Is There Anybody Out There? Substrate Availability Controls Microbial Activity outside of Hotspots in Subsoils
Abstract
:1. Introduction
- (i)
- Active microorganisms occupy small soil volumes, forming hotspots in subsoils.
- (ii)
- However, microorganisms also exist outside of subsoil hotspots, but are inactive due to energy limitations.
- (iii)
- Therefore, the strongest effect of glucose additions on enzyme activities will be induced outside of hotspots.
2. Materials and Methods
2.1. Study Site and Soil Sampling
2.2. Zymography
2.3. Incubation
2.4. Image Processing
2.5. Statistics
3. Results and Discussion
3.1. Distribution and Activity of Enzymes
3.2. Effect of Glucose on Enzyme Activities in Hotspots and Non-Hotspots
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Lützow, M.V.; Kögel-Knabner, I.; Ekschmitt, K.; Matzner, E.; Guggenberger, G.; Marschner, B.; Flessa, H. Stabilization of organic matter in temperate soils: Mechanisms and their relevance under different soil conditions—A review. Eur. J. Soil Sci. 2006, 57, 426–445. [Google Scholar] [CrossRef]
- Jobbágy, E.G.; Jackson, R.B. The vertical distribution of soil organic carbon and its relation to climated and vegetation. Ecol. Appl. 2000, 10, 423–436. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Torn, M.S.; Abiven, S.; Dittmar, T.; Guggenberger, G.; Janssens, I.A.; Kleber, M.; Kögel-Knabner, I.; Lehmann, J.; Manning, D.A.; et al. Persistence of soil organic matter as an ecosystem property. Nature 2011, 478, 49–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rumpel, C.; Kögel-Knabner, I. Deep soil organic matter—A key but poorly understood component of terrestrial C cycle. Plant Soil 2011, 338, 143–158. [Google Scholar] [CrossRef]
- Banfield, C.C.; Dippold, M.A.; Pausch, J.; Hoang, D.T.T.; Kuzyakov, Y. Biopore history determines the microbial community composition in subsoil hotspots. Biol. Fert. Soils 2017, 53, 573–588. [Google Scholar] [CrossRef]
- Hoang, D.T.; Bauke, S.L.; Kuzyakov, Y.; Pausch, J. Rolling in the deep: Priming effects in earthworm biopores in topsoil and subsoil. Soil Biol. Biochem. 2017, 114, 59–71. [Google Scholar] [CrossRef]
- Leinemann, T.; Mikutta, R.; Kalbitz, K.; Schaarschmidt, F.; Guggenberger, G. Small scale variability of vertical water and dissolved organic matter fluxes in sandy Cambisol subsoils as revealed by segmented suction plates. Biogeochemistry 2016, 131, 1–15. [Google Scholar] [CrossRef]
- Chabbi, A.; Kögel-Knabner, I.; Rumpel, C. Stabilised carbon in subsoil horizons is located in spatially distinct parts of the soil profile. Soil Biol. Biochem. 2009, 41, 256–261. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Blagodatskaya, E. Microbial hotspots and hot moments in soil: Concept review. Soil Biol. Biochem. 2015, 83, 184–199. [Google Scholar]
