Exploitation of Potentially New Antibiotics from Mangrove Actinobacteria in Maowei Sea by Combination of Multiple Discovery Strategies
Abstract
:1. Introduction
2. Results
2.1. Isolation and Diversity of Actinobacteria from Mangrove Soil of Maowei Sea
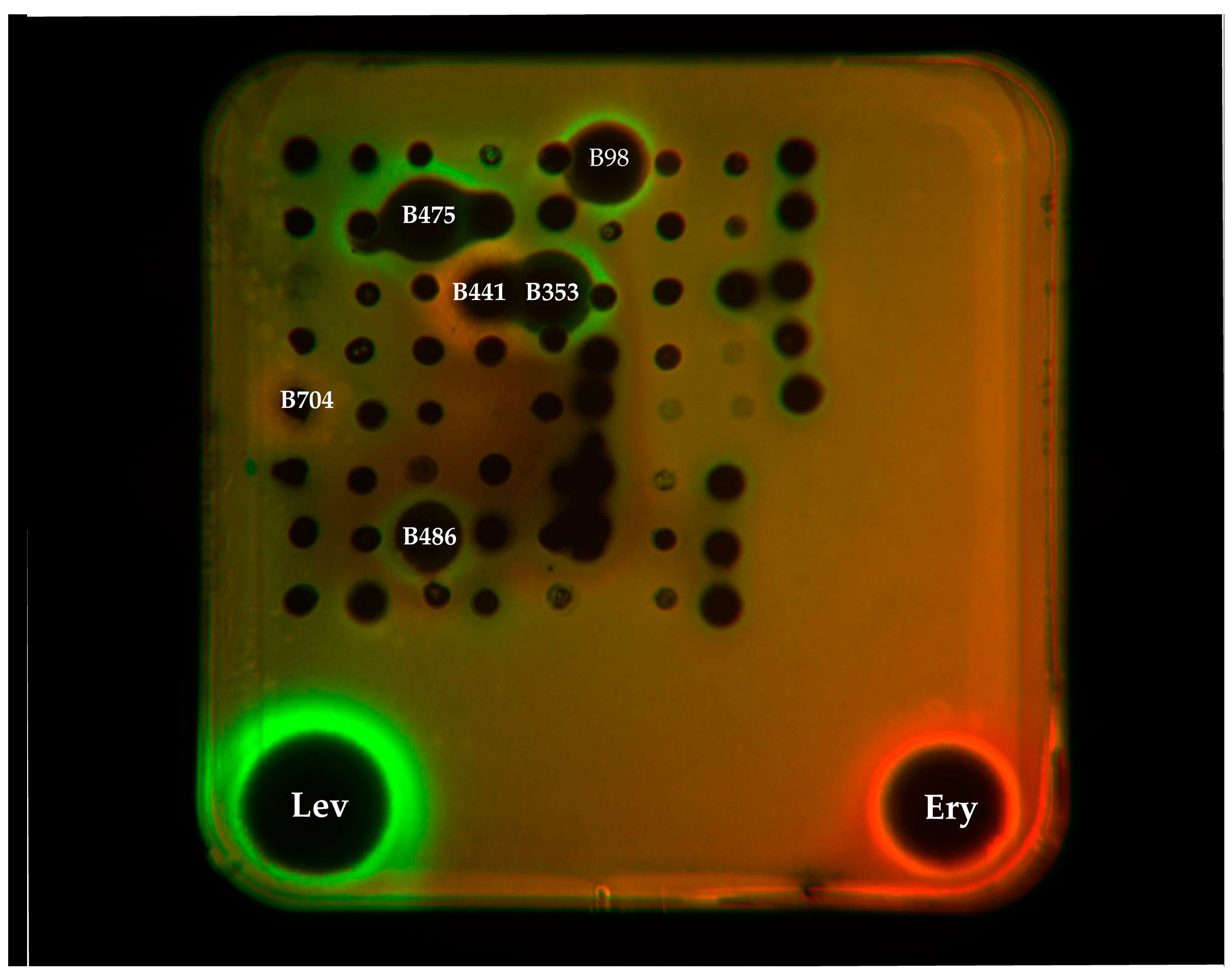
2.2. Antibacterial Assay and Mechanism Determination
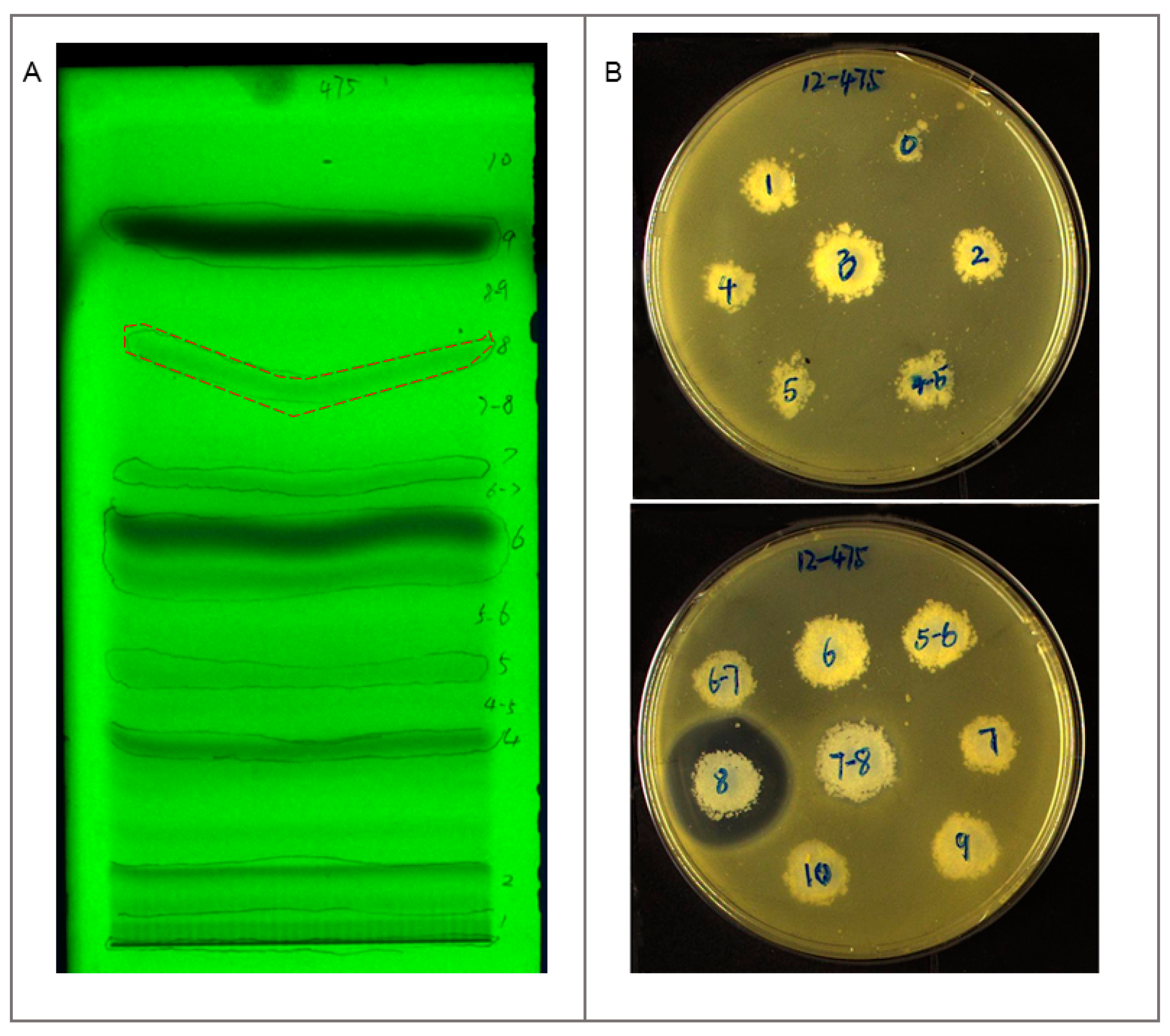
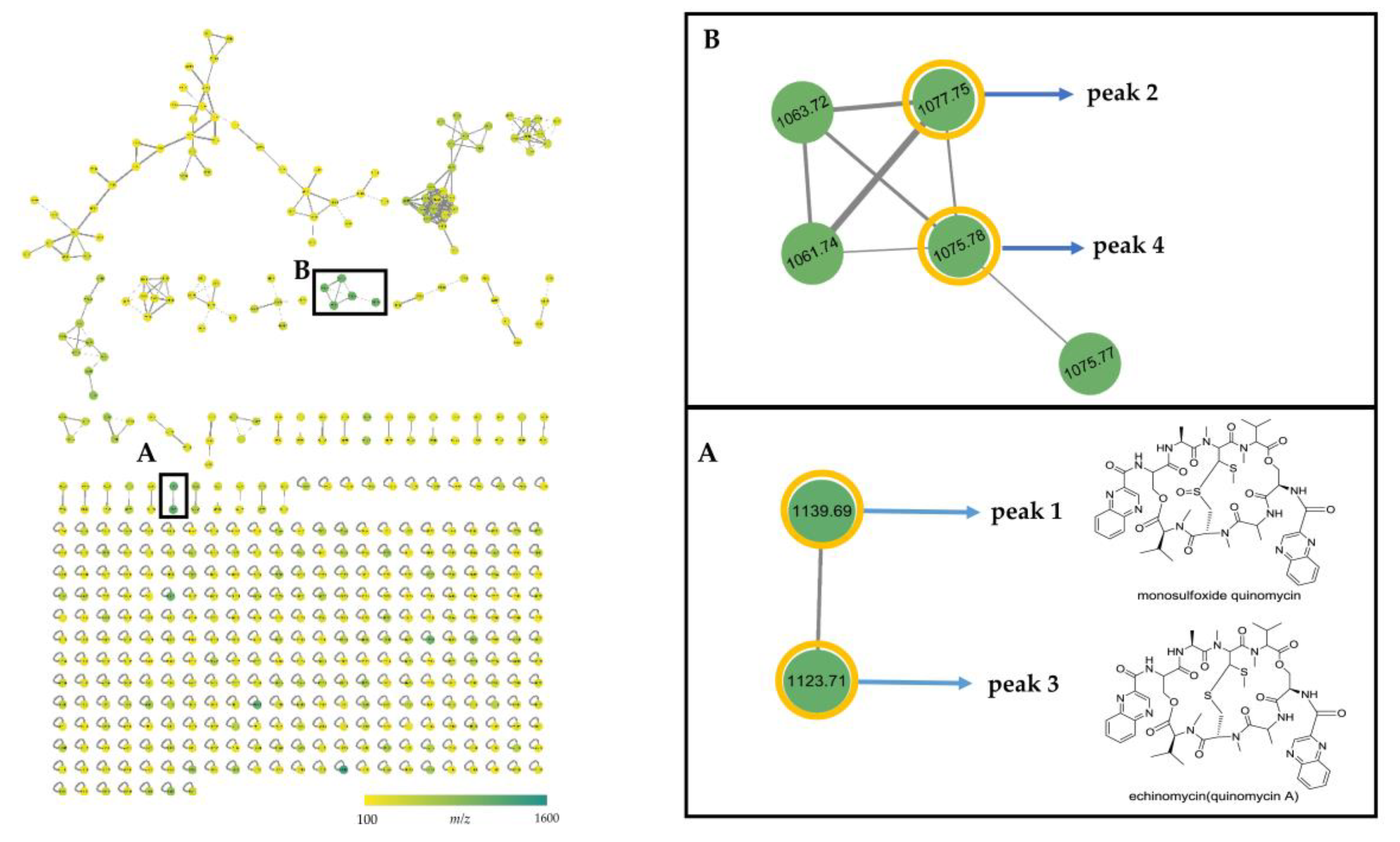
2.3. Identification and Dereplication of Antibacterial Components Produced by Strain B475
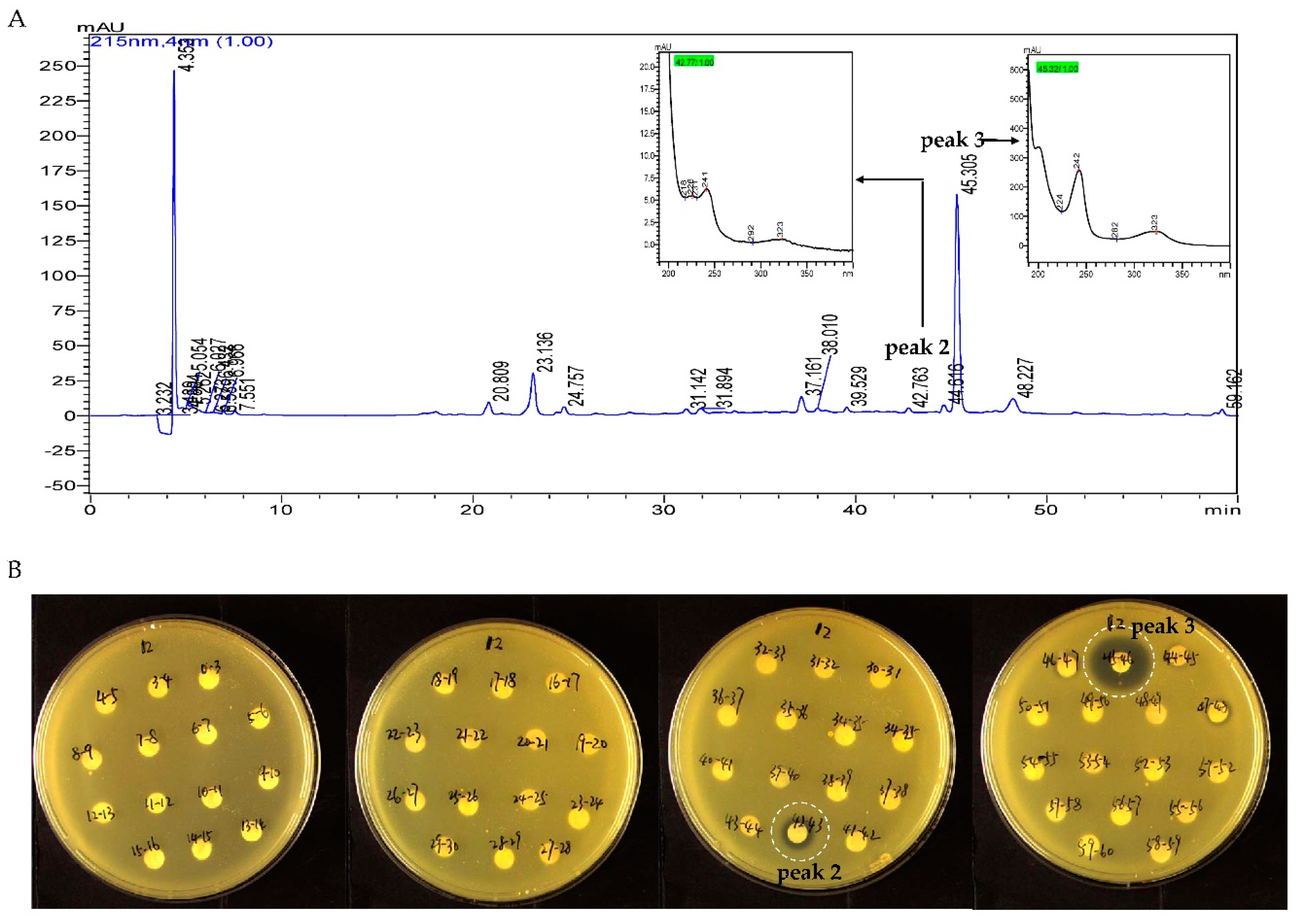
2.4. MS/MS Network-based Discovery of Potentially New Antibiotics Produced by Strain B475
3. Discussion
4. Materials and Methods
4.1. Collection of Mangrove Soil Sample
4.2. Isolation and Maintenance of Actinobacteria
4.3. PCR Amplification and Sequencing of 16S rRNA Gene
4.4. Sequence Analysis
4.5. Nucleotide Sequence Accession Numbers
4.6. Antibacterial Activity Screening
4.7. Mechanism of Action Determination
4.8. Identification of Bioactive Bands by Thin Layer Chromatography (TLC)
4.9. Identification of Bioactive Peaks by High Performance Liquid Chromatography (HPLC)
4.10. Dereplication and Molecular Network Analysis of Microbial Extracts
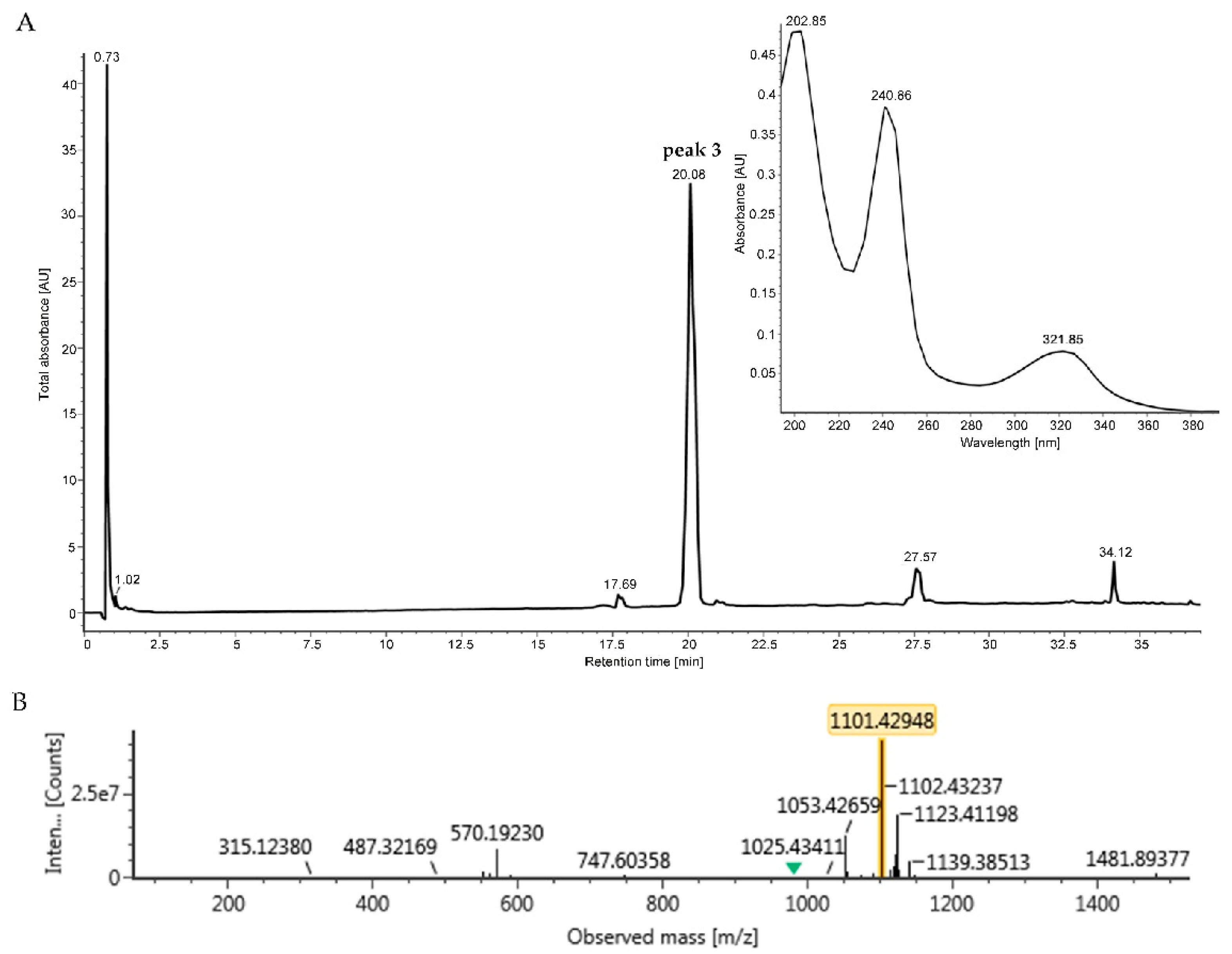
4.11. Accumulation and Measurement of Peak 3
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Magrini, N.; Kahlmeter, G.; Singh, N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; Volume 27, pp. 1–7. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2016, 2475067–2475075. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C. Opinion—Anti-infectives: Where will new antibiotics come from? Nat. Rev. Microbiol. 2003, 1, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Monciardini, P.; Iorio, M.; Maffioli, S.; Sosio, M.; Donadio, S. Discovering new bioactive molecules from microbial sources. Microb. Biotechnol. 2014, 7, 209–220. [Google Scholar] [CrossRef]
