatin dynamics in the nuclear interior. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.L.; Foisner, R. Lamin-binding proteins. Cold Spring Harb. Perspect. Biol. 2010, 2, a000554. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.; Bonne, G. “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 2007, 313, 2121–2133. [Google Scholar] [CrossRef] [PubMed]
- Melcer, S.; Gruenbaum, Y.; Krohne, G. Invertebrate lamins. Exp. Cell. Res. 2007, 313, 2157–2166. [Google Scholar] [CrossRef] [PubMed]
- Lyakhovetsky, R.; Gruenbaum, Y. Studying lamins in invertebrate models. Adv. Exp. Med. Biol. 2014, 773, 245–262. [Google Scholar] [PubMed]
- Lin, F.; Blake, D.L.; Callebaut, I.; Skerjanc, I.S.; Holmer, L.; McBurney, M.W.; Paulin-Levasseur, M.; Worman, H.J. Man1, an inner nuclear membrane protein that shares the lem domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 2000, 275, 4840–4847. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lee, K.K.; Segura-Totten, M.; Neufeld, E.; Wilson, K.L.; Gruenbaum, Y. Man1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in c. Elegans. Proc. Natl. Acad. Sci. USA 2003, 100, 4598–4603. [Google Scholar] [CrossRef] [PubMed]
- Morales-Martínez, A.; Dobrzynska, A.; Askjaer, P. Inner nuclear membrane protein lem-2 is required for proper nuclear separation and morphology. J. Cell Sci. 2015, in press. [Google Scholar]
- Sosa, B.A.; Kutay, U.; Schwartz, T.U. Structural insights into linc complexes. Curr. Opin. Struct. Biol. 2013, 23, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Meinke, P.; Schirmer, E.C. Linc’ing form and function at the nuclear envelope. FEBS Lett. 2015, 589, 2514–2521. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.I.; Birendra, K.C.; Roux, K.J. Making the linc: Sun and kash protein interactions. Biol. Chem. 2015, 396, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Crisp, M.; Burke, B. The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett. 2008, 582, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Tzur, Y.; Wilson, K.L.; Gruenbaum, Y. Sun-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat. Rev. Cell. Mol. Biol. 2006, 7, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; Starr, D.; Liu, J.; Cohen, M.; Han, M.; Wilson, K.; Gruenbaum, Y. Lamin-dependent localization of unc-84, a protein required for nuclear migration in c. Elegans. Mol. Biol. Cell 2002, 13, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Bone, C.R.; Tapley, E.C.; Gorjánácz, M.; Starr, D.A. The caenorhabditis elegans sun protein unc-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Mol. Biol. Cell 2014, 25, 2853–2865. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.J.; Fixsen, W.D.; Horvitz, H.R.; Han, M. Unc-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during c. Elegans development. Development 1999, 126, 3171–3181. [Google Scholar] [PubMed]
- Starr, D.A.; Fridolfsson, H.N. Interactions between nuclei and the cytoskeleton are mediated by sun-kash nuclear-envelope bridges. Annu. Rev. Cell. Dev. Biol. 2010, 26, 421–444. [Google Scholar] [CrossRef] [PubMed]
- Fridkin, A.; Mills, E.; Margalit, A.; Neufeld, E.; Lee, K.K.; Feinstein, N.; Cohen, M.; Wilson, K.L.; Gruenbaum, Y. Matefin, a caenorhabditis elegans germ line-specific sun-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci USA 2004, 101, 6987–6992. [Google Scholar] [CrossRef] [PubMed]
- Penkner, A.M.; Fridkin, A.; Gloggnitzer, J.; Baudrimont, A.; Machacek, T.; Woglar, A.; Csaszar, E.; Pasierbek, P.; Ammerer, G.; Gruenbaum, Y.; et al. Meiotic chromosome homology search involves modifications of the nuclear envelope protein matefin/sun-1. Cell 2009, 139, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.J.; Misner, L.; Le Bot, N.; Tsai, M.C.; Campbell, J.M.; Ahringer, J.; White, J.G. The c. Elegans hook protein, zyg-12, mediates the essential attachment between the centrosome and nucleus. Cell 2003, 115, 825–836. [Google Scholar] [CrossRef]
- Baudrimont, A.; Penkner, A.; Woglar, A.; Machacek, T.; Wegrostek, C.; Gloggnitzer, J.; Fridkin, A.; Klein, F.; Gruenbaum, Y.; Pasierbek, P.; et al. Leptotene/zygotene chromosome movement via the sun/kash protein bridge in caenorhabditis elegans. PLoS Genet. 2010, 6, e1001219. [Google Scholar] [CrossRef] [PubMed]
