Sleep Modelling across Physiological Levels
Abstract
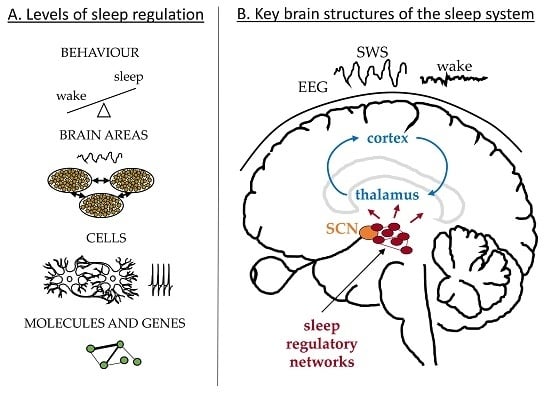
:1. Introduction
2. Behavioural Level
3. Brain Areas—Mean Field Dynamics
3.1. Large-Scale Brain Dynamics—Cortex and Thalamus
3.2. Sleep Regulatory Networks
4. Neuronal Interactions—Cellular Level
4.1. Large-Scale Brain Dynamics—Cortex and Thalamus
4.2. Sleep Regulatory Networks
5. Discussion
5.1. Links between Cellular and Behavioral Levels
5.2. Links between Cellular and Mean Field Levels
5.3. Links between Mean Field and Behavior Levels
5.4. Future Directions
Supplementary Materials
Funding
Conflicts of Interest
Abbreviations
| EEG | Electroencephalography |
| SWA | Slow wave activity |
| ipRGC | Intrinsic photosensitive retinal ganglion cell |
| SWS | Slow wave sleep |
| REMS | Rapid eye movement sleep |
| NREMS | Non-rapid eye movement sleep |
| SCN | Suprachiasmatic nucleus |
| fMRI | Functional magnetic resonance imaging |
| BOLD | Blood oxygen level dependent imaging |
References
- Brown, R.E.; Basheer, R.; McKenna, J.T.; Strecker, R.E.; McCarley, R.W. Control of sleep and wakefulness. Physiol. Rev. 2012, 92, 1087–1187. [Google Scholar] [CrossRef] [PubMed]
- Borbély, A.A.; Baumann, F.; Brandeis, D.; Strauch, I.; Lehmann, D. Sleep deprivation: Effect on sleep stages and EEG power density in man. Electroencephalogr. Clin. Neurophysiol. 1981, 51, 483–493. [Google Scholar] [CrossRef]
- Finelli, L.A.; Baumann, H.; Borbély, A.A.; Achermann, P. Dual electroencephalogram markers of human sleep homeostasis: Correlation between theta activity in waking and slow-wave activity in sleep. Neuroscience 2000, 101, 523–529. [Google Scholar] [CrossRef]
- Cajochen, C.; Brunner, D.P.; Kräuchi, K.; Graw, P.; Wirz-Justice, A. Power density in theta/alpha frequencies of the waking EEG progressively increases during sustained wakefulness. Sleep 1995, 18, 890–894. [Google Scholar] [CrossRef] [PubMed]
- McCormick, D.A.; Bal, T. Sleep and Arousal: Thalamocortical mechanisms. Annu. Rev. Neurosci. 1997, 20, 185–215. [Google Scholar] [CrossRef] [PubMed]
- Destexhe, A.; Sejnowski, T.J. Interactions between membrane conductances underlying thalamocortical slow-wave oscillations. Physiol. Rev. 2003, 83, 1401–1453. [Google Scholar] [CrossRef] [PubMed]
- Steriade, M. The corticothalamic system in sleep. Front. Biosci. 2003, 8, d878–d899. [Google Scholar] [CrossRef] [PubMed]
- Destexhe, A.; Sejnowski, T.J. Thalamocortical Assemblies: How Ion Channels, Single Neurons, and Large-Scale Networks Organize Sleep Oscillations; Oxford University Press: Oxford, UK, 2001; ISBN 0198524250. [Google Scholar]
- Saper, C.B.; Scammell, T.E.; Lu, J. Hypothalamic regulation of sleep and circadian rhythms. Nature 2005, 437, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Saper, C.B.; Fuller, P.M.; Pedersen, N.P.; Lu, J.; Scammell, T.E. Sleep state switching. Neuron 2010, 68, 1023–1042. [Google Scholar] [CrossRef] [PubMed]
- Borbély, A.A. A two process model of sleep regulation. Hum. Neurobiol. 1982, 1, 195–204. [Google Scholar] [PubMed]
- Daan, S.; Beersma, D.G.; Borbely, A.A. Timing of human sleep: Recovery process gated by a circadian pacemaker. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1984, 246, R161–R183. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Pittendrigh, C.S. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb. Symp. Quant. Biol. 1960, 25, 159–184. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.Y. Organization and function of a central nervous system circadian oscillator: the suprachiasmatic hypothalamic nucleus. Fed. Proc. 1983, 42, 2783–2789. [Google Scholar] [PubMed]
- Golombek, D.A.; Rosenstein, R.E. Physiology of circadian entrainment. Physiol. Rev. 2010, 90, 1063–1102. [Google Scholar] [CrossRef] [PubMed]
- Hattar, S.; Liao, H.W.; Takao, M.; Berson, D.M.; Yau, K.W. Melanopsin-containing retinal ganglion cells: Architecture, projections, and intrinsic photosensitivity. Science 2002, 295, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Gooley, J.J.; Lu, J.; Chou, T.C.; Scammell, T.E.; Saper, C.B. Melanopsin in cells of origin of the retinohypothalamic tract. Nat. Neurosci. 2001, 4, 1165. [Google Scholar] [CrossRef] [PubMed]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Stokkan, K.A.; Yamazaki, S.; Tei, H.; Sakaki, Y.; Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 2001, 291, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, J.C. Molecular bases for circadian clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef]
- King, D.P.; Takahashi, J.S. Molecular genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 2000, 23, 713–742. [Google Scholar] [CrossRef] [PubMed]