- Lewis, K. Antibiotics: Recover the lost art of drug discovery. Nature 2012, 485, 439–440. [Google Scholar] [CrossRef]
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 2008, 8, 557–563. [Google Scholar] [CrossRef]
- Zotchev, S.B. Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol. 2012, 158, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A. Strategies for discovering drugs from previously unexplored natural products. Drug Discov. Today 2000, 5, 294–300. [Google Scholar] [CrossRef]
- Genilloud, O. The re-emerging role of microbial natural products in antibiotic discovery. Antonie Van Leeuwenhoek 2014, 106, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Clardy, J.; Fischbach, M.A.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar] [CrossRef]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “ uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Rückert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef]
- Hubert, J.; Nuzillard, J.-M.; Renault, J.-H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem. Rev. 2017, 16, 55–95. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.R.; Crusemann, M.; Lechner, A.; Sarkar, A.; Li, J.; Ziemert, N.; Wang, M.; Bandeira, N.; Moore, B.S.; Dorrestein, P.C.; et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol. 2015, 22, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.X.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Perezvictoria, I.; Martin, J.; Reyes, F. Combined LC/UV/MS and NMR Strategies for the Dereplication of Marine Natural Products. Planta Med. 2016, 82, 857–871. [Google Scholar]
- Chervin, J.; Stierhof, M.; Tong, M.H.; Peace, D.; Hansen, K.O.; Urgast, D.S.; Andersen, J.H.; Yu, Y.; Ebel, R.; Kyeremeh, K.; et al. Targeted Dereplication of Microbial Natural Products by High-Resolution MS and Predicted LC Retention Time. J. Nat. Prod. 2017, 80, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nakashima, T. Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics 2018, 7, 45. [Google Scholar] [CrossRef]
- Sahoo, K.; Dhal, N. Potential microbial diversity in mangrove ecosystems: A review. Indian J. Mar. Sci. 2009, 38, 249–256. [Google Scholar]
- Nabeelah Bibi, S.; Fawzi, M.M.; Gokhan, Z.; Rajesh, J.; Nadeem, N.; Rengasamy, K.K.R.; Albuquerque, R.D.D.G.; Pandian, S.K. Ethnopharmacology, phytochemistry, and global distribution of mangroves―A comprehensive review. Mar. Drugs 2019, 17, 231. [Google Scholar] [CrossRef]
- Friess, D.A.; Rogers, K.; Lovelock, C.E.; Krauss, K.W.; Hamilton, S.E.; Lee, S.Y.; Lucas, R.; Primavera, J.; Rajkaran, A.; Shi, S. The state of the world’s mangrove forests: Past, present, and future. Annu. Rev. Environ. Resour. 2019, 44, 89–115. [Google Scholar] [CrossRef]
- Amrita, K.; Nitin, J.; Devi, C.S. Novel bioactive compounds from mangrove derived actinomycetes. Int. Res. J. Phar. 2012, 3, 25–29. [Google Scholar]
- Ancheeva, E.; Daletos, G.; Proksch, P. Lead compounds from mangrove-associated microorganisms. Mar. Drugs 2018, 16, 319. [Google Scholar] [CrossRef] [PubMed]