- Woglar, A.; Daryabeigi, A.; Adamo, A.; Habacher, C.; Machacek, T.; La Volpe, A.; Jantsch, V. Matefin/sun-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet. 2013, 9, e1003335. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The genetics of caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [PubMed]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of bacterially expressed dsrnas can produce specific and potent genetic interference in caenorhabditis elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef]
- Kamath, R.S.; Ahringer, J. Genome-wide rnai screening in caenorhabditis elegans. Methods 2003, 30, 313–321. [Google Scholar] [CrossRef]
- Jackman, M.; Lindon, C.; Nigg, E.A.; Pines, J. Active cyclin b1-cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003, 5, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Takizawa, C.G.; Morgan, D.O. Control of mitosis by changes in the subcellular location of cyclin-b1- cdk1 and cdc25c. Curr Opin Cell Biol 2000, 12, 658–665. [Google Scholar] [CrossRef]
- Zeiser, E.; Frøkjær-Jensen, C.; Jorgensen, E.; Ahringer, J. Mossci and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the c. Elegans germline. PLoS One 2011, 6, e20082. [Google Scholar] [CrossRef] [PubMed]
- Penkner, A.; Tang, L.; Novatchkova, M.; Ladurner, M.; Fridkin, A.; Gruenbaum, Y.; Schweizer, D.; Loidl, J.; Jantsch, V. The nuclear envelope protein matefin/sun-1 is required for homologous pairing in c. Elegans meiosis. Dev. Cell 2007, 12, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Minn, I.L.; Rolls M, M.; Hanna-Rose, W.; Malone, C.J. Sun-1 and zyg-12, mediators of centrosome-nucleus attachment, are a functional sun/kash pair in caenorhabditis elegans. Mol. Biol. Cell 2009, 20, 4586–4595. [Google Scholar] [CrossRef] [PubMed]
- Gönczy, P. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 2012, 13, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.T.; Bottrill, A.; Prosser, S.L.; Jayaraman, S.; Straatman, K.; Fry, A.M.; Shackleton, S. Mitotic phosphorylation of sun1 loosens its connection with the nuclear lamina while the linc complex remains intact. Nucleus 2014, 5, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Nahaboo, W.; Zouak, M.; Askjaer, P.; Delattre, M. Chromatids segregate without centrosomes during caenorhabditis elegans mitosis in a ran- and clasp-dependent manner. Mol. Biol. Cell 2015, 26, 2020–2029. [Google Scholar] [CrossRef] [PubMed]
- Meinke, P.; Mattioli, E.; Haque, F.; Antoku, S.; Columbaro, M.; Straatman, K.R.; Worman, H.J.; Gundersen, G.G.; Lattanzi, G.; Wehnert, M.; et al. Muscular dystrophy-associated sun1 and sun2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014, 10, e1004605. [Google Scholar] [CrossRef] [PubMed]
- Haque, F.; Lloyd, D.J.; Smallwood, D.T.; Dent, C.L.; Shanahan, C.M.; Fry, A.M.; Trembath, R.C.; Shackleton, S. Sun1 interacts with nuclear lamin a and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell Biol. 2006, 26, 3738–3851. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chi, Y.H.; Mutalif, R.A.; Starost, M.F.; Myers, T.G.; Anderson, S.A.; Stewart, C.L.; Jeang, K.T. Accumulation of the inner nuclear envelope protein sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 2012, 149, 565–577. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuela, N.; Gruenbaum, Y. Matefin/SUN-1 Phosphorylation on Serine 43 Is Mediated by CDK-1 and Required for Its Localization to Centrosomes and Normal Mitosis in C. elegans Embryos. Cells 2016, 5, 8. https://doi.org/10.3390/cells5010008
Zuela N, Gruenbaum Y. Matefin/SUN-1 Phosphorylation on Serine 43 Is Mediated by CDK-1 and Required for Its Localization to Centrosomes and Normal Mitosis in C. elegans Embryos. Cells. 2016; 5(1):8. https://doi.org/10.3390/cells5010008
Chicago/Turabian StyleZuela, Noam, and Yosef Gruenbaum. 2016. "Matefin/SUN-1 Phosphorylation on Serine 43 Is Mediated by CDK-1 and Required for Its Localization to Centrosomes and Normal Mitosis in C. elegans Embryos" Cells 5, no. 1: 8. https://doi.org/10.3390/cells5010008