- Welsh, D.K.; Logothetis, D.E.; Meister, M.; Reppert, S.M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 1995, 14, 697–706. [Google Scholar] [CrossRef]
- Herzog, E.D.; Takahashi, J.S.; Block, G.D. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat. Neurosci. 1998, 1, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Isejima, H.; Matsuo, T.; Okura, R.; Yagita, K.; Kobayashi, M.; Okamura, H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 2003, 302, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Tononi, G.; Cirelli, C. Sleep function and synaptic homeostasis. Sleep Med. Rev. 2006, 10, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Tononi, G.; Cirelli, C. Sleep and the price of plasticity: From synaptic and cellular homeostasis to memory consolidation and integration. Neuron 2014, 81, 12–34. [Google Scholar] [CrossRef] [PubMed]
- Obal, F.; Krueger, J.M. Biochemical regulation of non-rapid-eye-movement sleep. Front. Biosci. 2003, 8, d520–d550. [Google Scholar] [PubMed]
- Franken, P.; Chollet, D.; Tafti, M. The homeostatic regulation of sleep need is under genetic control. J. Neurosci. 2001, 21, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Goel, N. Neurobehavioral effects and biomarkers of sleep loss in healthy adults. Curr. Neurol. Neurosci. Rep. 2017, 17, 89. [Google Scholar] [CrossRef] [PubMed]
- Laing, E.E.; Möller-Levet, C.S.; Dijk, D.-J.; Archer, S.N. Identifying and validating blood mRNA biomarkers for acute and chronic insufficient sleep in humans: A machine learning approach. Sleep 2018, 42, zsy186. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.K.; Ang, J.E.; Revell, V.L.; Holmes, B.; Mann, A.; Robertson, F.P.; Cui, N.; Middleton, B.; Ackermann, K.; Kayser, M.; et al. Effect of sleep deprivation on the human metabolome. Proc. Natl. Acad. Sci. USA 2014, 111, 10761–10766. [Google Scholar] [CrossRef] [PubMed]
- Dijk, D.J.; Czeisler, C.A. Contribution of the circadian pacemaker and the sleep homeostat to sleep propensity, sleep structure, electroencephalographic slow waves, and sleep spindle activity in humans. J. Neurosci. 1995, 15, 3526–3538. [Google Scholar] [CrossRef] [PubMed]
- Trachsel, L.; Edgar, D.M.; Seidel, W.F.; Craig Heller, H. Sleep homeostasis in suprachiasmatic nuclei-lesioned rats: effects of sleep deprivation and triazolam administration. Brain Res. 1992, 589, 253–261. [Google Scholar] [CrossRef]
- Deboer, T. Sleep homeostasis and the circadian clock: Do the circadian pacemaker and the sleep homeostat influence each other’s functioning? Neurobiol. Sleep Circadian Rhythm. 2018, 5, 68–77. [Google Scholar] [CrossRef]
- Borbély, A.A.; Daan, S.; Wirz-Justice, A.; Deboer, T. The two-process model of sleep regulation: A reappraisal. J. Sleep Res. 2016, 25, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Franken, P. A role for clock genes in sleep homeostasis. Curr. Opin. Neurobiol. 2013, 23, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Burgess, H.J. Partial sleep deprivation reduces phase advances to light in humans. J. Biol. Rhythms 2010, 25, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Challet, E.; Turek, F.W.; Laute, M.; Van Reeth, O. Sleep deprivation decreases phase-shift responses of circadian rhythms to light in the mouse: role of serotonergic and metabolic signals. Brain Res. 2001, 909, 81–91. [Google Scholar] [CrossRef]
- Lazar, A.S.; Lazar, Z.I.; Dijk, D.-J. Circadian regulation of slow waves in human sleep: Topographical aspects. Neuroimage 2015, 116, 123–134. [Google Scholar] [CrossRef]
- Deboer, T.; Détári, L.; Meijer, J.H. Long term effects of sleep deprivation on the mammalian circadian pacemaker. Sleep 2007, 30, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Franken, P.; Dijk, D.J. Circadian clock genes and sleep homeostasis. Eur. J. Neurosci. 2009, 29, 1820–1829. [Google Scholar] [CrossRef] [PubMed]
- Borbély, A.A.; Achermann, P. Concepts and models of sleep regulation: An overview. J. Sleep Res. 1992, 1, 63–79. [Google Scholar] [CrossRef] [PubMed]
- Fuhr, L.; Abreu, M.; Pett, P.; Relógio, A. Circadian systems biology: When time matters. Comput. Struct. Biotechnol. J. 2015, 13, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Gonze, D. Modeling circadian clocks: From equations to oscillations. Open Life Sci. 2011, 6, 699. [Google Scholar] [CrossRef]
- Gonze, D. Modeling circadian clocks: Roles, advantages, and limitations. Open Life Sci. 2011, 6, 712. [Google Scholar] [CrossRef]
- Roenneberg, T.; Chua, E.J.; Bernardo, R.; Mendoza, E. Modelling biological rhythms. Curr. Biol. 2008, 18, R826–R835. [Google Scholar] [CrossRef] [PubMed]