- Azman, A.S.; Othman, I.; Velu, S.S.; Chan, K.G.; Lee, L.H. Mangrove rare actinobacteria: Taxonomy, natural compound, and discovery of bioactivity. Front. Microbiol. 2015, 6, 856. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.; Gao, A.H.; Xie, Q.Y.; Gao, H.G.; Zhuang, L.; Lin, H.P.; Yu, H.P.; Li, J.; Yao, X.S.; Goodfellow, M.; et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.B.; Ye, W.W.; Han, Y.; Deng, Z.X.; Hong, K. Natural products from mangrove actinomycetes. Mar. Drugs 2014, 12, 2590–2613. [Google Scholar] [CrossRef] [Green Version]
- Pejin, B.; Glumac, M. New cytotoxic natural products from the mangrove biome: Covering the period 2007–2015. Nat. Prod. Res. 2019, 33, 1624–1628. [Google Scholar] [CrossRef]
- Jiang, Z.K.; Tuo, L.; Huang, D.L.; Osterman, I.A.; Tyurin, A.P.; Liu, S.W.; Lukyanov, D.A.; Sergiev, P.V.; Dontsova, O.A.; Korshun, V.A.; et al. Diversity, novelty, and antimicrobial activity of endophytic actinobacteria from mangrove plants in Beilun Estuary National Nature Reserve of Guangxi, China. Front. Microbiol. 2018, 9, 868. [Google Scholar] [CrossRef]
- Li, F.N.; Liu, S.W.; Lu, Q.P.; Zheng, H.Y.; Osterman, I.A.; Lukyanov, D.A.; Sergiev, P.V.; Dontsova, O.A.; Liu, S.S.; Ye, J.J.; et al. Studies on antibacterial activity and diversity of cultivable actinobacteria isolated from mangrove soil in Futian and Maoweihai of China. Evid. Based Complement. Alternat. Med. 2019, 2019, 3476567. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.J.; Shao, L.; Wang, M.X.; Rao, M.; Ge, M.; Xu, Y.X. Two novel quinomycins discovered by UPLC-MS from Stretomyces sp. HCCB11876. J. Antibiot. 2019, 72, 164–168. [Google Scholar] [CrossRef]
- Zhao, W.; Jiang, B.Y.; Zuo, L.J.; Li, S.F.; Hu, X.X.; Yong, X.F.; Liu, H.Y.; Yu, L.Y.; Zuo, L.M.; Shang, G.Z.; et al. Identification of quinomycins as principal secondary metabolites from Streptomyces sp. CPCC 200497. Chin. Med. Biotechnol. 2016, 11, 21–26. [Google Scholar]