- Athmann, M.; Kautz, T.; Banfield, C.; Bauke, S.; Hoang, D.T.; Lüsebrink, M.; Pausch, J.; Amelung, W.; Kuzyakov, Y.; Köpke, U. Six months of L. terrestris L. activity in root-formed biopores increases nutrient availability, microbial biomass and enzyme activity. Appl. Soil Ecol. 2017, 120, 135–142. [Google Scholar] [CrossRef]
- Uksa, M.; Schloter, M.; Kautz, T.; Athmann, M.; Köpke, U.; Fischer, D. Spatial variability of hydrolytic and oxidative potential enzyme activities in different subsoil compartments. Biol. Fert. Soils 2015, 51, 517–521. [Google Scholar] [CrossRef]
- Hoang, D.T.; Razavi, B.S.; Kuzyakov, Y.; Blagodatskaya, E. Earthworm burrows: Kinetics and spatial distribution of enzymes of C-, N- and P- cycles. Soil Biol. Biochem. 2016, 99, 94–103. [Google Scholar] [CrossRef]
- Dworkin, J.; Shah, I.M. Exit from dormancy in microbial organisms. Nature reviews. Microbiology 2010, 8, 890–896. [Google Scholar] [PubMed]
- Blagodatskaya, E.; Kuzyakov, Y. Active microorganisms in soil: Critical review of estimation criteria and approaches. Soil Biol. Biochem. 2013, 67, 192–211. [Google Scholar] [CrossRef]
- Joergensen, R.G.; Wichern, F. Alive and kicking: Why dormant soil microorganisms matter. Soil Biol. Biochem. 2018, 116, 419–430. [Google Scholar] [CrossRef]
- Heitkötter, J.; Heinze, S.; Marschner, B. Relevance of substrate quality and nutrients for microbial C-turnover in top- and subsoil of a Dystric Cambisol. Geoderma 2017, 302, 89–99. [Google Scholar] [CrossRef]
- Fontaine, S.; Barot, S.; Barré, P.; Bdioui, N.; Mary, B.; Rumpel, C. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 2007, 450, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Wordell-Dietrich, P.; Don, A.; Helfrich, M. Controlling factors for the stability of subsoil carbon in a Dystric Cambisol. Geoderma 2016, 40–48. [Google Scholar] [CrossRef]
- Heitkötter, J.; Marschner, B. Soil zymography as a powerful tool for exploring hotspots and substrate limitation in undisturbed subsoil. Soil Biol. Biochem. 2018, (in press). [Google Scholar]
- Krueger, J.; Bachmann, J. New Field Sampling Method to Analyze Spatial Distribution of Small-Scale Soil Particle Surface Properties and Processes in Intact Soil. Vadose Zone J. 2017, 16. [Google Scholar] [CrossRef]