- Hogenesch, J.B.; Ueda, H.R. Understanding systems-level properties: Timely stories from the study of clocks. Nat. Rev. Genet. 2011, 12, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Forger, D. Multiscale complexity in the mammalian circadian clock. Curr. Opin. Genet. Dev. 2010, 20, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Goldbeter, A. Computational approaches to cellular rhythms. Nature 2002, 420, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Borbély, A.A.; Achermann, P. Sleep homeostasis and models of sleep regulation. J. Biol. Rhythms 1999, 14, 557–568. [Google Scholar] [PubMed]
- Beersma, D.G. Models of human sleep regulation. Sleep Med. Rev. 1998, 2, 31–43. [Google Scholar] [CrossRef]
- Booth, V.; Diniz Behn, C.G. Physiologically-based modeling of sleep-wake regulatory networks. Math. Biosci. 2014, 250C, 54–68. [Google Scholar] [CrossRef]
- Nakao, M.; Karashima, A.; Katayama, N. Mathematical models of regulatory mechanisms of sleep-wake rhythms. Cell. Mol. Life Sci. 2007, 64, 1236–1243. [Google Scholar] [CrossRef]
- Asgari-Targhi, A.; Klerman, E.B. Mathematical modeling of circadian rhythms. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, e1439. [Google Scholar] [CrossRef]
- Millius, A.; Ueda, H.R. Systems biology-derived discoveries of intrinsic clocks. Front. Neurol. 2017, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Forger, D.B.; Dean, D.A.; Gurdziel, K.; Leloup, J.C.; Lee, C.; von Gall, C.; Etchegaray, J.P.; Kronauer, R.E.; Goldbeter, A.; Peskin, C.S.; et al. Development and validation of computational models for mammalian circadian oscillators. Omi. A J. Integr. Biol. 2003, 7, 387–400. [Google Scholar] [CrossRef]
- Kim, J.K. Protein sequestration versus Hill-type repression in circadian clock models. IET Syst. Biol. 2016, 10, 125–135. [Google Scholar] [CrossRef]
- Beersma, D.G.M. Why and how do we model circadian rhythms? J. Biol. Rhythms 2005, 20, 304–313. [Google Scholar] [CrossRef]
- Raslear, T.G.; Hursh, S.R.; Van Dongen, H.P.A. Predicting cognitive impairment and accident risk. Prog. Brain Res. 2011, 190, 155–167. [Google Scholar]
- Dawson, D.; Ian Noy, Y.; Härmä, M.; Akerstedt, T.; Belenky, G. Modelling fatigue and the use of fatigue models in work settings. Accid. Anal. Prev. 2011, 43, 549–564. [Google Scholar] [CrossRef]
- Klerman, E.B.; St. Hilaire, M. Review: On mathematical modeling of circadian rhythms, performance, and alertness. J. Biol. Rhythms 2007, 22, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, H.P.A. Comparison of mathematical model predictions to experimental data of fatigue and performance. Aviat. Space. Environ. Med. 2004, 75, A15–A36. [Google Scholar] [PubMed]
- Daan, S.; Beersma, D.G. Circadian gating of human sleep-wake cycle. In Mathematical Models of the Circadian Sleep-Wake Cycle; Moore-Ede, M.C., Czeisler, C.A., Eds.; Raven Press: New York, NY, USA, 1984; pp. 129–158. [Google Scholar]
- Achermann, P.; Borbély, A.A. Simulation of human sleep: Ultradian dynamics of electroencephalographic slow-wave activity. J. Biol. Rhythms 1990, 5, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Achermann, P.; Dijk, D.J.; Brunner, D.P.; Borbély, A.A. A model of human sleep homeostasis based on EEG slow-wave activity: Quantitative comparison of data and simulations. Brain Res. Bull. 1993, 31, 97–113. [Google Scholar] [CrossRef]
- Franken, P.; Tobler, I.; Borbély, A.A. Sleep homeostasis in the rat: Simulation of the time course of EEG slow-wave activity. Neurosci. Lett. 1991, 130, 141–144. [Google Scholar] [CrossRef]
- Putilov, A.A. Timing of sleep modelling: Circadian modulation of the homeostatic process. Biol. Rhythm Res. 1995, 26, 1–19. [Google Scholar] [CrossRef]
- McCauley, P.; Kalachev, L.V.; Smith, A.D.; Belenky, G.; Dinges, D.F.; Van Dongen, H.P.A. A new mathematical model for the homeostatic effects of sleep loss on neurobehavioral performance. J. Theor. Biol. 2009, 256, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Rajdev, P.; Thorsley, D.; Rajaraman, S.; Rupp, T.L.; Wesensten, N.J.; Balkin, T.J.; Reifman, J. A unified mathematical model to quantify performance impairment for both chronic sleep restriction and total sleep deprivation. J. Theor. Biol. 2013, 331, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Postnova, S.; Lockley, S.W.; Robinson, P.A. Prediction of cognitive performance and subjective sleepiness using a model of arousal dynamics. J. Biol. Rhythms 2018, 33, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Akerstedt, T.; Folkard, S. The three-process model of alertness and its extension to performance, sleep latency, and sleep length. Chronobiol. Int. 1997, 14, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Bes, F.; Jobert, M.; Schulz, H. Modeling napping, post-lunch dip, and other variations in human sleep propensity. Sleep 2009, 32, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Klerman, E.B.; Butler, J.P. Modeling the adenosine system as a modulator of cognitive performance and sleep patterns during sleep restriction and recovery. PLoS Comput. Biol. 2017, 13, e1005759. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Wesensten, N.J.; Balkin, T.J.; Reifman, J. A Unified Model of Performance: Validation of its Predictions across Different Sleep/Wake Schedules. Sleep 2016, 39, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Jewett, M.E.; Dijk, D.J.; Kronauer, R.E.; Boivin, D.B.; Czeisler, C.A. Interaction of circadian- and sleep/wake homeostatic-processes modulate psychomotor vigilance test (PVT) performance. Sleep Res. 1997, 26, 759. [Google Scholar]
- Kawato, M.; Fujita, K.; Suzuki, R.; Winfree, A.T. A three-oscillator model of the human circadian system controlling the core temperature rhythm and the sleep-wake cycle. J. Theor. Biol. 1982, 98, 369–392. [Google Scholar] [CrossRef]
- Kronauer, R.E.; Czeisler, C.A.; Pilato, S.F.; Moore-Ede, M.C.; Weitzman, E.D. Mathematical model of the human circadian system with two interacting oscillators. Am. J. Physiol. Integr. Comp. Physiol. 1982, 242, R3–R17. [Google Scholar] [CrossRef] [PubMed]
- Strogatz, S.H. Human sleep and circadian rhythms: A simple model based on two coupled oscillators. J. Math. Biol. 1987, 25, 327–347. [Google Scholar] [CrossRef] [PubMed]
- St Hilaire, M.A.; Klerman, E.B.; Khalsa, S.B.S.; Wright, K.P.; Czeisler, C.A.; Kronauer, R.E. Addition of a non-photic component to a light-based mathematical model of the human circadian pacemaker. J. Theor. Biol. 2007, 247, 583–599. [Google Scholar] [CrossRef] [PubMed]
- Forger, D.B.; Jewett, M.E.; Kronauer, R.E. A simpler model of the human circadian pacemaker. J. Biol. Rhythms 1999, 14, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M.; McGinty, D.; Szymusiak, R.; Yamamoto, M. A thermoregulatory model of sleep control. Jpn. J. Physiol. 1995, 45, 291–309. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.H.; Swang, T.W.; Hamilton, I.M.; Best, J.A. State-dependent metabolic partitioning and energy conservation: A theoretical framework for understanding the function of sleep. PLoS ONE 2017, 12, e0185746. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.A.; Rennie, C.J.; Rowe, D.L.; O’Connor, S.C.; Gordon, E.; Connor, S.C.O.; Gordon, E. Multiscale brain modelling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.A.; Postnova, S.; Abeysuriya, R.G.; Kim, J.W.; Roberts, J.A.; McKenzie-Sell, L.; Karamjai, A.; Kerr, C.C.; Fung, F.; Anderson, R.; et al. A Multiscale “Working Brain” Model. In Validating Neuro-Computational Models of Neurobiological and Psychiatric Disorders; Bhattacharaya, B.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 107–140. [Google Scholar]
- Yamaguchi, I.; Kishi, A.; Togo, F.; Nakamura, T.; Yamamoto, Y. A robust method with high time resolution for sstimating the cortico-thalamo-cortical loop strength and the delay when using a scalp electroencephalography applied to the wake-sleep transition. Methods Inf. Med. 2018, 57, 122–128. [Google Scholar] [PubMed]
- Cona, F.; Lacanna, M.; Ursino, M. A thalamo-cortical neural mass model for the simulation of brain rhythms during sleep. J. Comput. Neurosci. 2014, 37, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.A.; Phillips, A.J.K.; Fulcher, B.D.; Puckeridge, M.; Roberts, J.A. Quantitative modelling of sleep dynamics. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2011, 369, 3840–3854. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kim, J.W.; Robinson, P.A. Slow-wave oscillations in a corticothalamic model of sleep and wake. J. Theor. Biol. 2015, 370, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Abeysuriya, R.G.; Rennie, C.J.; Robinson, P.A. Prediction and verification of nonlinear sleep spindle harmonic oscillations. J. Theor. Biol. 2014, 344, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Abeysuriya, R.G.; Rennie, C.J.; Robinson, P.A.; Kim, J.W. Experimental observation of a theoretically predicted nonlinear sleep spindle harmonic in human EEG. Clin. Neurophysiol. 2014, 125, 2016–2023. [Google Scholar] [CrossRef] [PubMed]