- Okpokwasili, G.C.; Ifenwanta, C.E.; Nweke, C.O. The microflora of eagle island mangrove swamp, southern Nigeria. J. Res. Biol. 2012, 2, 602–616. [Google Scholar]
- Jensen, P.R.; Dwight, R.; Fenical, W. Distribution of actinomycetes in near-shore tropical marine sediments. Appl. Environ. Microbiol. 1991, 57, 1102–1108. [Google Scholar] [PubMed]
- Hong, K. Actinomycetes from mangrove and their secondary metabolites. Wei Sheng Wu Xue Bao 2013, 53, 1131–1141. [Google Scholar] [PubMed]
- Sun, C.H.; Li, F.N.; Cai, Z.; Wu, L. Brief introduction of mangrove researches including drug discovery granted by National Natural Science Foundation of China from 1986 to 2016. Chin. J. Antibiot. 2017, 42, 241–248. [Google Scholar]
- Lee, L.H.; Zainal, N.; Azman, A.S.; Eng, S.K.; Goh, B.H.; Yin, W.F.; Ab Mutalib, N.S.; Chan, K.G. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014, 2014, 698178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arifuzzaman, M.; Khatun, M.; Rahman, H. Isolation and screening of actinomycetes from Sundarbans soil for antibacterial activity. Afr. J. Biotechnol. 2010, 9, 4615–4619. [Google Scholar]
- Gajdacs, M. The concept of an ideal antibiotic: Implications for drug design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osterman, I.A.; Komarova, E.S.; Shiryaev, D.I.; Korniltsev, I.A.; Khven, I.M.; Lukyanov, D.A.; Tashlitsky, V.N.; Serebryakova, M.V.; Efremenkova, O.V.; Ivanenkov, Y.A.; et al. Sorting out antibiotics mechanisms of action: A double fluorescent protein reporter for high-throughput screening of ribosome and DNA biosynthesis inhibitors. Antimicrob. Agents Chemother. 2016, 60, 7481–7489. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Masubuchi, M. Dereplication of microbial extracts and related analytical technologies. J. Antibiot. 2014, 67, 353–360. [Google Scholar] [CrossRef]