- Spohn, M.; Kuzyakov, Y. Spatial and temporal dynamics of hotspots of enzyme activity in soil as affected by living and dead roots—A soil zymography analysis. Plant Soil 2014, 379, 67–77. [Google Scholar] [CrossRef]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Olander, L.P.; Vitousek, P.M. Regulation of soil phosphatase and chitinase activityby N and P availability. Biogeochemistry 2000, 49, 175–191. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; FAO: Rome, Italy, 2014. [Google Scholar]
- Bundesanstalt für Bodenforschung. Geologische Übersichtskarte 1:200,000; Bundesanstalt für Bodenforschung: Hannover, Germany, 1973. [Google Scholar]
- Heinze, S.; Ludwig, B.; Piepho, H.P.; Mikutta, R.; Don, A.; Wordell-Dietrich, P.; Helfrich, M.; Hertel, D.; Leuschner, C.; Kirfel, K.; et al. Factors controlling the variability of organic matter in the top- and subsoil of a sandy Dystric Cambisol under beech forest. Geoderma 2018, 311, 37–44. [Google Scholar] [CrossRef]
- Angst, G.; John, S.; Mueller, C.W.; Kogel-Knabner, I.; Rethemeyer, J. Tracing the sources and spatial distribution of organic carbon in subsoils using a multi-biomarker approach. Sci. Rep. 2016, 6, 29478. [Google Scholar] [CrossRef] [PubMed]
- Tückmantel, T.; Leuschner, C.; Preusser, S.; Kandeler, E.; Angst, G.; Mueller, C.W.; Meier, I.C. Root exudation patterns in a beech forest: Dependence on soil depth, root morphology, and environment. Soil Biol. Biochem. 2017, 107, 188–197. [Google Scholar] [CrossRef]
- Quantum GIS Development Team. Quantum GIS Geographic Information System; Open Source Geospatial Foundation Project; 2017; Available online: https://qgis.org/en/site/about/index.html (accessed on 28 June 2017).
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2015. [Google Scholar]
- Köhl, M.; Magnussen, S.; Marchetti, M. Sampling Methods, Remote Sensing and GIS Multiresource Forest Inventory; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Loeppmann, S.; Blagodatskaya, E.; Pausch, J.; Kuzyakov, Y. Enzyme properties down the soil profile—A matter of substrate quality in rhizosphere and detritusphere. Soil Biol. Biochem. 2016, 103, 274–283. [Google Scholar] [CrossRef]
- Nunan, N.; Wu, K.; Young, I.M.; Crawford, J.W.; Ritz, K. Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil. FEMS Microbiol. Ecol. 2003, 44, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Tecon, R.; Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 2017, 41, 599–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leinemann, T.; Preusser, S.; Mikutta, R.; Kalbitz, K.; Cerli, C.; Höschen, C.; Mueller, C.W.; Kandeler, E.; Guggenberger, G. Multiple exchange processes on mineral surfaces control the transport of dissolved organic matter through soil profiles. Soil Biol. Biochem. 2018, 118, 79–90. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef] [Green Version]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging deeper to find unique microbial communities: The strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol. Biochem. 2012, 50, 58–65. [Google Scholar] [CrossRef]