- Abeysuriya, R.G.; Rennie, C.J.; Robinson, P.A. Physiologically based arousal state estimation and dynamics. J. Neurosci. Methods 2015, 253, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Abeysuriya, R.G.; Robinson, P.A. Real-time automated EEG tracking of brain states using neural field theory. J. Neurosci. Methods 2016, 258, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Assadzadeh, S.; Robinson, P.A. Necessity of the sleep-wake cycle for synaptic homeostasis: System-level analysis of plasticity in the corticothalamic system. R. Soc. Open Sci. 2018, 5, 171952. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.S.; Weigenand, A.; Ngo, H.-V.V.; Marshall, L.; Born, J.; Martinetz, T.; Claussen, J.C. A thalamocortical neural mass model of the EEG during NREM sleep and its response to auditory stimulation. PLoS Comput. Biol. 2016, 12, e1005022. [Google Scholar]
- Steyn-Ross, D.A.; Steyn-Ross, M.L.; Sleigh, J.W.; Wilson, M.T.; Gillies, I.P.; Wright, J.J. The sleep cycle modelled as a cortical phase transition. J. Biol. Phys. 2005, 31, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Weigenand, A.; Schellenberger Costa, M.; Ngo, H.-V.V.; Claussen, J.C.; Martinetz, T. Characterization of K-complexes and slow wave activity in a neural mass model. PLoS Comput. Biol. 2014, 10, e1003923. [Google Scholar] [CrossRef] [PubMed]
- Deco, G.; Cabral, J.; Saenger, V.M.; Boly, M.; Tagliazucchi, E.; Laufs, H.; Van Someren, E.; Jobst, B.; Stevner, A.; Kringelbach, M.L. Perturbation of whole-brain dynamics in silico reveals mechanistic differences between brain states. Neuroimage 2018, 169, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.S.; Born, J.; Claussen, J.C.; Martinetz, T. Modeling the effect of sleep regulation on a neural mass model. J. Comput. Neurosci. 2016, 41, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Robinson, P.A. A quantitative model of sleep-wake dynamics based on the physiology of the brainstem ascending arousal system. J. Biol. Rhythms 2007, 22, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Diniz Behn, C.G.; Brown, E.N.; Scammell, T.E.; Kopell, N.J. Mathematical model of network dynamics governing mouse sleep-wake behavior. J. Neurophysiol. 2007, 97, 3828–3840. [Google Scholar] [CrossRef] [PubMed]
- Rempe, M.J.; Best, J.; Terman, D. A mathematical model of the sleep/wake cycle. J. Math. Biol. 2010, 60, 615–644. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bose, A.; Mallick, B.N. A mathematical model towards understanding the mechanism of neuronal regulation of wake-NREMS-REMS states. PLoS ONE 2012, 7, e42059. [Google Scholar] [CrossRef] [PubMed]
- Dunmyre, J.R.; Mashour, G.A.; Booth, V. Coupled flip-flop model for REM sleep regulation in the rat. PLoS ONE 2014, 9, e94481. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Rangan, A. Role of the locus coeruleus in the emergence of power law wake bouts in a model of the brainstem sleep-wake system through early infancy. J. Theor. Biol. 2017, 426, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Sorooshyari, S.; Huerta, R.; de Lecea, L. A framework for quantitative modeling of neural circuits involved in sleep-to-wake transition. Front. Neurol. 2015, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Tamakawa, Y.; Karashima, A.; Koyama, Y.; Katayama, N.; Nakao, M. A quartet neural system model orchestrating sleep and wakefulness mechanisms. J. Neurophysiol. 2006, 95, 2055–2069. [Google Scholar] [CrossRef] [PubMed]
- Diniz Behn, C.G.; Booth, V. Simulating microinjection experiments in a novel model of the rat sleep-wake regulatory network. J. Neurophysiol. 2010, 103, 1937–1953. [Google Scholar] [CrossRef] [PubMed]
- Mosqueiro, T.; de Lecea, L.; Huerta, R. Control of sleep-to-wake transitions via fast aminoacid and slow neuropeptide transmission. New J. Phys. 2014, 16. [Google Scholar] [CrossRef] [PubMed]
- Jalewa, J.; Joshi, A.; McGinnity, T.M.; Prasad, G.; Wong-Lin, K.; Hölscher, C. Neural circuit interactions between the dorsal raphe nucleus and the lateral hypothalamus: An experimental and computational study. PLoS ONE 2014, 9, e88003. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Fulcher, B.D.; Robinson, P.A.; Klerman, E.B. Mammalian rest/activity patterns explained by physiologically based modeling. PLoS Comput. Biol. 2013, 9, e1003213. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Robinson, P.A. Sleep deprivation in a quantitative physiologically based model of the ascending arousal system. J. Theor. Biol. 2008, 255, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Puckeridge, M.; Fulcher, B.D.; Phillips, A.J.K.; Robinson, P.A. Incorporation of caffeine into a quantitative model of fatigue and sleep. J. Theor. Biol. 2011, 273, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, B.D.; Phillips, A.J.K.; Postnova, S.; Robinson, P.A. A physiologically based model of orexinergic stabilization of sleep and wake. PLoS ONE 2014, 9, e91982. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.P.; McKenzie-Sell, L.; Karanjai, A.; Robinson, P.A. Wake-sleep transition as a noisy bifurcation. Phys. Rev. E 2016, 94, 022412. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Chen, P.Y.; Robinson, P.A. Probing the mechanisms of chronotype using quantitative modeling. J. Biol. Rhythms 2010, 25, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Czeisler, C.A.; Klerman, E.B. Revisiting spontaneous internal desynchrony using a quantitative model of sleep physiology. J. Biol. Rhythms 2011, 26, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Postnova, S.; Lockley, S.W.; Robinson, P.A. Sleep propensity under forced desynchrony in a model of arousal state dynamics. J. Biol. Rhythms 2016, 31, 498–508. [Google Scholar] [CrossRef] [PubMed]
- Diniz Behn, C.G.; Kopell, N.; Brown, E.N.; Mochizuki, T.; Scammell, T.E. Delayed orexin signaling consolidates wakefulness and sleep: Physiology and modeling. J. Neurophysiol. 2008, 99, 3090–3103. [Google Scholar] [CrossRef] [PubMed]
- Gleit, R.D.; Diniz Behn, C.G.; Booth, V. Modeling interindividual differences in spontaneous internal desynchrony patterns. J. Biol. Rhythms 2013, 28, 339–355. [Google Scholar] [CrossRef] [PubMed]
- Booth, V.; Xique, I.; Diniz Behn, C.G. One-dimensional map for the circadian modulation of sleep in a sleep-wake regulatory network model for human sleep. SIAM J. Appl. Dyn. Syst. 2017, 16, 1089–1112. [Google Scholar] [CrossRef]
- Feinberg, I. Changes in sleep cycle patterns with age. J. Psychiatr. Res. 1974, 10, 283–306. [Google Scholar] [CrossRef]
- McCarley, R.W.; Hobson, J.A. Neuronal excitability modulation over the sleep cycle: A structural and mathematical model. Science 1975, 189, 58–60. [Google Scholar] [CrossRef] [PubMed]
- McCarley, R.W.; Massaquoi, S.G. A limit cycle mathematical model of the REM sleep oscillator system. Am. J. Physiol. Integr. Comp. Physiol. 1986, 251, R1011–R1029. [Google Scholar] [CrossRef] [PubMed]
- McCarley, R.W.; Massaquoi, S.G. Neurobiological structure of the revised limit cycle reciprocal interaction model of REM cycle control. J. Sleep Res. 1992, 1, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Massaquoi, S.G.; McCarley, R.W. Extension of the limit cycle reciprocal interaction model of REM cycle control. An integrated sleep control model. J. Sleep Res. 1992, 1, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Weber, F. Modeling the mammalian sleep cycle. Curr. Opin. Neurobiol. 2017, 46, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, A.L.; Huxley, A.F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952, 117, 500–544. [Google Scholar] [CrossRef] [PubMed]
- Bazhenov, M.; Timofeev, I.; Steriade, M.; Sejnowski, T.J. Model of thalamocortical slow-wave sleep oscillations and transitions to activated States. J. Neurosci. 2002, 22, 8691–8704. [Google Scholar] [CrossRef] [PubMed]
- Bonjean, M.; Baker, T.; Lemieux, M.; Timofeev, I.; Sejnowski, T.; Bazhenov, M. Corticothalamic feedback controls sleep spindle duration in vivo. J. Neurosci. 2011, 31, 9124–9134. [Google Scholar] [CrossRef] [PubMed]
- Bonjean, M.; Baker, T.; Bazhenov, M.; Cash, S.; Halgren, E.; Sejnowski, T. Interactions between core and matrix thalamocortical projections in human sleep spindle synchronization. J. Neurosci. 2012, 32, 5250–5263. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Krishnan, G.P.; Komarov, M.; Bazhenov, M. Differential roles of sleep spindles and sleep slow oscillations in memory consolidation. PLoS Comput. Biol. 2018, 14, e1006322. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Krishnan, G.P.; Bazhenov, M. Synaptic mechanisms of memory consolidation during sleep slow oscillations. J. Neurosci. 2016, 36, 4231–4247. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, G.P.; Chauvette, S.; Shamie, I.; Soltani, S.; Timofeev, I.; Cash, S.S.; Halgren, E.; Bazhenov, M. Cellular and neurochemical basis of sleep stages in the thalamocortical network. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.; Tononi, G. Modeling Sleep and Wakefulness in the Thalamocortical System. J. Neurophysiol. 2005, 93, 1671–1698. [Google Scholar] [CrossRef] [PubMed]
- Esser, S.K.; Hill, S.L.; Tononi, G. Sleep homeostasis and cortical synchronization: I. Modeling the effects of synaptic strength on sleep slow waves. Sleep 2007, 30, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Hoel, E.P.; Albantakis, L.; Cirelli, C.; Tononi, G. Synaptic refinement during development and its effect on slow-wave activity: a computational study. J. Neurophysiol. 2016, 115, 2199–2213. [Google Scholar] [CrossRef] [PubMed]