- Waring, M.; Wakelin, L. Echinomycin: A bifunctional intercalating antibiotic. Nature 1974, 252, 653–657. [Google Scholar] [CrossRef]
- Foster, B.J.; Clagettcarr, K.; Shoemaker, D.D.; Suffness, M.; Plowman, J.; Trissel, L.A.; Grieshaber, C.K.; Leylandjones, B. Echinomycin: The first bifunctional intercalating agent in clinical trials. Investig. New Drugs 1985, 3, 403. [Google Scholar] [CrossRef]
- Liu, H.M.; Qin, S.; Wang, Y.X.; Li, W.J.; Zhang, J. Insecticidal action of Quinomycin A from Streptomyces sp. KN-0647, isolated from a forest soil. World J. Microbiol. Biotechnol. 2008, 24, 2243–2248. [Google Scholar] [CrossRef]
- Zolova, O.E.; Mady, A.S.A.; Garneau-Tsodikova, S. Recent developments in bisintercalator natural products. Biopolymers 2010, 93, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Nakazawa, T.; Tsunematsu, Y.; Hotta, K.; Watanabe, K. Echinomycin biosynthesis. Curr. Opin. Chem. Biol. 2013, 17, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.; Malkinson, J.P.; Paumier, D.; Searcey, M. Bisintercalator natural products with potential therapeutic applications: Isolation, structure determination, synthetic and biological studies. Nat. Prod. Rep. 2007, 24, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-B.; Lee, G.-S.; Kim, Y.-B.; Kim, S.-K.; Kim, Y.H. In vitro antibacterial activity of echinomycin and a novel analogue, YK2000, against vancomycin-resistant Enterococci. Int. J. Antimicrob. Agents 2004, 24, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Shin, W.S.; Kim, S.K. In vitro and in vivo activities of echinomycin against clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 61, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Socha, A.M.; Laplante, K.L.; Russell, D.J.; Rowley, D.C. Structure-activity studies of echinomycin antibiotics against drug-resistant and biofilm-forming Staphylococcus aureus and Enterococcus faecalis. Bioorg. Med. Chem. Lett. 2009, 19, 1504–1507. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Kim, Y.H.; Park, J.Y.; Kim, S.K. Synthesis and biological activity of new quinoxaline antibiotics of echinomycin analogues. Bioorg. Med. Chem. Lett. 2004, 14, 541–544. [Google Scholar] [CrossRef]
- Watanabe, K.; Oguri, H.; Oikawa, H. Diversification of echinomycin molecular structure by way of chemoenzymatic synthesis and heterologous expression of the engineered echinomycin biosynthetic pathway. Curr. Opin. Chem. Biol. 2009, 13, 189–196. [Google Scholar] [CrossRef]
- Esposito, M.; Nothias, L.F.; Retailleau, P.; Costa, J.; Roussi, F.; Neyts, J.; Leyssen, P.; Touboul, D.; Litaudon, M.; Paolini, J. Isolation of premyrsinane, myrsinane, and tigliane diterpenoids from euphorbia pithyusa using a chikungunya virus cell-based assay and analogue annotation by molecular networking. J. Nat. Prod. 2017, 80, 2051–2059. [Google Scholar] [CrossRef]
- Kleigrewe, K.; Almaliti, J.; Tian, I.Y.; Kinnel, R.B.; Korobeynikov, A.; Monroe, E.A.; Duggan, B.M.; Di Marzo, V.; Sherman, D.H.; Dorrestein, P.C.; et al. Combining mass spectrometric metabolic profiling with genomic analysis: A powerful approach for discovering natural products from Cyanobacteria. J. Nat. Prod. 2015, 78, 1671–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molecular Networking. Available online: https://ccms-ucsd.github.io/GNPSDocumentation/massspecbackground/networkingtheory/ (accessed on 25 October 2019).