- He, S.; Guo, L.; Niu, M.; Miao, F.; Jiao, S.; Hu, T.; Long, M. Ecological diversity and co-occurrence patterns of bacterial community through soil profile in response to long-term switchgrass cultivation. Sci. Rep. 2017, 7, 3608. [Google Scholar] [CrossRef] [PubMed]
- Billings, S.A.; Ballantyne, F. How interactions between microbial resource demands, soil organic matter stoichiometry, and substrate reactivity determine the direction and magnitude of soil respiratory responses to warming. Glob. Chang. Biol. 2013, 19, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Blagodatskaya, E.; Kuzyakov, Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: Critical review. Biol. Fert. Soils 2008, 45, 115–131. [Google Scholar]
- Karhu, K.; Hilasvuori, E.; Fritze, H.; Biasi, C.; Nykänen, H.; Liski, J.; Vanhala, P.; Heinonsalo, J.; Pumpanen, J. Priming effect increases with depth in a boreal forest soil. Soil Biol. Biochem. 2016, 99, 104–107. [Google Scholar] [CrossRef]
- Heitkötter, J.; Niebuhr, J.; Heinze, S.; Marschner, B. Patterns of nitrogen and citric acid induced changes in C-turnover and enzyme activities are different in topsoil and subsoils of a sandy Cambisol. Geoderma 2017, 292, 111–117. [Google Scholar] [CrossRef]



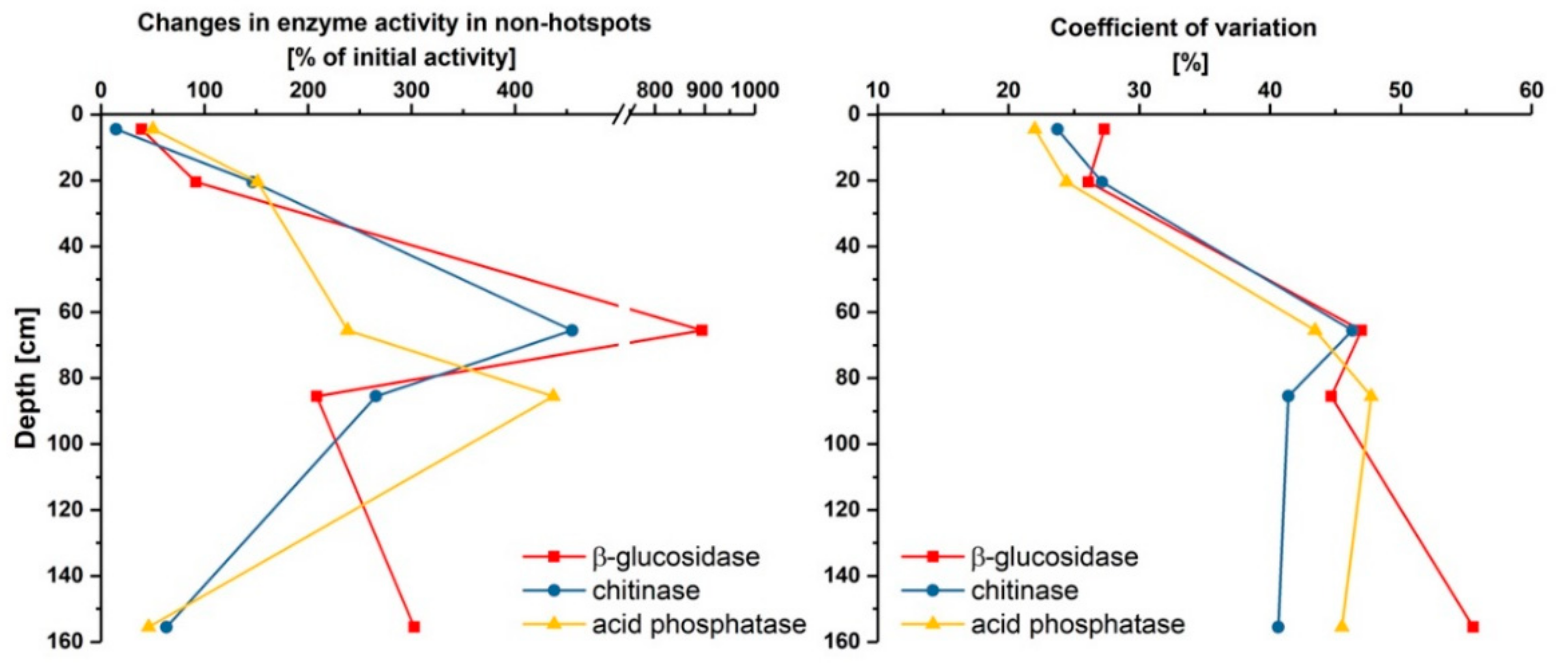
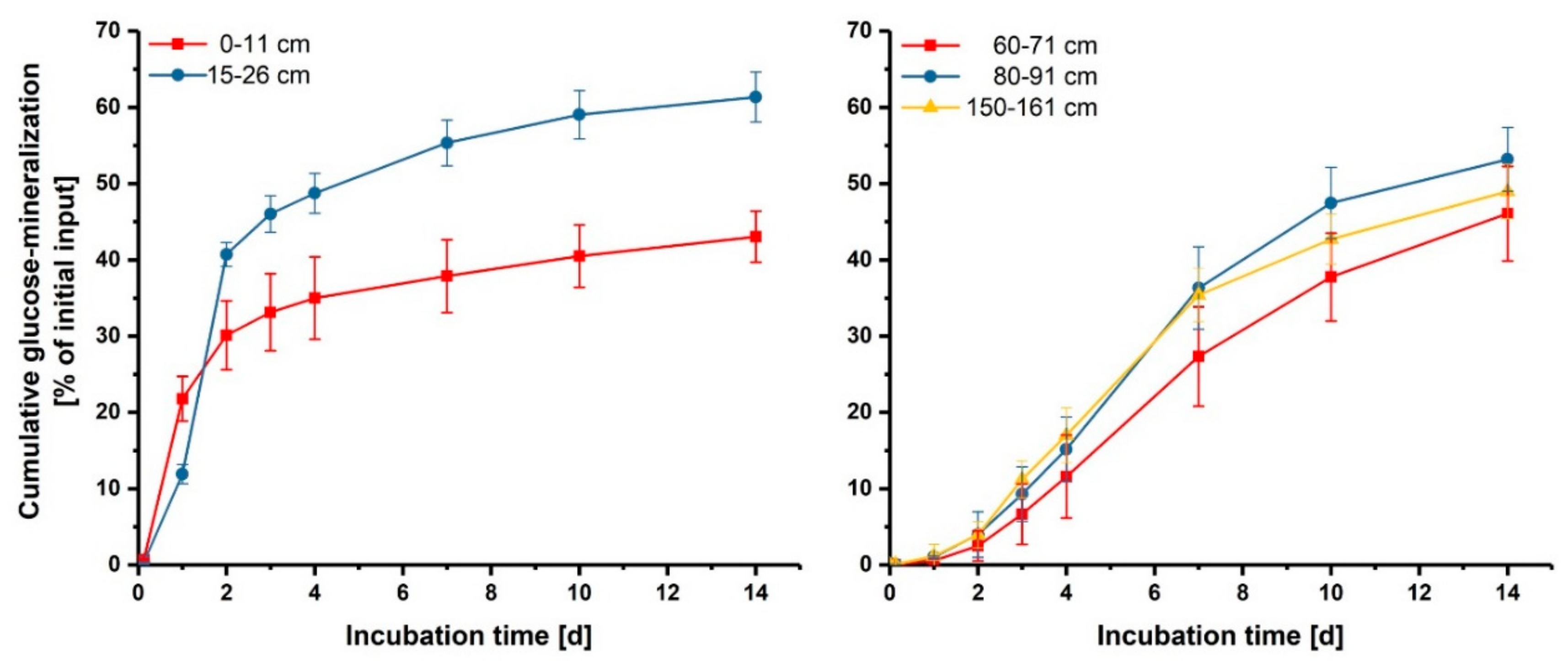
| Location | Parameter | β-Glucosidase | Chitinase | Acid Phosphatase |
|---|---|---|---|---|
| Initial hotspot | Mean | 2.0 | 6.4 | 39.9 |
| Std. Error | 13.7 | 35.0 | 34.7 | |
| p-value | 0.96 | 0.69 | 0.34 | |
| Initial non-hotspot | Mean | 307.2 | 189.1 | 184.6 |
| Std. Error | 198.4 | 101.9 | 93.6 | |
| p-value | 0.027 * | 0.032 * | 0.020 * |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heitkötter, J.; Marschner, B. Is There Anybody Out There? Substrate Availability Controls Microbial Activity outside of Hotspots in Subsoils. Soil Syst. 2018, 2, 35. https://doi.org/10.3390/soilsystems2020035
Heitkötter J, Marschner B. Is There Anybody Out There? Substrate Availability Controls Microbial Activity outside of Hotspots in Subsoils. Soil Systems. 2018; 2(2):35. https://doi.org/10.3390/soilsystems2020035
Chicago/Turabian StyleHeitkötter, Julian, and Bernd Marschner. 2018. "Is There Anybody Out There? Substrate Availability Controls Microbial Activity outside of Hotspots in Subsoils" Soil Systems 2, no. 2: 35. https://doi.org/10.3390/soilsystems2020035