- Olcese, U.; Esser, S.K.; Tononi, G. Sleep and synaptic renormalization: A computational study. J. Neurophysiol. 2010, 104, 3476–3493. [Google Scholar] [CrossRef] [PubMed]
- Nere, A.; Olcese, U.; Balduzzi, D.; Tononi, G. A neuromorphic architecture for object recognition and motion anticipation using burst-STDP. PLoS ONE 2012, 7, e36958. [Google Scholar] [CrossRef] [PubMed]
- Nere, A.; Hashmi, A.; Cirelli, C.; Tononi, G. Sleep-dependent synaptic down-selection (I): Modeling the benefits of sleep on memory consolidation and integration. Front. Neurol. 2013, 4, 143. [Google Scholar] [CrossRef] [PubMed]
- Krueger, J.M.; Rector, D.M.; Roy, S.; Van Dongen, H.P.A.; Belenky, G.; Panksepp, J. Sleep as a fundamental property of neuronal assemblies. Nat. Rev. Neurosci. 2008, 9, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Krueger, J.M.; Rector, D.M.; Wan, Y. A network model for activity-dependent sleep regulation. J. Theor. Biol. 2008, 253, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Deco, G.; Hagmann, P.; Hudetz, A.G.; Tononi, G. Modeling resting-state functional networks when the cortex falls asleep: Local and global changes. Cereb. Cortex 2014, 24, 3180–3194. [Google Scholar] [CrossRef] [PubMed]
- Tatsuki, F.; Sunagawa, G.A.; Shi, S.; Susaki, E.A.; Yukinaga, H.; Perrin, D.; Sumiyama, K.; Ukai-Tadenuma, M.; Fujishima, H.; Ohno, R.; et al. Involvement of Ca2+-dependent hyperpolarization in sleep duration in mammals. Neuron 2016, 90, 70–85. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Shi, S.; Ukai-Tadenuma, M.; Fujishima, H.; Ohno, R.-I.; Ueda, H.R. Leak potassium channels regulate sleep duration. Proc. Natl. Acad. Sci. USA 2018, 115, E9459–E9468. [Google Scholar] [CrossRef] [PubMed]
- Compte, A.; Sanchez-Vives, M.V.; McCormick, D.A.; Wang, X.J. Cellular and network mechanisms of slow oscillatory activity (<1 HZ) and wave propagations in a cortical network model. J. Neurophysiol. 2003, 89, 2707–2725. [Google Scholar] [PubMed]
- Destexhe, A.; Bal, T.; McCormick, D.A.; Sejnowski, T.J. Ionic mechanisms underlying synchronized oscillations and propagating waves in a model of ferret thalamic slices. J. Neurophysiol. 1996, 76, 2049–2070. [Google Scholar] [CrossRef] [PubMed]
- Destexhe, A.; Contreras, D.; Sejnowski, T.J.; Steriade, M. A model of spindle rhythmicity in the isolated thalamic reticular nucleus. J. Neurophysiol. 1994, 72, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Komarov, M.; Krishnan, G.; Chauvette, S.; Rulkov, N.; Timofeev, I.; Bazhenov, M. New class of reduced computationally efficient neuronal models for large-scale simulations of brain dynamics. J. Comput. Neurosci. 2018, 44, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.; Cauller, L.J.; Llano, D.A. Presence of a chaotic region at the sleep-wake transition in a simplified thalamocortical circuit model. Front. Comput. Neurosci. 2016, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Holmgren Hopkins, N.; Sanz-Leon, P.; Roy, D.; Postnova, S. Spiking patterns and synchronization of thalamic neurons along the sleep-wake cycle. Chaos Interdiscip. J. Nonlinear Sci. 2018, 28, 106314. [Google Scholar] [CrossRef] [PubMed]
- Postnova, S.; Voigt, K.; Braun, H.A. Modelling the hypothalamic control of thalamic synchronization along the sleep-wake cycles. In Advances in Cognitive Neurodynamics (II); Springer: Dordrecht, The Netherlands, 2011; pp. 563–570. [Google Scholar]
- de Lecea, L.; Kilduff, T.S.; Peyron, C.; Gao, X.; Foye, P.E.; Danielson, P.E.; Fukuhara, C.; Battenberg, E.L.; Gautvik, V.T.; Bartlett, F.S.; et al. The hypocretins: Hypothalamus-specific peptides with neuroexcitatory activity. Proc. Natl. Acad. Sci. USA 1998, 95, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Amemiya, A.; Ishii, M.; Matsuzaki, I.; Chemelli, R.M.; Tanaka, H.; Williams, S.C.; Richardson, J.A.; Kozlowski, G.P.; Wilson, S.; et al. Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 1998, 92, 573–585. [Google Scholar] [CrossRef]
- Postnova, S.; Voigt, K.; Braun, H.A. A mathematical model of homeostatic regulation of sleep-wake cycles by hypocretin/orexin. J. Biol. Rhythms 2009, 24, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Patriarca, M.; Postnova, S.; Braun, H.A.; Hernández-García, E.; Toral, R. Diversity and noise effects in a model of homeostatic regulation of the sleep-wake cycle. PLoS Comput. Biol. 2012, 8, e1002650. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.E.; Brill, J.; Bonnavion, P.; Huguenard, J.R.; Huerta, R.; de Lecea, L. Mechanism for Hypocretin-mediated sleep-to-wake transitions. Proc. Natl. Acad. Sci. USA 2012, 109, E2635–E2644. [Google Scholar] [CrossRef] [PubMed]