- Zhou, S.Q.; Huang, X.L.; Huang, D.Y.; Hu, X.W.; Chen, J.L. A rapid method for extracting DNA from actinomycetes by Chelex-100. Biotechnol. Bull. 2010, 2, 123–125. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]




| Genus | Total Number of Actinobacterial Strains | ||||
|---|---|---|---|---|---|
| Isolation | Assay | Positive in Assay | RFP | Katushka2S | |
| Micromonospora | 126 | 35 | 14 | 0 | 1 |
| Streptomyces | 38 | 19 | 10 | 4 | 1 |
| Agromyces | 24 | 5 | 3 | 0 | 0 |
| Microbispora | 15 | 1 | 0 | 0 | 0 |
| Actinomadura | 13 | 4 | 0 | 0 | 0 |
| Rhodococcus | 11 | 3 | 0 | 0 | 0 |
| Nocardia | 9 | 3 | 1 | 0 | 0 |
| Mycobacterium | 6 | 1 | 0 | 0 | 0 |
| Kitasatospora | 4 | 1 | 1 | 0 | 0 |
| Paenarthrobacter | 4 | 2 | 0 | 0 | 0 |
| Sinomonas | 3 | 1 | 0 | 0 | 0 |
| Pseudarthrobacter | 1 | 1 | 0 | 0 | 0 |
| Micrococcus | 1 | 1 | 0 | 0 | 0 |
| Intrasporangium | 1 | 1 | 1 | 0 | 0 |
| Gordonia | 1 | 1 | 0 | 0 | 0 |
| Nakamurella | 1 | 1 | 1 | 0 | 0 |
| Actinocorallia | 1 | 1 | 0 | 0 | 0 |
| Arthrobacter | 1 | 1 | 0 | 0 | 0 |
| Mycolicibacterium | 1 | 1 | 1 | 0 | 0 |
| Total number | 261 | 83 | 32 | 4 | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Q.-P.; Ye, J.-J.; Huang, Y.-M.; Liu, D.; Liu, L.-F.; Dong, K.; Razumova, E.A.; Osterman, I.A.; Sergiev, P.V.; Dontsova, O.A.; et al. Exploitation of Potentially New Antibiotics from Mangrove Actinobacteria in Maowei Sea by Combination of Multiple Discovery Strategies. Antibiotics 2019, 8, 236. https://doi.org/10.3390/antibiotics8040236
Lu Q-P, Ye J-J, Huang Y-M, Liu D, Liu L-F, Dong K, Razumova EA, Osterman IA, Sergiev PV, Dontsova OA, et al. Exploitation of Potentially New Antibiotics from Mangrove Actinobacteria in Maowei Sea by Combination of Multiple Discovery Strategies. Antibiotics. 2019; 8(4):236. https://doi.org/10.3390/antibiotics8040236
Chicago/Turabian StyleLu, Qin-Pei, Jing-Jing Ye, Yong-Mei Huang, Di Liu, Li-Fang Liu, Kun Dong, Elizaveta A. Razumova, Ilya A. Osterman, Petr V. Sergiev, Olga A. Dontsova, and et al. 2019. "Exploitation of Potentially New Antibiotics from Mangrove Actinobacteria in Maowei Sea by Combination of Multiple Discovery Strategies" Antibiotics 8, no. 4: 236. https://doi.org/10.3390/antibiotics8040236