- de Lecea, L.; Huerta, R. Hypocretin (orexin) regulation of sleep-to-wake transitions. Front. Pharmacol. 2014, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.S.; Diniz Behn, C.G. Dynamic interactions between orexin and dynorphin may delay onset of functional orexin effects: A modeling study. J. Biol. Rhythms 2011, 26, 171–181. [Google Scholar] [CrossRef] [PubMed]
- van Albada, S.J.; Kerr, C.C.; Chiang, A.K.I.; Rennie, C.J.; Robinson, P.A. Neurophysiological changes with age probed by inverse modeling of EEG spectra. Clin. Neurophysiol. 2010, 121, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.A.; Robinson, P.A. Quantitative theory of driven nonlinear brain dynamics. Neuroimage 2012, 62, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Müller, E.J.; van Albada, S.J.; Kim, J.W.; Robinson, P.A. Unified neural field theory of brain dynamics underlying oscillations in Parkinson’s disease and generalized epilepsies. J. Theor. Biol. 2017, 428, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Leon, P.; Robinson, P.A.; Knock, S.A.; Drysdale, P.M.; Abeysuriya, R.G.; Fung, F.K.; Rennie, C.J.; Zhao, X. NFTsim: Theory and simulation of multiscale neural field dynamics. PLoS Comput. Biol. 2018, 14, e1006387. [Google Scholar] [CrossRef] [PubMed]
- Olbrich, E.; Claussen, J.C.; Achermann, P. The multiple time scales of sleep dynamics as a challenge for modelling the sleeping brain. Philos. Trans. Math. Phys. Eng. Sci. 2011, 369, 3884–3901. [Google Scholar] [CrossRef] [PubMed]
- Postnova, S.; Layden, A.; Robinson, P.A.; Phillips, A.J.K.; Abeysuriya, R.G. Exploring sleepiness and entrainment on permanent shift schedules in a physiologically based model. J. Biol. Rhythms 2012, 27, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Postnova, S.; Robinson, P.A.; Postnov, D.D. Adaptation to shift work: Physiologically based modeling of the effects of lighting and shifts’ start time. PLoS ONE 2013, 8, e53379. [Google Scholar] [CrossRef] [PubMed]
- Postnova, S.; Postnov, D.D.; Seneviratne, M.; Robinson, P.A. Effects of rotation interval on sleepiness and circadian dynamics on forward rotating 3-shift systems. J. Biol. Rhythms 2014, 29, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Skeldon, A.; Derks, G.; Dijk, D.J. Modelling changes in sleep timing and duration across the lifespan: Changes in circadian rhythmicity or sleep homeostasis? Sleep Med. Rev. 2015, 28, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Skeldon, A.C.; Dijk, D.J.; Derks, G. Mathematical models for sleep-wake dynamics: Comparison of the two-process model and a mutual inhibition neuronal model. PLoS ONE 2014, 9, e103877. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, B.D.; Phillips, A.J.K.; Robinson, P.A. Quantitative physiologically based modeling of subjective fatigue during sleep deprivation. J. Theor. Biol. 2010, 264, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Kang, H.; Xu, Q.; Chen, M.J.; Liao, Y.; Thiyagarajan, M.; O’Donnell, J.; Christensen, D.J.; Nicholson, C.; Iliff, J.J.; et al. Sleep drives metabolite clearance from the adult brain. Science 2013, 342, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Vanderheyden, W.M.; Goodman, A.G.; Taylor, R.H.; Frank, M.G.; Van Dongen, H.P.A.; Gerstner, J.R. Astrocyte expression of the Drosophila TNF-alpha homologue, Eiger, regulates sleep in flies. PLoS Genet. 2018, 14, e1007724. [Google Scholar] [CrossRef] [PubMed]
- Achermann, P.; Borbely, A.A. Combining different models of sleep regulation. J. Sleep Res. 1992, 1, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Robinson, P.A.; Kedziora, D.J.; Abeysuriya, R.G. Mammalian sleep dynamics: How diverse features arise from a common physiological framework. PLoS Comput. Biol. 2010, 6, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.K.; Robinson, P.A.; Klerman, E.B. Arousal state feedback as a potential physiological generator of the ultradian REM/NREM sleep cycle. J. Theor. Biol. 2013, 319, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, P.F.; Rinzel, J. Intrinsic and network rhythmogenesis in a reduced Traub model for CA3 neurons. J. Comput. Neurosci. 1994, 1, 39–60. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Postnova, S. Sleep Modelling across Physiological Levels. Clocks & Sleep 2019, 1, 166-184. https://doi.org/10.3390/clockssleep1010015
Postnova S. Sleep Modelling across Physiological Levels. Clocks & Sleep. 2019; 1(1):166-184. https://doi.org/10.3390/clockssleep1010015
Chicago/Turabian StylePostnova, Svetlana. 2019. "Sleep Modelling across Physiological Levels" Clocks & Sleep 1, no. 1: 166-184. https://doi.org/10.3390/clockssleep1010015